Abstract
The splicing factor Transformer-2 (Tra2) is expressed almost ubiquitously in Drosophila adults, but participates in the tissue-specific regulation of splicing in several RNAs. In somatic tissues Tra2 participates in the activation of sex-specific splice sites in doublesex and fruitless pre-mRNAs. In the male germline it affects splicing of other transcripts and represses removal of the M1 intron from its own pre-mRNA. Here we test the hypothesis that the germline specificity of M1 repression is determined by tissue-specific differences in Tra2 concentration. We find that Tra2 is expressed at higher levels in primary spermatocytes of males than in other cell types. Increased Tra2 expression in other tissues reduces viability in a manner consistent with known dose-dependent effects of excessive Tra2 expression in the male germline. Somatic cells were found to be competent to repress M1 splicing if the level of Tra2 transcription was raised above endogenous concentrations. This suggests not only that M1 repression is restricted to the germline by a difference in Tra2 transcription levels but also that the protein's threshold concentration for M1 regulation differs from that of doublesex and fruitless RNAs. We propose that quantitative differences in regulator expression can give rise to cell-type-specific restrictions in splicing.
INTRODUCTION
Alternative pre-mRNA splicing is an important mechanism affecting gene function. The utilization of alternative splice sites is regulated by a variety of proteins that interact with elements in the pre-mRNA to affect the function and/or assembly of spliceosomal complexes on them. Perhaps the best understood of these regulators come from the SR superfamily of splicing factors (1). Proteins from this family are critical components in the earliest steps of constitutive splice site recognition and often bind specifically near alternative splice sites to favor their selection.
Transformer-2 (Tra2) is an SR superfamily protein required for sex-specific splicing in several mRNAs involved in sexual differentiation in Drosophila (2). Mammalian orthologues of Tra2 have been reported to affect alternative splicing of a number of RNAs, including transcripts implicated in breast cancer (3), neuropathies (4–9) and sexual differentiation (10). The targets of Tra2 regulation appear to be restricted by tissue type. Drosophila Tra2 is expressed in most somatic tissues where it is required along with Transformer (Tra) and other SR proteins to activate the female-specific utilization of an alternative 3′ splice site in the doublesex pre-mRNA (11,12). This leads to the formation of an mRNA encoding the Dsx-F protein which specifies female differentiation in these tissues. In the nervous system Tra2 and Tra similarly direct female-specific utilization of an alternative 5′ splice site in fruitless pre-mRNA resulting in sex-specific expression of Fruitless protein isoforms which in turn specify sex-specific behaviors (13). A third function of Tra2 is restricted to the male germline where it represses removal of the M1 intron from its own pre-mRNA (14,15). The presence of the intron prevents the translation of functional Tra2 protein isoforms and thus splicing repression serves as a negative feedback mechanism (16). This mechanism affects a significant fraction of germline Tra2 transcripts where ∼50% retain the M1 intron in a manner dependent on functional Tra2 protein. Studies on an altered tra2 gene lacking the M1 intron indicate that its retention in these RNAs prevents excess or inappropriate Tra2 expression that would otherwise reduce fertility (17).
The mechanism by which M1 repression is restricted to the male germline is not known. Tra2 is translated and functions in a variety of somatic tissues in the adult, but Tra2-dependent repression of M1 splicing does not occur in these cells (18). Rather only a small fraction of Tra2 transcripts retain M1 and this intron retention is independent of Tra2 protein (19,20). It has previously been suggested that Tra2-dependent M1 retention requires additional germline-specific splicing regulators but such factors have not yet been identified. Moreover, nuclear extracts from somatically derived Schneider 2 cells have been found to support Tra2-dependent repression of M1 splicing (15). This suggests that a germline-specific factor is not required for M1 repression.
Here we test an alternate hypothesis that the tissue specificity of M1 repression results from differences in Tra2 concentration levels in the soma and germline. This idea is suggested by the observations that the highest levels of Tra2 expression are found in adult spermatocytes and that the fraction of M1 retaining RNA in these cells is highly sensitive to gene dose (17). Our results support this hypothesis and suggest that splicing factors are able to recognize different sets of targets at different cellular concentration levels.
MATERIALS AND METHODS
Plasmid constructs
The plasmid pActTra2 was constructed by inserting a 533 nt fragment containing the Actin5C distal promoter upstream of a Tra2 cDNA insert encoding the Tra2-P2 (previously called ‘Tra2-226’) protein isoform. The plasmid pActTra2-Nae+1 was generated by using the same Actin promoter fragment to replace the region upstream of the ApaI site in the plasmid pTZ3.9A-ORF3 (18). This places the Actin promoter and initiation site 23 nt upstream of the natural male germline transcription start which is internal to exon 3. The plasmid contains 3.7 kb of genomic sequences including all downstream tra2 exons and introns. The plasmid pUAS-MycTra2 was generated from the vector pTMW (Drosophila Genomic Resource Center and T. D. Murphy, unpublished data) by inserting a Tra2-P2 encoding cDNA at the translation start codon to form an N-terminal fusion with a 6× Myc epitope tag. We subsequently determined that the provided version of this vector contains only two UAS elements. We therefore replaced its UAS cassette with the equivalent region from the plasmid pUAST which has five copies of the UAS (21).
