Abstract
The early post-pollination phase of maize (Zea mays) development is particularly sensitive to water deficit stress. Using cDNA microarray, we studied transcriptional profiles of endosperm and placenta/pedicel tissues in developing maize kernels under water stress. At 9 d after pollination (DAP), placenta/pedicel and endosperm differed considerably in their transcriptional responses. In placenta/pedicel, 79 genes were significantly affected by stress and of these 89% were up-regulated, whereas in endosperm, 56 genes were significantly affected and 82% of these were down-regulated. Only nine of the stress-regulated genes were in common between these tissues. Hierarchical cluster analysis indicated that different sets of genes were regulated in the two tissues. After rewatering at 9 DAP, profiles at 12 DAP suggested that two regulons exist, one for genes responding specifically to concurrent imposition of stress, and another for genes remaining affected after transient stress. In placenta, genes encoding recognized stress tolerance proteins, including heat shock proteins, chaperonins, and major intrinsic proteins, were the largest class of genes regulated, all of which were up-regulated. In contrast, in endosperm, genes in the cell division and growth category represented a large class of down-regulated genes. Several cell wall-degrading enzymes were expressed at lower levels than in controls, suggesting that stress delayed normal advance to programmed cell death in the central endosperm. We suggest that the responsiveness of placenta to whole-plant stress factors (water potential, abscisic acid, and sugar flux) and of endosperm to indirect factors may play key roles in determining the threshold for kernel abortion.
Water deficit during pollination and grain formation causes severe losses in crop production. In maize (Zea mays), the early reproductive stages of kernel development have long been recognized as being particularly vulnerable to water deficit (Claassen and Shaw, 1970); however, the mechanistic bases of cellular response are still not fully understood. Stresses that occur soon after pollination coincide with the period of endosperm cell division. This phase is particularly sensitive to water deficit, whereas later phases of kernel development, when starch and zein synthesis are at their maximum, are usually less affected (Grant et al., 1989; Artlip et al., 1995; Mambelli and Setter, 1998). Water deficit during the first few days after pollination inhibits endosperm cell proliferation, which is well correlated with kernel size at maturity (Nicolas et al., 1985; Ober et al., 1991). During this period, the development of placenta and vascular tissue of the pedicel creates capacity for influx of sugar and signaling molecules. Such development also occurs during the late phases of floral growth, which is also highly sensitive to stresses (Otegui et al., 1995; Edmeades et al., 2000).
Previous studies of maize have indicated that reproductive abortion in stress environments involves the plant hormone abscisic acid (ABA) and inadequate sugar supply to growing tissue (Schussler and Westgate, 1995; Zinselmeier et al., 1999; Setter et al., 2001). ABA accumulates dramatically in both endosperm and placenta during water stress and returns to normal after rewatering (Setter et al., 2001; Wang et al., 2002). Sugar flux into endosperm is also decreased by water stress (Schussler and Westgate, 1995), especially in the apical region of the ear where kernel abortion is most severe (Setter et al., 2001). The role of sugar supply is supported by studies that have shown that kernel set in water-stressed maize plants can be partially restored with stem infusion of supplemental Suc (Zinselmeier et al., 1999).
Studies of ABA synthesis mutants that were subjected to water stress in the early post-pollination stage have shown that the source of ABA, which accumulates in endosperm at the early phase, is maternal tissues (Ober and Setter, 1992). Other studies have shown that ABA is transported from leaves to sink organs via phloem (Ober and Setter, 1990; Zhang et al., 1996). Hence, it is possible that tissues in the transport pathway between the site of phloem unloading in placenta, to the sites of photosynthate import in endosperm, might play important roles during stress in affecting the flux of both ABA and sugar into the endosperm.
Other processes may also be involved. A large body of research in other abiotic stress systems indicates that stress can affect expression of numerous gene products, including dehydrins, oxidant protectants, heat shock proteins (HSPs), compatible solute synthetic pathways, and senescence-related proteins (Ingram and Bartels, 1996; Shinozaki et al., 1998; Zhu, 2002). Therefore, a global assessment of gene expression is needed to understand the whole-system response.
Microarray provides an analytical tool by which thousands of genes can be studied at one time. cDNA microarray has recently been used to monitor global gene expression in response to several abiotic stresses in higher plants. In Arabidopsis, Seki et al. (2001, 2002) monitored expression of genes in response to cold, drought, and salt stress; Gong et al. (2001) used 84 salt-regulated cDNAs to profile transcription of wild type and the salt-hypersensitive mutant sos3; and Fowler and Thomashow (2002) profiled transcripts responding to cold acclimation. In rice (Oryza sativa; Kawasaki et al., 2001) and barley (Hordeum vulgare; Ozturk et al., 2002), cDNA microarray was used to study transcriptional profiling in response to salt and drought stress. In maize kernels and immature ears, Zinselmeier et al. (2002) used cDNA microarrays to monitor expression of 384 genes in response to shade stress, and used oligonucleotide microarrays to examine expression of 1,502 genes in response to water stress. These studies have provided new insight into the transcriptomes involved in responses to these stresses and are contributing to our understanding of the function of the responding genes.
To advance our understanding of maize kernel response to water deficit, we monitored gene expression of developing endosperm and placenta/pedicel tissues under water deficit and rewatering using cDNA microarray slides containing about 2,500 unique cDNAs from immature maize ear tissue. The goals of this work were to identify genes whose expression in endosperms and placenta/pedicel tissues of maize kernels was affected by water deficit at the early period after pollination, and to gain insight into the processes involved in stress responses by analyzing their expression profiles.
RESULTS
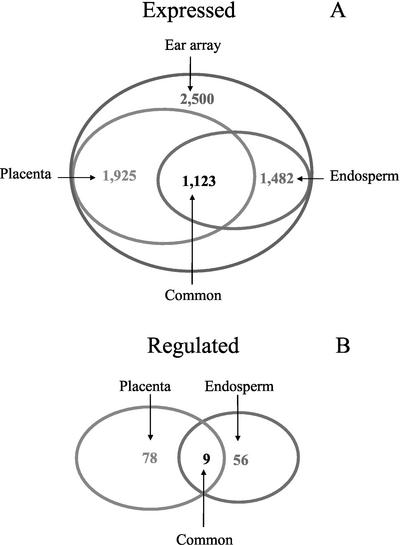
To monitor expression in maize endosperm and placenta/pedicel tissues during post-pollination development, we used cDNA microarray slides from the Maize Gene Discovery Project (Fernandes et al., 2002) containing expressed sequence tags (ESTs) from immature ear tissue. Our focus was on the developmental time frame of early post-pollination when placenta/pedicel are still actively growing and differentiating, while endosperm is in its phase of most rapid cell division (Kiesselbach, 1949; Kowles and Phillips, 1988; McSteen et al., 2000). The placenta/pedicel (hereafter called “placenta” for the collective structure), which is a part of the maize ear (female inflorescence), has an abundance of vascular tissue, as do the other parts of the ear. We expected that these tissues would share a considerable proportion of their transcriptomes. An overall assessment of the number of expressed genes detected with the ear array indicated that this assumption was valid (Fig. 1A). The ear array contained more than 5,000 ESTs, composing 2,500 tentatively unique genes based on sequence homology in overlapping regions (Fernandes et al., 2002). Of these, we observed that placenta expressed 1,925 of them or about 77%. We expected that the ear array would also be satisfactory for assessing expression in endosperms because at the sampled time frame, endosperm cells are highly proliferative, as are the immature ear tissues that were used as the basis of the ear array. We found that endosperm sampled at 9 d after pollination (DAP) expressed 1,482 of the ear array unique genes or about 60% of the total. This is similar to the findings of Fernandes et al. (2002) that 57% of the ear array genes were expressed in 10- to 14-DAP endosperm. Of the expressed genes, 1,123 were in common between endosperm and placenta (Fig. 1A). Thus, the ear array was an appropriate tool with which to compare gene expression in the two tissues, because it contained a substantial number of genes that were expressed in common as well as genes expressed specifically in one tissue or the other.