Transfection of Schneider 2 cells
For each transfection, 1.0 × 106 cells were seeded in 100 mm plates. Cells were transfected using the cellfectin reagent (Invitrogen, CA) and a total of 20 μg plasmid DNA. The DNA mixture included 5 μg of pTZ-Tra2Nae+1 in all cases except where noted as well as 0, 1.5 or 5 μg of either pSK-ActTra2 or pSK-ActβGal construct using cellfectin reagent according to the manufacturer's manual. Cells were harvested 48 h after transfection for RNA preparation.
RNA isolation and RT–PCR
RNA from S2 cells was extracted using the Trizol reagent (Invitrogen). Reverse transcription was performed using Superscript-II reverse transcriptase (Invitrogen) and oligo(dT) primers on 20 μg total RNA. One-fourth of the reverse transcription product was used for low-cycle PCR with a sense primer from the Actin–Tra2 junction (Act/T2×3 5′-CTACCGTTTGAGGGCCCTTTC-3′) and an antisense primer overlapping the 7 nt insertion in exon 4 (Nae+1 5′-GACCGCTGGTGCCGATCCCCG-3′). PCR conditions were 5 min at 94°C, followed by 20 cycles, each consists of 30 sec at 94°C, 60 sec at 58°C and 60 sec at 72°C, with a final extension for 10 min. To detect PCR products,10 μl of the amplification was separated on a 1% agarose gel, blotted and hybridized with a 32P kinase-labeled oligonucleotide X3-1182 (5′-ACCCGATTTCATTTCATTGGAAG-3′) in the Rapid-Hyb buffer (Amersham Biosciences). The blots were exposed to a phosphoimager screen and Kodak 1-D software was used to quantify the data.
RNA from adult flies was produced by collecting 50–100 females as described previously (16). Amplification of endogenous RNA from Drosophila adults was carried out using a procedure similar to that described above except that a sense primer from within exon 3 (X3S, 5′-CTCAGCCGATTCAGCTGGTGCTCTTG) and an antisense primer from the exon 5/6 junction (X5/X6, 5′-CGCTGTGTTTGTGCGTCAATCA) were used. To detect products the blot was probed with a 32P kinase-labeled oligonucleotide from exon 4 (Nae WT, 5′-TGGTGCCG GCGACTG).
Fly crosses
Transgenic flies were generated by P element mediated germline transformation (22). The plasmid pUAS-MycTra2 was injected directly into Drosophila embryos of the genotype w1118/BsY; tra2b/CyO. Transformed lines were identified in the G1 progeny of injectees by w+ eye pigmentation. Once established these strains were crossed with various GAL4 strains and adult progeny recovered at 23 or 25°C. Heat-shock induction was carried out at 37°C for 1 h by immersing vials in a water bath. Viability of adults carrying both Gal4 and UAS transgenes was measured by counting adult progeny. Percent viabilities are given in relation to the recovery of siblings of other genotypes and expected Mendelian ratios. Rescue of viability by loss-of-function tra2 mutations was determined by crossing w1118/Y; Bl tra21/+; P{w+mC, GAL4-Hsp70.PB} 89-2-1/+ males with y1 w67c23; P{w+, UAS-MycTra2}, tra2b/CyO, P{Ubi-GFP.S65T} (Line 1) females. Clones of cells expressing MycTra2 were produced by crossing w1118/Y; P{w+, UAS-MycTra2}, tra2b/CyO; P{w+, CZP-ORF3} males with females of the genotype P{hs-FLP}; P{w+mC,AyGAL4}, P{UAS-eGFP}. Progeny were subjected to 37°C heat shock for 30 min at between 24 and 36 h of development. This induced FLP recombinase expression and the formation of random GAL4 expressing clones by excision of the FRT flanked yellow gene which separates the Actin 5C promoter from GAL4 coding sequences in P{AyGAL4}. Other fly strains were obtained from the Bloomington Drosophila Stock Center.
Western blot and immunofluorescent staining of larval tissues
Western blots were carried out with a 1:1000 dilution the mouse anti-Myc antibody 9E10 (Santa Cruz) at 4°C overnight. The secondary antibody was a 1:1000 dilution HRP-conjugated goat anti-mouse IgG (Pierce Biotechnology, Inc.), incubated at room temperature for 1 h. Immunofluorescent staining of larvae was carried out after sorting males and females at third instar stage. These were dissected and fixed with 4% formaldehyde in BPT buffer (BPS+ 0.1% Triton X-100). The primary antibodies were mouse anti-GFP and rabbit anti-β-galactosidase (Molecular Probes), used at 1:300 dilution at 4°C overnight. The secondary antibodies were goat anti-mouse IgG Alexa fluor 488 and goat anti-moust IgG Alexa fluor 546 (Molecular Probes), incubated at 1:100 dilution at room temperature for 2 h. Samples were mounted with Vectashield and observed under a Zeiss LSM 510 confocal microscope.
RESULTS
M1 retention is induced by increased Tra2 expression in Schneider 2 cells
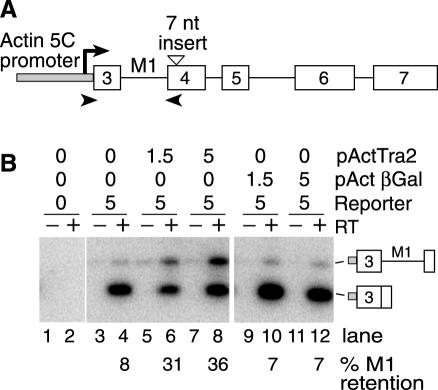
It has previously been shown that in vitro splicing of the M1 intron in Schneider 2 (S2) cell nuclear extracts is repressed by the addition of recombinant Tra2 protein (15). Because this line of cells is thought to be of somatic origin (23), we investigated whether a similar Tra2-dependent effect on M1 splicing could be observed in vivo by transfection. Transfection experiments were carried out using two different plasmids in which Tra2 transcripts are driven by the Actin 5C promoter. pActTra2 which contains a Tra2 cDNA encoding the Tra2 protein isoform Tra2-P2 (Figure 1A) serves as the primary source of Tra2 protein. This isoform is normally expressed in both the soma and male germline and is known to be sufficient for M1 repression in the germline. The second plasmid pActTra2Nae+1 contains genomic coding sequences altered by a 7 nt insertion introduced into exon 4. This small insertion causes a shift in reading frame, disrupting expression of all Tra2 protein isoforms competent for repression of M1 (18). Importantly, this insertion also allows the reporter transcripts to be distinguished from those of pActTra2 and the endogenous tra2 gene in RT–PCR assays. The Actin 5C promoter in pActTra2-Nae+1 is positioned to initiate transcription in exon 3 because this is the position of the transcription start site used in endogenous male germline transcripts. When no Tra2 expressing plasmid was introduced, only 8% of the reporter transcripts retained the M1 intron (Figure 1B, lane 4). This fraction increased to as much as 36% with increasing amounts of co-transfected pActTra2 (Figure 1B, lanes 6 and 8). The effects are specific to the Tra2 protein as a plasmid driving β-galactosidase from the same actin promoter pActβGal had no effect on M1 retention in the reporter RNA (Figure 1B, lanes 10 and 12). Together these results indicate that Tra2-dependent splicing repression is supported by cultured cells of somatic origin.
Figure 1.
Drosophila S2 cells support Tra2 induced M1 retention. (A) A schematic diagram of the M1 splicing reporter pActTra2-Nae+1 is shown. The unfilled boxes indicate exons and the solid lines indicate introns deriving from the tra2 gene. The grey box indicates promoter sequences from the Actin 5C. The position of the predicted transcription start site is indicated (arrow). The position of the fusion of actin sequences with exon 3 is nearly coincident with the natural male germline transcription start site. The position of the 7 nt insertion in exon 4 used to disrupt translation is also indicated by a triangle. The positions of primers used for RT–PCR experiments are shown as arrowheads. (B) RT–PCR results on the M1 splicing reporter in transfected S2 cells. For each sample, both reaction with (+) and without (−) reverse transcriptase (RT) are shown. The amount of M1 retention is indicated below the gel lanes. The amount (micrograms) of pActTra2, pActβGal and pActTra2-Nae+1 (reporter) used in each transfection are indicated above the gel. The total amount of DNA used in each transfection was kept constant at 20 μg using empty vector plasmid for the balance. The PCR products of M1 retaining and M1-spliced forms are indicated with diagrams on the side.
The Tra2 promoter is most active in the male germline
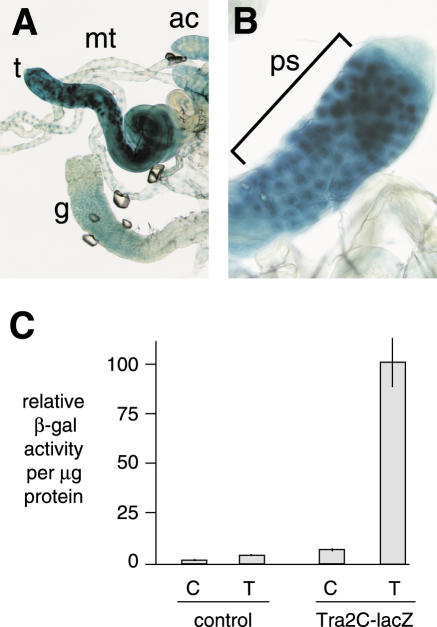
Based on the above results we hypothesized that the germline restriction of M1 repression in male flies does not require tissue-specific splicing regulators but rather that higher levels of Tra2 in this tissue are responsible. Specific antibodies for the Tra2 protein are not available so to estimate the relative level of its expression in the male germline and other tissues we examined a tra2 reporter (C-lacZ), which contains tra2 genomic sequences fused with the Escherichia coli β-galactosidase gene at the C-terminal end of Tra2 protein-coding region (24). This transgene contains all required upstream regulatory sequences from the tra2 gene including both the somatic and male germline promoters. It has been shown to robustly rescue somatic female sexual differentiation, suggesting that it produces protein at or above normal endogenous levels. Tra2 is expressed in most tissues of both male and female adults (12), but as shown in Figure 2A, testes from flies carrying the transgene stained more strongly with X-gal than did other male tissues indicating that the highest levels of protein are expressed there. Within the testis, X-gal staining was most concentrated in primary spermatocytes where Tra2 function is required (Figure 2B) (25). To more quantitatively determine the extent of this difference in expression, we prepared extracts from dissected testes and carcasses of adult males and measured β-galactosidase activity levels in relation to total protein concentration. As shown in Figure 2C, the specific activity in testes was found to be over 14 times higher than that in the remaining carcass. The above results confirm that Tra2 expression is significantly higher in the germline than in other tissues and are consistent with previous direct comparisons of mRNA levels in wild-type and germline-deficient flies (12,26).
Figure 2.