Figure 1.
Shared expression of cDNAs between endosperm and placenta tissues at 9 DAP. A, The number of transcripts whose average fluorescence exceeded the negative-control threshold (Expressed) in each tissue. B, The number of transcripts that were differentially expressed (Regulated) in control versus water stress for each tissue. Fluorescence data from four replicate microarray slides for each tissue were used to calculate averages. Regulated genes were defined as those whose expression in controls was significantly different from water stress according to SAM analysis and had a minimum change of 1.6-fold.
Stress Treatments and ABA Kinetics
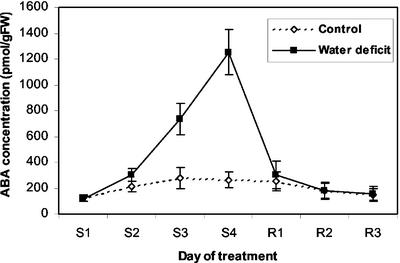
Water deficit treatment was started at 5 DAP. After 2 to 3 d of withholding water (7–8 DAP), visible signs of stress such as leaf rolling and glaucous leaf blade coloration appeared on the lower part of the plant, and progressed to upper leaves as stress continued. Kernels accumulated ABA from 2 to 5 d after withholding water and reached about 4- to 5-fold higher ABA levels than controls at the time of sampling for microarray analysis (9 DAP; Fig. 2). After sampling at 9 DAP, the stressed plants were rewatered; 1 d later, leaves unrolled, and ABA levels returned to normal (Fig. 2, R1). At 3 d after rewatering (12 DAP), plants had recovered water status and the sampled apical kernels had resumed growth (data not shown).
Figure 2.
ABA accumulation in maize kernels during water deficit and rewatering. Water was withheld from whole plants beginning at 4 DAP (S0) until soil water was depleted to the gravimetrically defined stress set point, which was reached between S2 and S3. Daily irrigations maintained the stress until 9 DAP (S4) when plants were rewatered. Averages ± sd of four replicates are shown.
Microarray Statistical Analysis
In microarray experiments, a complete set of treatments was imposed on each of four sequential batches of replicate plants. Each replicate was exposed to somewhat different greenhouse solar irradiance and temperature environments (see “Materials and Methods” for details) such that the experiment as a whole emphasizes the most reproducible differences among treatments. We analyzed the fluorescence data from microarray slides with the statistical analysis of microarrays (SAM) procedure (Tusher et al., 2001). This statistical method determines whether expression of each gene is significantly affected by a treatment based on the average change relative to the sd of repeated measurements of plant replicates. The method calculates a relative difference statistic, di, whose absolute value increases as the observed difference between compared treatments exceeds the experimental variability. Within a slide, triplicate spots of each clone almost always showed good agreement (data not shown), whereas biological variability and differences in extraction and labeling were the main sources of error variance. Triplicate spots were averaged before data were subjected to statistical analysis. We used normalized fluorescence signals rather than ratios in the tests of statistical significance, as recommended (Nadon and Shoemaker, 2002).
RNA Gel-Blot Analysis
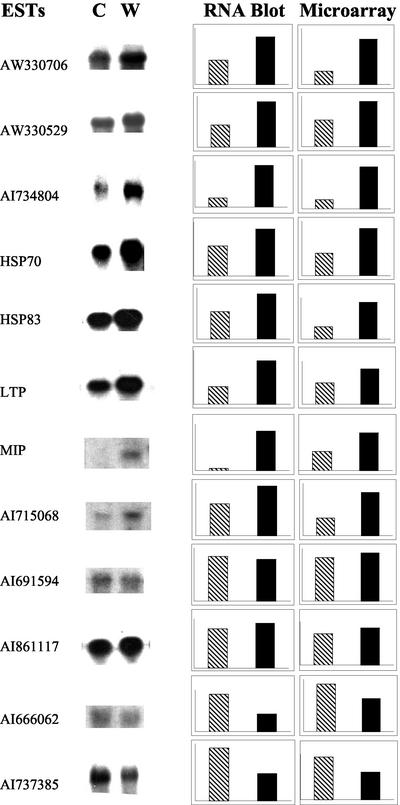
To confirm the microarray quantification, we randomly selected 12 ESTs as probes for RNA gel-blot analysis. Microarray hybridization indicated that among these transcripts, eight were up-regulated, two were down-regulated, and two were unchanged in response to water deficit in placenta tissues. Among them, several are known stress-responding genes, such as HSPs (HSP70 and HSP83), lipid transfer protein (LTP), and plasma membrane intrinsic protein, a member of the major intrinsic protein (MIP) family. The rest are unknown ESTs or ESTs with uncertain annotations. Figure 3 shows the RNA gel-blot analysis and microarray quantification of these 12 ESTs or EST contigs in control and stressed tissues. In general, microarray quantification showed good agreement with RNA gel blots, with a few differing slightly between the two methods. For instance, MIP showed up-regulation under water stress in both the RNA gel-blot and microarray analyses, but it had modest signal intensity in the control channel in microarray analysis, whereas in the RNA gel blot, the control signal of MIP was nearly zero. Nevertheless, taken as a whole, the comparison indicates that microarray quantification was reliable and comparable with RNA gel-blot results.
Figure 3.
Comparison of transcripts quantified by conventional RNA gel-blot versus cDNA microarray methods. Selected cDNA ESTs that microarray analysis indicated were up-regulated, down-regulated, or unchanged by water stress were used as probes in gel blots with 5 μg of total RNA from placenta tissue. The gene names or GenBank accession numbers of ESTs are presented with images of control (C) and water stress (W) 32P signal. Plots indicate the digitized signals (arbitrary scale) obtained for control (gray) and water stress (black) samples using RNA gel-blot and microarray methods.