Tra2 is expressed at highest levels in the germline of male adults. Expression monitored using a β-galactosidase reporter transgene (C-lacZ) under the control of the tra2 promoter is shown. (A) Shows X-gal staining of testes (t) in comparison to malphigian tubules (mt), accessory glands (ac) and gut (g). A closeup view of the testis showing indicating staining of primary spermatocytes (bracket) is shown in (B). (C) Shows results from quantitative determination of β-galactosidase activity in extracts from various testes (T) and remaining carcass (C) in both normal w1118 male adults (control) and transgenic flies carrying the reporter in the same genetic background. Standard errors are indicated by the lines.
Increased Tra2 expression through the Gal4-UAS system reduces viability
To test whether an increase in Tra2 protein outside the male germline would stimulate retention of the M1 intron we employed the GAL4-UAS system which allows the expression of proteins under the control of inducible and tissue-specific drivers (21). A P element based plasmid was constructed with a cDNA encoding the Tra2-P2 protein preceded by UAS elements and an in-frame N-terminal repeat of six Myc epitope tags (Figure 3A). This construct UAS-MycTra2 was then introduced into the fly genome by P element mediated transformation. Three insertions of this transgene were obtained at independent sites and used to produce stable stocks.
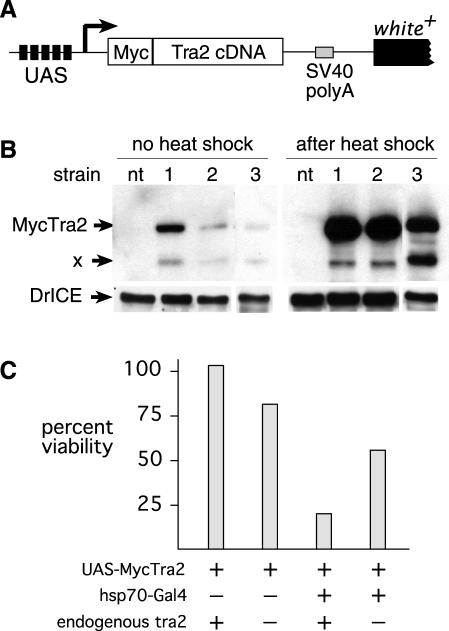
Figure 3.
Increased expression of Tra2 under control of the hsp70 promoter reduces viability. A schematic diagram of the UAS-MycTra2 transgene is shown (A). The position of the 6×Myc tag at the protein's N-terminus and the position of five UAS elements is indicated. The arrow indicates the position expected for transcription initiation. Not shown are P element sequences from the transformation vector. When anti-Myc antibodies are used to probe a western blot on extracts (B) from female adults carrying the hsp70-Gal4 transgene and any of three UAS-MycTra2 insertions, a prominent band is observed at the expected size for the fusion protein (∼48 kDa). This protein is not present extracts of non-transgenic control females (nt). Lower molecular weight proteins (X) are also observed in transgenics and may represent degradation products or alternate initiations within the 6×Myc tag. Extracts prepared after heat shock and recovery have sharply increased levels of Myc-Tra2. As a loading control, the same blot was probed with an antibody against Drosophila drICE. (C) Shows that viability at 25° is reduced in flies carrying both hsp70-Gal4 and UAS-MycTra2 (line 1) as measured in relation to their siblings. Homozygosity for a tra2 loss-of-function mutation (tra2b) causes a slight reduction in measured viability of control flies (UAS-MycTra2), but the same mutation increases viability of flies driving expression from hsp70-Gal4 (P < 0.0001).
As an initial test of the function of UAS-MycTra2 transgene insertions, we crossed each strain to various driver strains expressing yeast GAL4 with different tissue distributions (Table 1). However, adult flies carrying the combination of GAL4 and UAS-MycTra2 transgenes were not recovered in these crosses due to lethality at earlier developmental stages. To overcome this, we generated flies in which Tra2 expression was driven by the heat-inducible heat shock protein 70 (hsp70) promoter which is active at a low level in a wide variety of somatic tissues even when flies are cultured at 23°C without heat shock. This low level of GAL4 activity allowed us to recover from 41 to 70% of the expected individuals carrying both hsp70-Gal4 and UAS-MycTra2 as viable adults (Table 1). The number of survivors was diminished to <1% when flies were subjected to heat shock at various times during larval development. Western-blot analysis of total protein from adults raised without heat shock confirmed that they express a protein product of the expected size for the MycTra2 fusion protein (Figure 3B). A small amount of another product detected specifically in transgenic flies migrated at a smaller size. This may be either a product of proteolysis or due to translation initiation from one of several AUG initiation codons within the Myc tag repeats near the protein's N-terminus. In either case, the levels of functional MycTra2 expression were sufficient to partially rescue sexual differentiation defects in the soma of diplo-X (chromosomally female) individuals homozygous for a tra2 loss-of-function mutation as indicated by feminization of their genitalia, leg bristles and abdominal pigmentation (data not shown). Thus, the MycTra2 protein is functional in the soma.
Table 1.
Viability of adults expressing UAS-MycTra2
| GAL4 driver line | Tissues | UAS-MycTra2 line | Adult viability (%) |
|---|---|---|---|
| None | 1 | 91 | |
| 2 | 94 | ||
| 3 | 98 | ||
| Actin 5C | Ubiquitous | 1 | 0 |
| 2 | 0 | ||
| 3 | 0 | ||
| T76 | Imaginal discs, testes, CNS | 2 | 0 |
| T80 | Imaginal discs, gonads, CNS | 1 | 0 |
| 2 | 0 | ||
| 3 | 0 | ||
| Hsp70 | Ubiquitous | 1 (23°C) | 41 |
| 1 (hs) | 1 | ||
| 2 (23°C) | 70 | ||
| 2 (hs) | 0 | ||
| 3 (23°C) | 48 | ||
| 3 (hs) | 1 |
Even when expressed from the hsp70-GAL4 driver at 23°C UAS-MycTra2 caused some reduction in viability relative to Mendelian expectations (Table 1). Such lethality might result either from an excess of protein in tissues where it is normally expressed or from an aberrant activity conferred by the Myc tag. In the former case we would expect that viability would be improved if Tra2 expression from the endogenous gene was reduced in transgenic flies. Consistent with this we observed that transgenic individuals homozygous for a tra2 loss-of-function mutation were significantly more viable (P < 0.0001) than their tra2+ siblings (Figure 3C). We conclude that excess Tra2 is deleterious to survival.
Somatic tissues are competent to carry out Tra2-dependent M1 repression
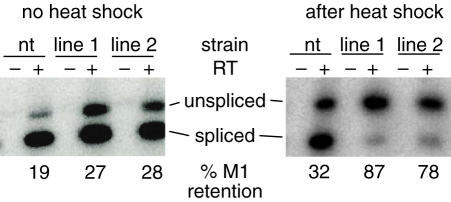
To determine if increased levels of Tra2 lead to M1 repression in transcripts expressed outside of the male germline we examined M1 splicing in endogenous tra2 transcripts isolated from females expressing MycTra2. We allowed diplo-X flies carrying both hsp70-GAL4 and UAS-MycTra2 to develop without heat induction and then isolated RNA after subjecting them to a brief heat shock as adults. Somatic tissues produced a low level of endogenous RNA retaining the M1 intron; however, this retention occurs independently of the Tra2 protein and may reflect transcripts that have simply not completed processing (20). As shown in Figure 4, the basal levels of MycTra2 expressed without heat shock caused a small increase in the fraction of RNA retaining the M1 intron. As expected heat shock increased MycTra2 expression dramatically (Figure 3B) and further increased M1 retention from the endogenous Tra2 gene (Figure 4). In these experiments we also observed a small heat-shock-induced increase in M1 retention in individuals without the transgene. The latter effect is presumably due to disruption of RNA splicing by heat-shock conditions. In summary, these results indicate that the expression of Tra2 at high levels outside the male germline results in ectopic M1 intron retention.
Figure 4.
Increased Tra2 expression in females results in increased M1 retention. Low cycle RT–PCR analysis of M1 splicing in non-transgenic (nt) female adults and females carrying both hsp70-Gal4 and either of two UAS-MycTra2 insertions (lines 1 and 2) are shown. The percent of products with the M1 intron retained is indicated below the gel. The level of M1 retention is increased slightly without heat shock and more dramatically after heat shock and recovery. Reactions were performed both with (+) and without (−) reverse transcriptase. RNA samples are from the same individuals used for the western blot in Figure 3B.
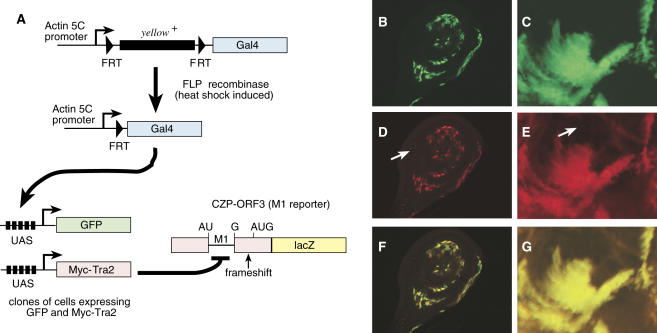
To verify that M1 repression occurs when Tra2 levels are increased in a specific somatic tissue, we next generated clones of cells overexpressing MycTra2 in larval imaginal discs and monitored M1 splicing levels using a splicing reporter transgene CZP-ORF3 that expresses β-galactosidase only from transcripts containing the intron (18). Patches of clonally derived somatic cells expressing Gal4 were generated using a strategy in which heat-shock-induced expression of FLP recombinase causes the ubiquitously active Actin 5C promoter to fuse with Gal4-coding sequences in random cells during development (see Materials and Methods for details). The Gal4 expressing cells were detected by activation of a UAS-GFP transgene that was carried by the same flies as a marker. As shown in Figure 5, GFP-positive cells that also express MycTra2 had significantly elevated levels of β-galactosidase expression in relation to other cells in the same disc (see arrows in panels D and E) which did not have an activated Gal4 gene and therefore did not express GFP or MycTra2. These results indicate that the induction of MycTra2 expression leads to an increase in M1 intron retention. Control assays which lacked the reporter transgene or the β-galactosidase primary antibody showed no such signal in the GFP-positive cells (data not shown).
Figure 5.