Microarray Comparison of Placenta and Endosperm
Comparison of the stress effects revealed that tissues differed considerably in their transcriptional responses. In placenta, 79 genes were affected by stress, and in endosperm, 56 genes were affected, with only nine genes in common between the tissues (Fig. 1B). More detailed assessment of the changes indicated that tissues differed both in the general pattern of response and in specific genes involved (Tables I and II). In placenta, 89% of the affected genes were up-regulated, whereas in endosperm, 82% of the affected genes were down-regulated. The categories of genes affected also differed in the two tissues. In placenta, genes associated with stress, including HSPs, chaperonins, and major intrinsic proteins constituted the largest class of genes regulated, representing 20 of the 56 classified to a known functional category (Table I). All of these stress-related genes were up-regulated in response to stress. In placenta, genes were up-regulated in all categories, except a part of those under “metabolism” and “cell division and growth.” In contrast, in endosperm, only four stress-related genes were up-regulated, whereas genes in the cell division and growth category, including histones, cyclin-dependent kinase, and DNA replication licensing factor, represented a substantial class, with seven of eight of them down-regulated (Table II). Also, among genes classified as “metabolism,” several down-regulated genes were related to growth processes such as Suc utilization (Suc synthase) and cell wall breakdown (β-1,3-glucanase, β-d-glucan exohydrolase, β-galactosidase, and endoxylanase). In endosperms, down-regulated genes predominated in all categories, except genes classified as stress response.
Table I.
Genes differentially expressed in placenta tissue in response to whole-plant water deficit
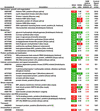 |
The expression of mRNA extracted from placenta was determined by cDNA microarray hybridization. Relative fluorescence signals are shown for those genes whose 9-DAP comparison was statistically significant according to SAM procedures. Shown are the average fluorescence ratios of four replicate microarray slides for comparisons of water stress with control placentas sampled at 9 DAP (WS/C), and for comparisons of placentas sampled 3 d after rewatering (12 DAP; Recovery from stress) with placentas sampled at 9 DAP from water stressed plants (R/WS). The SAM relative difference statistic, di, is shown for the WS/C comparison. Values of WS/C are highlighted in red or green if expression was significantly up- or down-regulated, respectively. Values of R/WS are highlighted in red or green if expression differs by ≥ 1.4-fold, indicating up- or down-regulation, respectively. The control fluorescence signal is shown for 9-DAP placentas.
Table II.
Genes differentially expressed in endosperm tissue in response to whole-plant water deficit
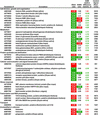 |
The expression of mRNA extracted from endosperm was determined by cDNA microarray hybridization. Relative fluorescence signals are shown for those genes whose 9-DAP comparison was statistically significant according to SAM procedures. Shown are the average fluorescence ratios of four replicate microarray slides for comparisons of water stress with control endosperms sampled at 9 DAP (WS/C), and for comparisons of endosperms sampled 3 d after rewatering (12 DAP; Recovery from stress) with endosperms sampled at 9 DAP from water stressed plants (R/WS). The SAM relative difference statistic, di, is shown for the WS/C comparison. Values of WS/C are highlighted in red or green if expression was significantly up- or down-regulated, respectively. Values of R/WS are highlighted in red or green if expression differs by ≥ 1.4-fold, indicating up- or down-regulation, respectively. The control fluorescence signal is shown for 9-DAP endosperms.
We also examined the response to rewatering. These data are potentially useful in identifying whether stress permanently affected expression, as might be expected if it aborted or arrested development, or whether altered expression was tied to concurrent imposition of the stress condition. We examined the response to rewatering by comparing transcription levels at 12 DAP in rewatered plants that had previously been stressed, relative to the transcript levels in stressed plants at 9 DAP. The rewatered to water stress ratios (R/WS) are shown in Tables I and II for those genes that had significantly responded at 9 DAP to the stress. In placenta, a substantial fraction of the genes that were up-regulated by stress at 9 DAP, returned back toward control levels after rewatering, as indicated by R/WS ratios less than 1:1 (Table I). This was most evident for genes in the stress response category, where 11 of 20, or 55% had R/WS ratios ≤0.7, whereas as a whole, 43% of stress up-regulated genes returned toward control levels upon rewatering. Thus in placenta, a substantial share of the genes responded in a pattern similar to that observed for ABA (Fig. 2) with apparent regulation based on concurrent stress condition.
Although only 6% of the stress up-regulated genes in placenta increased further after rewatering (R/WS ≥ 1.4), about one-half of them remained up-regulated after rewatering (R/WS about 1:1; Table I). This suggests that at least two regulons exist: one for genes responding specifically to factors concurrent with imposition of stress and another for genes remaining affected after a transient imposition of stress. It is plausible that the latter category might be involved in altering the timing or type of tissue development such as toward senescence or hastened differentiation. However, relatively few senescence-related gene products were up-regulated by stress, and none of these were among those with prolonged up-regulation. Also, in endosperm, zein and starch-pathway enzymes, whose expression would indicate hastened development, were not among those up-regulated. To the contrary, all of the genes that were down-regulated by stress in placenta and most of them in endosperm either remained down-regulated or were decreased even further after rewatering (R/WS ≤ 0.7; Tables I and II). Rather than hastened differentiation, many of the genes with sustained down-regulation were those associated with cell proliferation (α-tubulin and ribonucleotide reductase in placenta; cyclin-dependent kinase, β-tubulin, and DNA licensing factor in endosperm) or cell wall degradation associated with programmed cell death that occurs in the central endosperm beginning at late phases of proliferation (β-d-glucan exohydrolase, β-galactosidase, and endoxylanase in endosperm), consistent with arrested or retarded growth.
To obtain a broader basis for assessing tissue specificity in expression, we performed hierarchical cluster analysis for those genes up-regulated in either placenta or endosperm. In this analysis, we focused on those genes identified with the SAM procedure as significantly affected by stress at 9 DAP. For these, we extended our analysis to determine their response to rewatering in both placenta and endosperm using a relaxed criterion of 1.4-fold change in either an up or down direction. This analysis confirmed that for the most part, water deficit and rewatering altered the expression of a different set of genes in each tissue (Fig. 4). Among the 70 genes identified as up-regulated in placenta, only eight of them also increased in endosperm (Fig. 4A). And only 4 of 14 of the genes identified as up-regulated in endosperm also increased in placenta (Fig. 4B). Among these were several that have expected stress tolerance roles in stabilization of protein and membrane structure during stress (HSP70, DnaJ, and LTP), including trehalose-6-phosphate synthase (TPS). Trehalose, a disaccharide of Glc, is considered a compatible solute with roles in stabilization of macromolecular structure during stress. Also up-regulated in both tissues was a plasma membrane intrinsic protein (AI770766), which may provide aquaporin function.
Figure 4.
Cluster analysis of genes found significantly affected by stress or rewatering in placenta (A) and endosperm (B). Genes were clustered using hierarchical clustering, and expression ratios in columns are shown for: placenta WS/C (1), endosperm WS/C (2), placenta R/WS (3), and endosperm R/WS (4), where WS is tissue sampled from water-stressed plants at 9 DAP, C is tissue sampled from control plants at 9 DAP, and R is tissue sampled from rewatered plants at 12 DAP. Ratios greater than 1:1 are indicated in red, ratios less than 1:1 are green, and ratios equal to 1:1 are black. A gene was included in A or B of this figure if its SAM score indicated expression significantly affected by one or more treatment in placenta or endosperm, respectively. Color bands to the left of each panel group together various response patterns.