M1 retention in Drosophila somatic tissues expressing UAS-MycTra2. (A) A schematic diagram is shown of the strategy for producing Tra2 overexpressing clones. Induced expression of yeast FLP recombinase from a FLP transgene driven by the hsp70 promoter causes fusion of ubitquitously active actin 5C promoter with Gal4 coding sequences in random cells. Clones of GAL4 positive cells are detected by activation of UAS-GFP and also express UAS-MycTra2. These clones were generated in flies also carrying a Tra2-β-galactosidase reporter transgene (CZP-ORF3) which produces RNA ubiquitously, but only expresses β-galactosidase if the M1 intron is retained. The reporter contains a frameshift mutation blocking translation from open reading frames used when the M1 intron is removed and initiating in exons 2 or 3. Expression of the reporter protein in intron-retaining RNA results from translation initiation at an AUG codon located downstream of M1 in exon 4. Shown are results from whole mount immunofluorescent staining of a larval imaginal disc (B, D and F) or brain tissue (C, E and G) that include clones of cells overexpressing UAS-MycTra2. Green staining (GFP) marks cells in which UAS-Myc-Tra2 is activated by Gal4 (B and C) and Red staining (β-galactosidase) marks cells with increased M1 retention (D and E). A merge of the red and green channels is also shown (F and G). Arrows in (D and E) indicate examples of large areas where GAL4 and Myc-Tra2 are not expressed and the reporter is not induced. Note that GFP staining is cytoplasmic and the Tra2-β-galactosidase fusion protein from the reporter is nuclear.
DISCUSSION
A number of known RNA splicing regulators have been shown to be widely expressed within organisms but to vary spatiotemporally in concentration (10,27–33). In some cases qualitative differences in alternative splicing can be elicited by manipulating the levels of such factors either in vitro or in vivo. This suggests that natural quantitative variations in expression levels have functional significance. For instance, the selection of alternative splice sites in several genes is sensitive to the amount of the SR protein SF2/ASF in relation to hnRNP A1 (28,34–36). Likewise, artificially increased expression of several splicing factors in Drosophila transgenic strains has been shown to affect the splicing of specific RNAs or the viability and development of the organism (28,34–37). Such observations suggest the possibility that there are threshold concentrations in vivo at which such factors become able to influence the splicing of different RNA targets.
The studies presented here argue that a threshold exists in the amount of Tra2 that is required for repression of M1 splicing and that somatic levels of Tra2 expression are below the threshold required for M1 repression while levels in the germline exceed it. Tra2 RNA is expressed in a non-sex-specific manner at significantly lower levels in the soma than in the male germline (12,18). Analysis of steady-state Tra2 transcript levels indicates that over half of all Tra2 RNA in adult males derives from the germline being most concentrated in primary spermatocytes. The remaining half of the mRNA is distributed in a relatively large number of cells in a variety of somatic tissues that undergo and maintain sexual differentiation. An analysis of Tra2 proteins expressed in wild-type and mutant flies also suggests that levels of Tra2 in the testes are higher than in the soma (38). These observations are further supported by our analysis of transgenic flies expressing a Tra2/β-galactosidase fusion protein under direction of the tra2 promoter and other flanking regions. In these experiments we observed fusion protein expression in a number of different somatic tissues but in all cases at a significantly lower level than that found in spermatocytes. Moreover, the relative cellular concentration of fusion protein (as measured in relation to total protein) is several fold higher in the testis than in the soma. Interestingly, the human Tra2β protein has also recently been reported to be most prominently expressed in testes where it regulates the male germline-specific inclusion of the T exon of the HipK3 gene in a concentration dependent manner (10).
Although it is expressed at lower levels, Tra2 has clear effects on RNA splicing in somatic tissues where it acts with Tra and other SR factors to direct selection of female-specific alternative splice sites within the dsx and fru mRNAs (11,13,39,40). Strong tra2 loss-of-function mutations result in selection of the alternative male-specific dsx and fru splice sites indicating that Tra2 is required in most or all tissues where these RNAs are produced. We propose that these RNAs must have a lower threshold concentration at which Tra2 is able to interact with them than does M1. Consistent with this idea, we found that increasing the level of Tra2 expression induced M1 retention in somatic tissues. These cells are thus able to support Tra2-dependent repression of the intron's splicing provided sufficient levels of Tra2 protein are present. These results explain why Tra2 is observed to specifically repress M1 splicing in extracts derived from Drosophila S2 cells after they are supplemented with recombinant protein (15).
The difference in Tra2 levels needed for the repression of M1 splicing versus those needed to activate splice sites in dsx and fru seems likely to reflect differences in the affinity of the protein for its RNA target. In the case of dsx and fru, Tra2 interacts with an exonic splicing enhancer and behaves as a splicing activator (40–42). In the M1 intron Tra2 binds instead to an intronic splicing silencer (ISS) and interferes with early steps in spliceosome assembly [(15), J. Qi, S. Su, and W. Mattox, manuscript submitted]. In the dsx splicing enhancer binding of Tra2 is facilitated by cooperative interactions with other SR proteins such as Tra and Rbp1. However, the sequences specifying Tra2 binding in the ISS differ from those identified in the dsx splicing enhancer and similar cooperative interactions have not been observed (15). Thus, the threshold at which an RNA responds to Tra2 may be determined by arrangement and sequence of the regulatory elements within each RNA.
As feedback regulation through M1 splicing is simply a means of limiting Tra2 expression, it does not in of itself account for why expression is higher in the germline. It seems likely that the higher levels of Tra2 present in the germline are required for fertility, but this has not been rigorously tested. Other genes producing alternatively processed pre-mRNAs in the germline are known to be Tra2 dependent (16) and it is possible that these targets, similar to the M1 intron, require a relatively high level of Tra2 for proper processing.