None of the genes that were down-regulated in placenta also decreased in endosperm (Fig. 4A), and only eight of the 42 genes down-regulated in endosperm also decreased in placenta (Fig. 4B). Among them were two involved in amino acid synthesis (acetyl-Glu kinase and Met synthase). In addition to Met synthase, other members of the S-adenosyl-Met cycle were also down-regulated in either placenta (S-adenosyl homo-Cys hydrolase; Table I) or endosperm (S-adenosyl Met synthase; Table II). A substantial portion of the flux through the S-adenosyl Met cycle is directed toward methyl-donor reactions in the synthesis of pectin, lignin precursors, choline, and numerous other products. This suggests that down-regulation of this pathway is related to decreased growth activities in these tissues. However, S-adenosyl homo-Cys hydrolase responded to stress in opposite directions in the two tissues. It decreased in endosperm, and increased in placenta (Fig. 4B), indicating that different roles may be played by the S-adenosyl Met cycle in each of these tissues.
Responses of Regulatory Transcripts
Several transcription factors were among the stress-regulated genes (Table I and II). Both of the transcription factors that were affected by stress in endosperm, were correspondingly affected in placenta (TATA binding protein AI881681 and AP1-like MADS box protein AI881560; Fig. 4B); and one identified in placenta correspondingly changed in endosperm (zinc finger protein AI881804; Fig. 4A). This suggests that some transcription factors have common stress roles in both tissues. But three additional transcription factors that were identified in placenta (Table I) were unchanged in endosperm (Fig. 4B). And none of the 10 stress-responsive transcripts classified as having roles in signal transduction (Table I and II) were correspondingly affected in the alternate tissue (Fig. 4, A and B). Thus for the most part, each tissue employed a different suite of regulatory factors to respond to stress.
DISCUSSION
Placenta and Endosperm Respond Differently to Water Deficit and Recovery
The transcript profiles of the two tissues examined in this study, endosperm and placenta, differed considerably in response to water deficit and recovery. Whereas most of the significantly responding genes in placenta involved up-regulation, most of the affected genes in endosperm involved down-regulation. Furthermore, stress-related genes such as HSPs and chaperonins were the largest class of genes in placenta, whereas these were a relatively minor category of affected genes in endosperm. A possible basis for this difference is a variable extent to which each tissue experienced low water potential during the stress episode. Whereas placenta is highly vascularized such that its water status can equilibrate with whole-plant water potential during a stress episode, endosperm is remote from vasculature and is hydraulically isolated to some extent. Such isolation was documented in studies where whole-plant water deficit substantially lowered water potential in leaves and floral tissues, whereas water potentials of whole maize kernels or embryos remained the same as controls (Westgate and Thomson Grant, 1989; Ober and Setter, 1990). Steep downhill gradients in water potential were found from pedicel phloem to the grain in wheat (Triticum aestivum), consistent with hydraulic isolation between vasculature and surrounding tissues (Fisher and Cash-Clark, 2000). Hence in the present study, placenta may have experienced a more pronounced lowering of tissue water status than did endosperm, and this may be a partial explanation for the greater up-regulation of stress genes in placenta than in endosperm.
Another factor that may have contributed to the greater up-regulation of stress genes in placenta than endosperm is that placenta accumulates greater concentrations of ABA during stress than does endosperm (Setter et al., 2001). Studies have indicated that when stress is imposed at the early post-pollination stage, the source of ABA that accumulates in endosperm is maternal tissues (Ober and Setter, 1992), and ABA is transported from leaves to sink organs via phloem (Ober and Setter, 1990; Zhang et al., 1996). Moreover, placenta is positioned along the transport pathway between phloem unloading and endosperm cells, where it intercepts the flux of ABA during stress and, by enzymatic hydroxylation of ABA to inactive catabolites, it modulates the levels of ABA reaching the endosperm (Wang et al., 2002). Also related to a transport role, studies indicate that the diminished flux of photosynthate into developing kernels is an important factor in determining the developmental fate of maize kernels during stress (Zinselmeier et al., 1999). By its direct linkage via phloem with whole-plant carbohydrate status, the placenta may respond to a greater extent than endosperm to carbohydrate deprivation or to the interplay of ABA- and sugar-signaling interactions (Finkelstein and Gibson, 2001).
A Large Group of Stress-Related Genes Were Up-Regulated
A large proportion of the genes that were up-regulated by water stress encode proteins that assist in protein folding and stabilize macromolecular structure, such as chaperonins and HSPs. In placenta, eight HSPs were up-regulated, including representatives of several HSP families: two HSP70s, five HSP90s (80–94 kD), and one small HSP (15–30 kD; Tables I and II). Studies indicate that the function of HSP70s in protein folding and stabilization involve interaction with DnaJ and other cochaperonins (Netzer and Hartl, 1998). Two genes encoding proteins of the DnaJ family were among those up-regulated in placenta. Although HSPs have important chaperonin roles during high-temperature stress, they are also expressed at low levels in non-stress conditions, because they are involved in folding newly translated polypeptides (Netzer and Hartl, 1998; Krishna, 2000). In addition to thermal stress, studies in plants have indicated that HSPs are up-regulated in response to low water potential and to exogenously applied ABA (Pareek et al., 1995; Coca et al., 1999; Sun et al., 2001), consistent with the present findings. Other genes up-regulated by water stress in both placenta and endosperm were LTP and cyclophilins (Tables I and II). LTP has been shown to have remarkable stability to a variety of stress conditions, including denaturants and heat up to 100°C (Lindorff and Winther, 2001), consistent with its elevated expression during stress. Cyclophilins, a protein family of which one member was up-regulated during water stress in both placenta and endosperm, are also chaperonins that have been shown to interact with HSPs (Andreeva et al., 1999). Thus, the current study indicates that proteins with involvement in protein folding and stabilization were the most numerous of the up-regulated genes in water-stressed kernels.
Three genes encoding plasma membrane aquaporins, which are members of the MIP family, were up-regulated during water stress. Two of those up-regulated in placenta and the one in endosperm encode plasma membrane MIPs of the ZmPIP1–3 subfamily (AI770766 and AI734741), whereas the third one in placenta is in the ZmPIP2–1 subfamily (AI734803; Chaumont et al., 2001). Aquaporins serve as membrane water channels that increase the permeability for liquid water movement. Some studies have indicated that ABA and stress increase aquaporin transcript abundance (Mariaux et al., 1998; Barrieu et al., 1999), whereas others have shown decreased transcript levels (Smart et al., 2001). Aquaporins are often highly expressed in meristematic and rapidly expanding regions, consistent with the need for water flux during growth (Chaumont et al., 1998). Studies of rice seedlings exposed to salinity stress indicated that MIP transcript levels increase at advanced phases of a stress cycle, perhaps indicating recovery as plants acclimate to the stress (Kawasaki et al., 2001). Consistent with this interpretation, all of the MIP transcripts that were up-regulated during stress in the present study, remained at elevated levels after rewatering, when rapid cell rehydration and growth resume.