In agreement with our findings in the soma, in vivo studies on M1 splicing in the male germline previously indicated that the extent of M1 repression is highly dependent on the level of Tra2 present there (17). For instance males heterozygous for a point mutation disrupting the protein-coding region produce a smaller fraction of germline M1 retaining RNA than do males with two wild-type alleles. The dose sensitivity of M1 splicing and its relatively high threshold seems logical in view of the known role for M1 splicing repression in the negative feedback regulation of Tra2. In normal flies over half of all germline transcripts retain the intron in wild-type individuals (18). This prevents excessive expression of the protein that would result if the intron were removed from all endogenous germline transcripts (17,38). Although it is presently unclear why such an excess of transcripts is produced it appears that regulatory elements in the intron are ‘calibrated’ to undergo repression when the concentration of Tra2 protein approaches deleterious levels.
Beyond the observed effects on M1 splicing, we found that artificially increasing somatic Tra2 expression had negative effects on viability. This may indicate that inappropriate concentrations of Tra2 alter the splicing pattern of additional somatic RNAs which are important for the survival of the organism. Similar observations have been made after overexpression of the splicing factors B52 and FNE (28,43). Thus concentration may have a vital role in determining the targets that are affected by multiple splicing regulators. This dose-sensitive behavior of splicing regulators is not universal as some have been reported to have little or no significant adverse effects when overexpressed (44,45). As in the germline, negative feedback regulation of Tra2 through M1 splicing potentially provides a mechanism for the fly to curb deleterious effects if a natural circumstance occurred in which excessive Tra2 transcripts were produced. However, normal endogenous levels of expression do not lead to Tra2-dependent M1 retention in any of several somatic tissues we have examined so far.
The ability of Tra2 to induce qualitatively different effects on alternative splicing at different concentration levels in the soma is interesting in light of the large number of transcripts now known to undergo alternative splicing in complex organisms (46) and the relatively small number of known splicing regulators. It is already clear that SR proteins and other factors can control the processing of multiple pre-mRNA targets. By activating or repressing different splice sites at different concentration levels, such regulators are likely to expand the diversity of tissue-specific splicing patterns that they can specify.
Acknowledgments
We are grateful to Hank Adams for assistance with confocal microscopy, Andreas Bergmann for antibodies and to Rong Dong for excellent preparation of Drosophila media. This work was supported by NIH grant GM070892 to W.M. DNA sequencing was carried out at the U.T. M.D. Anderson DNA Analysis Facility and supported by grant CA16672 (DAF) from the National Cancer Institute. Funding to pay the Open Access publication charges for this article was provided by U.T. M.D. Anderson.
Conflict of interest statement. None declared.
REFERENCES
- 1.Graveley B.R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lopez A.J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 3.Watermann D.O., Tang Y., Zur Hausen A., Jager M., Stamm S., Stickeler E. Splicing factor Tra2-beta1 is specifically induced in breast cancer and regulates alternative splicing of the CD44 gene. Cancer Res. 2006;66:4774–4780. doi: 10.1158/0008-5472.CAN-04-3294. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2) Proc. Natl Acad. Sci. USA. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofmann Y., Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum. Mol. Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- 6.Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2beta1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 7.Jiang Z., Tang H., Havlioglu N., Zhang X., Stamm S., Yan R., Wu J.Y. Mutations in tau gene exon10 associated with FTDP-17 alter the activity of an exonic splicing enhancer to interact with Tra2beta. J. Biol. Chem. 2003;278:18997–19007. doi: 10.1074/jbc.M301800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stoilov P., Daoud R., Nayler O., Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum. Mol. Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Wang J., Gao L., Lafyatis R., Stamm S., Andreadis A. Tau exons 2 and 10, which are misregulated in neurodegenerative diseases, are partly regulated by silencers which bind a SRp30c·SRp55 complex that either recruits or antagonizes htra2beta1. J. Biol. Chem. 2005;280:14230–14239. doi: 10.1074/jbc.M413846200. [DOI] [PubMed] [Google Scholar]
- 10.Venables J.P., Bourgeois C.F., Dalgliesh C., Kister L., Stevenin J., Elliott D.J. Up-regulation of the ubiquitous alternative splicing factor Tra2beta causes inclusion of a germ cell-specific exon. Hum. Mol. Genet. 2005;14:2289–2303. doi: 10.1093/hmg/ddi233. [DOI] [PubMed] [Google Scholar]
- 11.Nagoshi R.N., McKeown M., Burtis K.C., Belote J.M., Baker B.S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 12.Mattox W., Palmer M.J., Baker B.S. Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 1990;4:789–805. doi: 10.1101/gad.4.5.789. [DOI] [PubMed] [Google Scholar]
- 13.Ryner L.C., Goodwin S.F., Castrillon D.H., Anand A., Villella A., Baker B.S., Hall J.C., Taylor B.J., Wasserman S.A. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 14.Mattox W., Ryner L., Baker B.S. Autoregulation and multifunctionality among trans-acting factors that regulate alternative pre-mRNA processing. J. Biol. Chem. 1992;267:19023–19026. [PubMed] [Google Scholar]