TPS was significantly up-regulated in placenta (Table I), and was apparently elevated in endosperm as well (Fig. 4B). Given that TPS was the only osmolyte-synthesizing enzyme up-regulated in the current study, this finding suggests a unique role for this disaccharide. Although trehalose is known to be an important compatible solute that contributes to stress tolerance and macromolecular stability in yeasts and other organisms, its importance in plants has been uncertain. Recent findings that genes encoding enzymes leading to trehalose synthesis are widespread in plants and that trehalose accumulation enhances plant water stress or salinity tolerance have suggested that it also plays important functions in plants (Garcia et al., 1997; Romero et al., 1997; Eastmond et al., 2002). However, the quantity of trehalose accumulated during stress has generally been considered too small to contribute to plant osmoprotection (Garcia et al., 1997; Romero et al., 1997). Nevertheless, the recent discovery that an Arabidopsis mutation in TPS, tps1, is embryo lethal suggests that trehalose metabolism or regulatory functions of TPS are essential, at least in some tissues or growth stages (Eastmond et al., 2002). Thus further study of the functions of trehalose and TPS in maize kernels is warranted.
In contrast to many studies involving several-day periods of stress, only a few senescence-related genes were up-regulated in the current study. In endosperm, stress increased the expression of a 20S proteosome β-subunit and a ubiquitin proteosome-associate protein (OsRAD23), both of which are a components of the ubiquitin system for proteolytic turnover. In placenta, stress up-regulated an uncharacterized senescence-associated protein (AI714517). But contrary to these effects, several cell wall breakdown enzymes, often associated with plant cell senescence and programmed cell death, were down-regulated by stress. Included were endoxylanosidase, β-galactosidase, β-d-glucan exohydrolase, and β-1,3-glucanase. A possible explanation is that these latter enzymes are expressed as a normal event in endosperm development, which stress delayed, thereby lowering the observed ratio of their expression relative to controls. In support of this interpretation, studies have indicated that midway through endosperm development, the central cells of the endosperm engage in programmed cell death, resulting in nDNA degradation, cell lysis and collapse (Young and Gallie, 2000). Furthermore, these studies employed ABA mutants and exogenous treatments to show that ABA delays the onset and decreases the rate of programmed cell death, consistent with the present observation of an association between high-ABA levels during stress and lowered expression of cell wall degrading enzymes. The current studies also show that a gene encoding the ethylene synthesis pathway enzyme, 1-aminocyclopropane 1-carboxylic acid oxidase, was down-regulated in endosperm. Previous study of maize endosperm had shown that a peak in ethylene production occurs at about 16 DAP (Young et al., 1997). Ethylene was shown to have a regulatory role in programmed cell death because ethylene treatment of kernels hastened and amplified endosperm cell breakdown and nDNA fragmentation (Young et al., 1997). The decreased expression of 1-aminocyclopropane 1-carboxylic acid oxidase in water-stressed kernels is consistent with the proposed model of delayed development. Thus, taken as a whole, the current and cited studies suggest that response to water stress may involve both increased ubiquitin-directed turnover of proteins, perhaps those damaged by stress or not part of the expression profile during a stress episode, and delayed development, including normal programmed cell death, relative to controls.
Genes Related to Cell Division and Growth Were Down-Regulated in Endosperm
Several cell cycle-related genes were down-regulated in endosperms during stress. These included two histone H2Bs, a β-tubulin, and a cyclin-dependent kinase. This result agrees with previous findings that water stress during the early period of endosperm development decreases transcription of members of these the gene families concomitant with decreased rates of cell division and nDNA endoreduplication (Setter and Flannigan, 2001). These transcripts have known roles in the cell cycle, and are known to be expressed at highest levels in actively proliferating cells. Also down-regulated were replication protein A1 (RPA1), DNA replication licensing factor Mcm5, and ribonucleotide reductase. RPA1 is involved in stabilizing single-stranded DNA during DNA replication. In rice, its expression is highest in tissues containing dividing cells, and in stem tissues its expression is stimulated with exogenous gibberellin, which also increases the rate of cell division and growth (van der Knapp et al., 1997). Mcm5, a member of a family of minichromosome maintenance factors that were first discovered in yeast, is a component of DNA replication licensing factor, which is believed to be responsible for restriction of DNA synthesis to once per cell cycle (Tye, 1999). Mcm proteins are highly expressed in cells engaged in cell proliferation. In plants, this has been demonstrated for the Arabidopsis gene, PROLIFERA, an Mcm7 homolog required for megagametophyte and embryo development that is expressed in dividing cells throughout the plant (Springer et al., 1995). Ribonucleotide reductase catalyzes the biosynthesis of deoxyribonucleotides. It is the rate-limiting enzyme for DNA synthesis in many systems, and accordingly its expression is specific to proliferating cells and the S phase of the cell cycle (Chaboute et al., 1998). Thus, the down-regulation of these seven transcripts in endosperm during stress and the significant recovery upon rewatering by three of them provide strong evidence that the previously observed stress inhibition of cell proliferation (Ober et al., 1991; Artlip et al., 1995; Setter and Flannigan, 2001) involves down-regulation of a large suite of cell cycle genes.
Although stress down-regulated the above-mentioned cell proliferation genes in endosperm, stress up-regulated a member of the histone H2A family. Such contrary expression may indicate that this H2A is a histone variant that is uniquely expressed during stress, analogous to the situation reported for a water stress and ABA up-regulated histone H1 variant in tomato (Scippa et al., 2000). Studies indicate that variants of histone H2A have specialized roles through alterations they create in chromatin stability and folding (Ausio and Abbott, 2002). Thus the contrary trend in the present case may reflect this H2A's specialized role during stress when stability properties may be important.
Response of Signaling and Transcription Factors to Water Deficit
Several signaling and transcription factors were among the stress-regulated genes in the present study. Previous study of abiotic stress has shown that some transcription factors respond within an hour of stress imposition, and many of these responses are transient (Fowler and Thomashow, 2002; Seki et al., 2002). Although sampling after 2 to 5 d of stress in the current study may have missed such early events, several potential signaling factors were identified. In placenta, all of the regulated genes classified as functioning in cellular communication and signal transduction and all except one classified as transcription and RNA processing involved increased transcript levels during water stress. In contrast, in endosperm, most of the regulated genes classified as signal transduction were down-regulated, whereas there was one transcription factor up- and one down-regulated.
Calcium-Dependent Protein Kinases
In both endosperm and placenta, a calcium-dependent protein kinase was up-regulated. Several studies in rice and other species have indicated that members of the CDPK family are involved in signal transduction pathways of several stress responses (Saijo et al., 2000; Cheng et al., 2002). When we aligned the two CDPKs in the current study with others (Saijo et al., 2000; Cheng et al., 2002; Ozturk et al., 2002), we found that they differ from each other and from those reported in previous studies. OsCDPK7 is homologous to the type-I CDPKs of Arabidopsis (Cheng et al., 2002) and is up-regulated in response to salt and salt stress (Saijo et al., 2000; Kawasaki et al., 2001; Ozturk et al., 2002). Saijo et al. (2000) found that overexpression of OsCDPK in rice conferred both cold and salt/drought tolerance in rice seedlings. Whether the water stress-up-regulated genes observed in the current study have overlapping signaling targets with this and other CDPKs or have unique tissue specificity awaits further analysis.