- 15.Chandler D.S., Qi J., Mattox W. Direct repression of splicing by transformer-2. Mol. Cell. Biol. 2003;23:5174–5185. doi: 10.1128/MCB.23.15.5174-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattox W., McGuffin M.E., Baker B.S. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics. 1996;143:303–314. doi: 10.1093/genetics/143.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGuffin M.E., Chandler D., Somaiya D., Dauwalder B., Mattox W. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics. 1998;149:1477–1486. doi: 10.1093/genetics/149.3.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattox W., Baker B.S. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 1991;5:786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- 19.Amrein H., Hedley M.L., Maniatis T. The role of specific protein–RNA and protein–protein interactions in positive and negative control of pre-mRNA splicing by transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 20.Chandler D.S., McGuffin M.E., Mattox W. Functionally antagonistic sequences are required for normal autoregulation of Drosophila tra-2 pre-mRNA splicing. Nucleic Acids Res. 2001;29:3012–3019. doi: 10.1093/nar/29.14.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 22.Spradling A.C., Rubin G.M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 23.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 24.Dauwalder B., Mattox W. Analysis of the functional specificity of RS domains in vivo. EMBO J. 1998;17:6049–6060. doi: 10.1093/emboj/17.20.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belote J.M., Baker B.S. The dual functions of a sex determination gene in Drosophila melanogaster. Dev. Biol. 1983;95:512–517. doi: 10.1016/0012-1606(83)90054-4. [DOI] [PubMed] [Google Scholar]
- 26.Amrein H., Maniatis T., Nothiger R. Alternatively spliced transcripts of the sex-determining gene tra-2 of Drosophila encode functional proteins of different size. EMBO J. 1990;9:3619–3629. doi: 10.1002/j.1460-2075.1990.tb07573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton J.G., Mayer S.A., Tempst P., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 28.Kraus M.E., Lis J.T. The concentration of B52, an essential splicing factor and regulator of splice site choice in vitro, is critical for Drosophila development. Mol. Cell. Biol. 1994;14:5360–5370. doi: 10.1128/mcb.14.8.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flickinger T.W., Salz H.K. The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 1994;8:914–925. doi: 10.1101/gad.8.8.914. [DOI] [PubMed] [Google Scholar]
- 30.Hanamura A., Caceres J.F., Mayeda A., Franza B.R., Jr, Krainer A.R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 31.Nayler O., Cap C., Stamm S. Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics. 1998;53:191–202. doi: 10.1006/geno.1998.5471. [DOI] [PubMed] [Google Scholar]
- 32.Pollard A.J., Sparey C., Robson S.C., Krainer A.R., Europe-Finner G.N. Spatio-temporal expression of the trans-acting splicing factors SF2/ASF and heterogeneous ribonuclear proteins A1/A1B in the myometrium of the pregnant human uterus: a molecular mechanism for regulating regional protein isoform expression in vivo. J. Clin. Endocrinol. Metab. 2000;85:1928–1936. doi: 10.1210/jcem.85.5.6537. [DOI] [PubMed] [Google Scholar]
- 33.Bronstein N.B., Kishore R., Ismail Z., Zhang Q., Taylor T., Newman S.A. cDNA cloning and spatiotemporal expression during avian embryogenesis of hnRNP A1, a regulatory factor in alternative splicing. Gene Expr. Patterns. 2003;3:285–295. doi: 10.1016/s1567-133x(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 34.Caceres J.F., Stamm S., Helfman D.M., Krainer A.R. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 35.Gallego M.E., Gattoni R., Stevenin J., Marie J., Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard A.J., Krainer A.R., Robson S.C., Europe-Finner G.N. Alternative splicing of the adenylyl cyclase stimulatory G-protein G alpha(s) is regulated by SF2/ASF and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and involves the use of an unusual TG 3′-splice Site. J. Biol. Chem. 2002;277:15241–15251. doi: 10.1074/jbc.M109046200. [DOI] [PubMed] [Google Scholar]
- 37.Quinn L.M., Dickins R.A., Coombe M., Hime G.R., Bowtell D.D., Richardson H. Drosophila Hfp negatively regulates dmyc and stg to inhibit cell proliferation. Development. 2004;131:1411–1423. doi: 10.1242/dev.01019. [DOI] [PubMed] [Google Scholar]
- 38.Unni E., Su S., Mattox W. Analysis of a null mutation in the Drosophila splicing regulator Tra2 suggests its function is restricted to sexual differentiation. Genesis. 2003;37:76–83. doi: 10.1002/gene.10234. [DOI] [PubMed] [Google Scholar]
- 39.Hedley M.L., Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- 40.Heinrichs V., Ryner L.C., Baker B.S. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 1998;18:450–458. doi: 10.1128/mcb.18.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian M., Maniatis T. A splicing enhancer complex controls alternative splicing of doublesex pre-mRNA. Cell. 1993;74:105–114. doi: 10.1016/0092-8674(93)90298-5. [DOI] [PubMed] [Google Scholar]
- 42.Lynch K.W., Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 43.Samson M.L., Chalvet F. Found in neurons, a third member of the Drosophila elav gene family, encodes a neuronal protein and interacts with elav. Mech. Dev. 2003;120:373–383. doi: 10.1016/s0925-4773(02)00444-6. [DOI] [PubMed] [Google Scholar]
- 44.Zu K., Sikes M.L., Haynes S.R., Beyer A.L. Altered levels of the Drosophila HRB87F/hrp36 hnRNP protein have limited effects on alternative splicing in vivo. Mol. Biol. Cell. 1996;7:1059–1073. doi: 10.1091/mbc.7.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hammond L.E., Rudner D.Z., Kanaar R., Rio D.C. Mutations in the hrp48 gene, which encodes a Drosophila heterogeneous nuclear ribonucleoprotein particle protein, cause lethality and developmental defects and affect P-element third-intron splicing in vivo. Mol. Cell. Biol. 1997;17:7260–7267. doi: 10.1128/mcb.17.12.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]