ABA-Insensitive 1/2 (ABI1/ABI2) Protein Phosphatase 2C
An ortholog of the two closely related Arabidopsis genes ABI1 and ABI2 was up-regulated in placenta. This finding agrees with previous studies in Arabidopsis where ABI1 was up-regulated by low temperature, drought, high salt, and ABA (Tahtiharju and Palva, 2001). An ABI1 ortholog was also up-regulated in kernels of maize plants that were subjected to shade stress (Zinselmeier et al., 2002). ABI1 and ABI2 are now recognized as negative regulators of ABA signaling (Merlot et al., 2001; Shen et al., 2001). Thus, the observed ABA-induced up-regulation of ABI1/2 protein phosphatases might be part of a signal-attenuating feedback loop of the ABA signal transduction pathway (Tahtiharju and Palva, 2001).
Protein Kinase PK4
A protein kinase of the PK4 family of SNF1-related proteins (SnRK3 subgroup) was up-regulated in placenta. Studies of closely related PK4s in rice (OsPK4) and wheat (WPK4) indicate that their transcription is increased when Suc level is decreased (Ikeda et al., 1999). This suggests a possible signaling role in the abundant phloem of the placenta, whereby decreased availability of Suc during stress might initiate signaling and metabolic regulation via PK4.
Homeodomain Leu Zipper (HD-Zip) Transcription Factor
In the current study, stress up-regulated an HD-Zip with 93% nucleotide identity with ZmOCL5, an HD-Zip from maize (Ingram et al., 2000). Previous work showed that in maize and rice, a family of HD-Zips related to the Arabidopsis GLABRA2 gene (GL2) are expressed in an epidermis-specific pattern during early development of embryos and other organs (Ingram et al., 2000; Ito et al., 2002). In situ hybridizations show that ZmOCL5 expression is most prominent in the abaxial face of protodermal embryo layer, but expression is also found in the endosperm and in young floral tissues. Thus it is conceivable that stress up-regulation of the HD-Zip, reported currently, provides tissue-specific stress responses in kernels.
Other Members of Gene Families and Unknowns
In addition to those discussed above, several other putative signaling factors were regulated by stress. Many of these are members of large gene families for which a detailed understanding is known for only a few representatives. For example, Goff et al. (2002) estimated that in the rice genome, there are 156 MYB and 160 zinc finger transcription factor genes. Also included in the current findings are 24 transcripts whose function is unknown based on available sequence and comparison with published information. Thus, by identifying those transcripts that are significantly regulated by stress in maize kernels, the present work provides a valuable starting point for further elucidation of the roles played by these gene products.
In summary, we have shown that water deficit elicited substantially different gene expression profiles in placenta and endosperm. Although the predominate response in placenta was increased expression of stress tolerance proteins, endosperm responded with an expression profile indicating arrested growth, down-regulation of cell cycle genes, and slowed developmental advance. These responses may help improve kernel survival during stress. In placenta, increased expression of stress tolerance proteins may enhance the likelihood of tissue survival in the face of decreasing water potentials and may maintain phloem function. In endosperm, arrested growth and development will decrease demand for limited supplies of photosynthate and may poise it for rapid resumption of growth after rewatering. When stress becomes more prolonged or severe, the placenta, with its vascular connection and responsiveness to the whole-plant status of water, photosynthate, and ABA may play a key role in determining the threshold for kernel abortion and conveying signals to endosperm. The information obtained in the present study will help point the way to factors that regulate such development.
MATERIALS AND METHODS
Plant Material and Stress Treatments
Maize (Zea mays cv Pioneer Brand 39K72) was grown in a greenhouse with supplemental lighting and hourly irrigation as described by Setter et al. (2001). Four batches of plants, grown in different times of the year, were used in the study. Average day/night during the stress periods were 24.4°C/15.6°C, 26.6°C/18.4°C, 25.3°C/15.2°C, and 24.6°C/15.6°C for batches 1 to 4, respectively. Average daily photon flux was 33, 43, 43, and 17 mol photons (400–700 nm wavelength) m−2 d−1 for batches 1 to 4, respectively. Treatments (control and stress) were randomly assigned to paired equivalent plants in each batch. Plants were subjected to water deficit treatment beginning at 5 DAP. These plants were fully irrigated and allowed to drain, and then the mass of plants and soil was obtained. Irrigation was withheld until plants depleted water to a set point of 50% of initial weight of plant + pot. The set point was maintained by periodic addition of irrigation solution until sampling at 9 DAP. The stressed plants were then rewatered and regular irrigation was continued until 12 DAP.
ABA Measurement
ABA was measured according to Setter et al. (2001). In brief, maize kernels from stressed and control plants were dissected, weighed, and placed immediately in cold 80% (v/v) methanol on ice. Tissues were macerated to extract ABA and stored at −20°C. The ABA extract was fractionated by C18 reverse-phase chromatography, and the ABA fractions were assayed by enzyme-linked immunosorbant assay (Setter et al., 2001).
RNA Extraction and Labeling
Endosperm and placenta/pedicel tissues in the apical region of the ear, the upper 33% with respect to ear length, were dissected free of embryo, nucellus, and pericarp and frozen immediately in liquid nitrogen until RNA extraction. Total RNA was extracted using a kit that employs guanidine isothiocyanate and a silica gel-based membrane (Qiagen USA, Valencia, CA) according to the manufacture's procedure. RNA targets were labeled with aminoallyl dUTP via first-strand cDNA synthesis followed by coupling of the aminoallyl groups to either Cyanine 3 or Cyanine 5 fluorescent molecules, according to the protocol of Hasseman (2001).
Microarray Processing and Data Analysis
Slides of the maize immature ear tissue 606 microarray were obtained from the microarray laboratory of the Maize Gene Discovery project as described by Fernandes et al. (2002). Labeled cDNA was hybridized to these slides according to the protocol recommended (Fernandes et al., 2002; details at http://zmdb.iastate.edu/zmdb/microarray/protocols.html). After washing, the microarray slides were dried briefly by centrifugation. They were then scanned by a laser scanner (ScanArray 5000, GSI Lumonics, Wilmington, MA) for both channel 1 (Cy3) and 2 (Cy5) at 10-μm resolution. The channel 1 and channel 2 images were analyzed using ScanAlyze software (v2.35, Stanford University, http://genome-ww4.stanford.edu/Microarray/SMD/restech.html; Eisen et al., 1998) to obtain average signal for each spot and to screen out spots with poor uniformity or in regions with high background. Microarray data were then analyzed using Microsoft Excel (Microsoft, Redmond, WA). Local median background was subtracted from the total channel intensity of each spot. The net channel intensities were used for calculating ratios after normalization. Normalization was done according to Pérez-Amador et al. (2001).
Normalized data from triplicate spots within each slide were first averaged to obtain each gene's fluorescence value, and then values from four replicates of each treatment/tissue combination from four different batches of plants were analyzed by SAM, a statistical analysis tool (Tusher et al., 2001). The treatments were randomly assigned to plants in the four batches, as in a randomized complete block design, and each slide was hybridized with a Cy3/Cy5-labeled pair of cDNA from a batch of plants. We reversed the assignment of Cy3/Cy5 dyes for stress/control treatment pairs between batches. For the analysis, normalized fluorescence data for each pair of treatments in four replicate slides were input, and for each gene, a relative difference statistic, di, was calculated (observed di), as well a balanced set of permutations of the replicate data for that gene, representing random variability (expected di). Genes were called significant at a false discovery rate set at 11% for placenta and at 15% for endosperm. After analysis, significant genes with ratios between treatments of >1.6 (up-regulated) or <0.7 (down-regulated) were selected and subjected to hierarchical cluster analysis with J-Express v2.1 (http://www.molmine.com).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Footnotes
This work was supported by the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture (grant no. 00–35100–9279).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.014365.
LITERATURE CITED
- Andreeva L, Heads R, Green CJ. Current status review: cyclophilins and their possible role in the stress response. Int J Exp Pathol. 1999;80:305–315. doi: 10.1046/j.1365-2613.1999.00128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artlip TS, Madison JT, Setter TL. Water deficit in developing endosperm of maize: cell division and nuclear endoreduplication. Plant Cell Environ. 1995;18:1034–1040. [Google Scholar]
- Ausio J, Abbott DW. The many tales of a tail: carboxyl-terminal tail herterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry. 2002;41:5945–5949. doi: 10.1021/bi020059d. [DOI] [PubMed] [Google Scholar]
- Barrieu F, Marty MD, Thomas D, Chaumont F, Charbonnier M, Marty F. Desiccation and osmotic stress increase the abundance of mRNA of the tonoplast aquaporin BobTIP26–1 in cauliflower cells. Planta. 1999;209:77–86. doi: 10.1007/s004250050608. [DOI] [PubMed] [Google Scholar]
- Chaboute M, Edith CB, Clement B, Gigot C, Philipps G. Molecular characterization of tobacco ribonucleotide reductase RNR1 and RNR2 cDNAs and cell cycle-regulated expression in synchronized plant cells. Plant Mol Biol. 1998;38:797–806. doi: 10.1023/a:1006083318906. [DOI] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 1998;117:1143–1152. doi: 10.1104/pp.117.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001;125:1206–1215. doi: 10.1104/pp.125.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129:469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claassen MM, Shaw RH. Water deficit effects on corn: II. Grain components. Agron J. 1970;62:652–655. [Google Scholar]
- Coca MA, Almoguera C, Thomas TL, Jordano J. Differential regulation of small heat-shock genes in plants: analysis of a water-stress-inducible and developmentally activated sunflower promoter. Plant Mol Biol. 1999;31:863–876. doi: 10.1007/BF00019473. [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJH, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JDG, Smeekens SC, Graham IA. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 2002;29:225–235. doi: 10.1046/j.1365-313x.2002.01220.x. [DOI] [PubMed] [Google Scholar]
- Edmeades GO, Bolanos J, Elings A, Ribaut JM, Banziger JM, Westgate ME. The role and regulation of the anthesis-silking interval in maize. In: Westgate ME, Boote KJ, editors. Physiology and Modeling Kernel Set in Maize. Madison, WI: Crop Science Society of America; 2000. pp. 43–73. [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Brendel V, Gai X, Lal S, Chandler VL, Elumalai RP, Galbraith DW, Pierson EA, Walbot V. Comparison of RNA expression profiles based on maize expressed sequence tag frequency analysis and micro-array hybridization. Plant Mol Biol. 2002;128:896–910. doi: 10.1104/pp.010681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gibson SI. ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol. 2001;5:26–32. doi: 10.1016/s1369-5266(01)00225-4. [DOI] [PubMed] [Google Scholar]
- Fisher DB, Cash-Clark CE. Gradients in water potential and turgor pressure along the translocation pathway during grain filling in normally watered and water-stressed wheat plants. Plant Physiol. 2000;123:139–148. doi: 10.1104/pp.123.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AB, De Almeida EJ, Iyer S, Gerats T, Van Montagu M, Caplan AB. Effects of osmoprotectants upon NaCl stress in rice. Plant Physiol. 1997;115:159–169. doi: 10.1104/pp.115.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Gong Z, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu J, Cushman JC, Bressan RA et al. Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol. 2001;126:363–375. doi: 10.1104/pp.126.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RF, Jackson BS, Kiniry KR, Arkin GF. Water deficit timing effects on yield components in maize. Agron J. 1989;81:61–65. [Google Scholar]
- Hasseman J (2001) Aminoallyl labeling of RNA for microarrays. The Institute for Genomic Research. http://atarrays.tigr.org (August 2, 2001)
- Ikeda Y, Koizumi N, Kusano T, Sano H. Sucrose and cytokinin modulation of WPK4, a gene encoding a SNF1-related protein kinase from wheat. Plant Physiol. 1999;121:813–820. doi: 10.1104/pp.121.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram GC, Boisnard-Lorig C, Dumas C, Rogowsky PM. Expression patterns of genes encoding HD-ZipIV homeo domain proteins define specific domains in maize embryos and meristems. Plant J. 2000;22:401–414. doi: 10.1046/j.1365-313x.2000.00755.x. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Ito M, Sentoku N, Nishimura A, Hong S-K, Sato Y, Matsuoka M. Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J. 2002;29:497–507. doi: 10.1046/j.1365-313x.2002.01234.x. [DOI] [PubMed] [Google Scholar]
- Kawasaki S, Borchert C, Deyholos M, Wang H, Brazille S, Kawai K, Galbraith D, Bohnert HJ. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell. 2001;13:889–905. doi: 10.1105/tpc.13.4.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiesselbach TA. The Structure and Reproduction of Corn. Lincoln: University of Nebraska Press; 1949. [Google Scholar]
- Kowles RV, Phillips RL. Endosperm development in maize. Int Rev Cytol. 1988;112:97–136. [Google Scholar]
- Krishna P. Plant Hsp90 and its partner proteins. J Plant Biochem Biotechnol. 2000;9:53–56. [Google Scholar]
- Lindorff LK, Winther JR. Surprisingly high stability of barley lipid transfer protein, LTP1, towards denaturant, heat and proteases. FEBS Lett. 2001;488:145–148. doi: 10.1016/s0014-5793(00)02424-8. [DOI] [PubMed] [Google Scholar]
- Mambelli S, Setter TL. Inhibition of maize endosperm cell division and endoreduplication by exogenously applied abscisic acid. Physiol Plant. 1998;104:266–272. [Google Scholar]
- Mariaux JB, Bockel C, Salamini F, Bartels D. Desiccation- and abscisic acid-responsive genes encoding major intrinsic protein (MIPs) from the resurrection plant Craterostigma plantagineum. Plant Mol Biol. 1998;38:1089–1099. doi: 10.1023/a:1006013130681. [DOI] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J. A floret by any other name: control of meristem identity in maize. Trends Plant Sci. 2000;5:61–66. doi: 10.1016/s1360-1385(99)01541-1. [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat JAD. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Nadon R, Shoemaker J. Statistical issues with microarrays: processing and analysis. Trends Genet. 2002;18:265–271. doi: 10.1016/s0168-9525(02)02665-3. [DOI] [PubMed] [Google Scholar]
- Netzer WJ, Hartl FU. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- Nicolas ME, Gleadow RM, Dalling MJ. Effect of post-anthesis drought on cell division and starch accumulation in developing wheat grains. Ann Bot. 1985;55:433–444. [Google Scholar]
- Ober ES, Setter TL. Timing of kernel development in water stressed maize: water potentials and abscisic acid concentrations. Ann Bot. 1990;66:665–672. [Google Scholar]
- Ober ES, Setter TL. Water deficit induces abscisic acid accumulation in endosperm of maize viviparous mutants. Plant Physiol. 1992;98:353–356. doi: 10.1104/pp.98.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober ES, Setter TL, Madison JT, Thompson JF, Shapiro PS. Influence of water deficit on maize endosperm development: enzyme activities and RNA transcripts of starch and zein synthesis, abscisic acid, and cell division. Plant Physiol. 1991;97:154–164. doi: 10.1104/pp.97.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui ME, Andrade FH, Suero EE. Growth, water use, and kernel abortion of maize subjected to drought stress at silking. Field Crops Res. 1995;40:87–94. [Google Scholar]
- Ozturk ZN, Talamé V, Deyholos M, Michalowski CB, Galbraith DW, Gozukirmizi N, Tuberosa R, Bohnert HJ. Monitoring large-scale changes in transcript abundance in drought- and salt-stressed barley. Plant Mol Biol. 2002;48:551–573. doi: 10.1023/a:1014875215580. [DOI] [PubMed] [Google Scholar]
- Pareek A, Singla SL, Grover A. Immunological evidence for accumulation of two-high-molecular-weight (104 and 90 kDa) HSPs in response to different stresses in rice and in response to high temperature stress in diverse plant genera. Plant Mol Biol. 1995;29:293–301. doi: 10.1007/BF00043653. [DOI] [PubMed] [Google Scholar]
- Pérez-Amador MA, Lidder P, Johnson MA, Landgraf J, Wisman, Green PJ. New molecular phenotypes in the dst mutants of Arabidopsis revealed by DNA microarray analysis. Plant Cell. 2001;13:2703–2717. doi: 10.1105/tpc.010295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero C, Belles JM, Vaya JL, Serrano R, Culianez-Macia FA. Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta. 1997;201:293–297. doi: 10.1007/s004250050069. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Schussler JR, Westgate ME. Assimilate flux determines kernel set at low water potential in maize. Crop Sci. 1995;35:1074–1080. [Google Scholar]
- Scippa GS, Griffiths A, Chiatante D, Bray EA. The H1 histone variant of tomato, H1-S, is targeted to the nucleus and accumulates in chromatin in response to water-deficit stress. Planta. 2000;211:173–181. doi: 10.1007/s004250000278. [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Abe H, Kasuga M, Yamaguchi-Shinozaki K, Carninci P, Hayashizaki Y, Shinozaki K. Monitoring the expression pattern of 1,300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell. 2001;13:61–72. doi: 10.1105/tpc.13.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al. Monitoring the expression profiles of 7,000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31:279–292. doi: 10.1046/j.1365-313x.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- Setter TL, Flannigan BA. Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. J Exp Bot. 2001;52:1401–1408. doi: 10.1093/jexbot/52.360.1401. [DOI] [PubMed] [Google Scholar]
- Setter TL, Flannigan BA, Melkonian J. Loss of kernel set due to water deficit and shade in maize: carbohydrate supplies, abscisic acid, and cytokinins. Crop Sci. 2001;41:1530–1540. [Google Scholar]
- Shen Q, Gomez-Cadenas A, Zhang P, Walker-Simmons MK, Sheen J, Ho THD. Dissection of abscisic acid signal transduction pathways in barley aleurone layers. Plant Mol Biol. 2001;47:437–448. doi: 10.1023/a:1011667312754. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Mizoguchi T, Urao T, Katagiri T, Nakashima K, Abe H, Ichimura K, Liu Q, Nanjyo T et al. Molecular responses to water stress in Arabidopsis thaliana. J Plant Res. 1998;111:345–351. [Google Scholar]
- Smart L, Moskal WA, Cameron KD, Bennett AB. MIP genes are down-regulated under drought stress in Nicotiana glauca. Plant Cell Physiol. 2001;42:686–693. doi: 10.1093/pcp/pce085. [DOI] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA. Gene trap tagging of PROLIFERA, an essential MCM2–3-5-like gene in Arabidopsis. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Sun W, Bernard C, van de Cotte B, Van Montagu M, Verbruggen N. At-HSP17.6A, encoding a small heat-shock protein in Arabidopsis, can enhance osmotolerance upon overexpression. Plant J. 2001;27:407–415. doi: 10.1046/j.1365-313x.2001.01107.x. [DOI] [PubMed] [Google Scholar]
- Tahtiharju S, Palva T. Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis thaliana. Plant J. 2001;26:461–470. doi: 10.1046/j.1365-313x.2001.01048.x. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK. MCM proteins in DNA replication. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- van der Knapp E, Jagoueix S, Kende H. Expression of an ortholog of replication protein A1 (RPA1) is induced by gibberellin in deepwater rice. Proc Natl Acad Sci USA. 1997;94:9979–9983. doi: 10.1073/pnas.94.18.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Mambelli S, Setter TL. Abscisic acid catabolism in maize kernels in response to water deficit at early endosperm development. Ann Bot. 2002;90:623–630. doi: 10.1093/aob/mcf239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westgate ME, Thomson Grant DL. Water deficits and reproduction in maize. Response of the reproductive tissue to water deficits at anthesis and mid-grain fill. Plant Physiol. 1989;91:862–867. doi: 10.1104/pp.91.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TE, Gallie DR. Regulation of programmed cell death in maize endosperm by abscisic acid. Plant Mol Biol. 2000;42:397–414. doi: 10.1023/a:1006333103342. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR, DeMason DA. Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiol. 1997;115:737–751. doi: 10.1104/pp.115.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Hartung W, Komor E, Schobert C. Phloem transport of abscisic acid in Ricinus communis L. seedlings. Plant Cell Environ. 1996;19:471–477. [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeier C, Jeong BR, Boyer JS. Starch and the control of kernel number in maize at low water potentials. Plant Physiol. 1999;121:25–35. doi: 10.1104/pp.121.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinselmeier C, Sun Y, Helentjaris T, Beatty M, Yang S, Smith H, Habben J. The use of gene expression profiling to dissect the stress sensitivity of reproductive development in maize. Field Crop Res. 2002;75:111–121. [Google Scholar]