Abstract
The identification of promoters and their regulatory elements is one of the major challenges in bioinformatics and integrates comparative, structural, and functional genomics. Many different approaches have been developed to detect conserved motifs in a set of genes that are either coregulated or orthologous. However, although recent approaches seem promising, in general, unambiguous identification of regulatory elements is not straightforward. The delineation of promoters is even harder, due to its complex nature, and in silico promoter prediction is still in its infancy. Here, we review the different approaches that have been developed for identifying promoters and their regulatory elements. We discuss the detection of cis-acting regulatory elements using word-counting or probabilistic methods (so-called “search by signal” methods) and the delineation of promoters by considering both sequence content and structural features (“search by content” methods). As an example of search by content, we explored in greater detail the association of promoters with CpG islands. However, due to differences in sequence content, the parameters used to detect CpG islands in humans and other vertebrates cannot be used for plants. Therefore, a preliminary attempt was made to define parameters that could possibly define CpG and CpNpG islands in Arabidopsis, by exploring the compositional landscape around the transcriptional start site. To this end, a data set of more than 5,000 gene sequences was built, including the promoter region, the 5′-untranslated region, and the first introns and coding exons. Preliminary analysis shows that promoter location based on the detection of potential CpG/CpNpG islands in the Arabidopsis genome is not straightforward. Nevertheless, because the landscape of CpG/CpNpG islands differs considerably between promoters and introns on the one side and exons (whether coding or not) on the other, more sophisticated approaches can probably be developed for the successful detection of “putative” CpG and CpNpG islands in plants.
Arabidopsis, and probably most plants, encode an exceptionally large number of DNA-binding proteins, potentially acting as transcription factors (TFs). In fact, more than 3,000 genes have been anticipated to be involved in transcription, more than one-half of which were expected to encode TFs (Arabidopsis Genome Initiative, 2000), corresponding to more than 5% of the Arabidopsis genes, and approximately twice the ratio observed for yeast and animal genomes (Riechmann et al., 2000). These TFs bind to the DNA on specific cis-acting regulatory elements (CAREs) and orchestrate the initiation of transcription, which is one of the most important control points in the regulation of gene expression. CAREs are short conserved motifs of five up to 20 nucleotides usually found in the vicinity of the 5′ end of genes in what is called the promoter. The promoter sequence is usually located upstream from the transcription start site (TSS), but regulatory elements can also be located downstream, for example, in the first intron of the gene itself (Zhang et al., 1994; Gidekel et al., 1996; de Boer et al., 1999; Dorsett, 1999). The promoter can roughly be divided in two parts: a proximal part, referred to as the core, and a distal part. The proximal part is believed to be responsible for correctly assembling the RNA polymerase II complex at the right position and for directing a basal level of transcription (Nikolov et al., 1996; Nikolov and Burley, 1997; Berk, 1999). It is mediated by elements, such as TATA and Initiator boxes through the binding of the TATA box-binding protein, and other general TFs specific for the RNA polymerase II (Featherstone, 2002). The distal part of the promoter is believed to contain those elements that regulate the spatio-temporal expression (Tjian and Maniatis, 1994; Fessele et al., 2002). How far upstream (or downstream) such a distal part reaches is not defined. In addition to the proximal and distal parts, somewhat isolated, regulatory regions have also been described, mainly in animals, that contain enhancer and/or repressors elements (Barton et al., 1997; Bagga et al., 2000). The latter elements can be found from a few kilobase pairs upstream from the TSS, in the introns, or even at the 3′ side of the genes they regulate (Larkin et al., 1993; Wasserman et al., 2000). Lastly, eukaryotic genomes can be organized into domains of transcriptional activity or transcriptional silencing, encompassing one or more genes (Oki and Kamakaka, 2002).
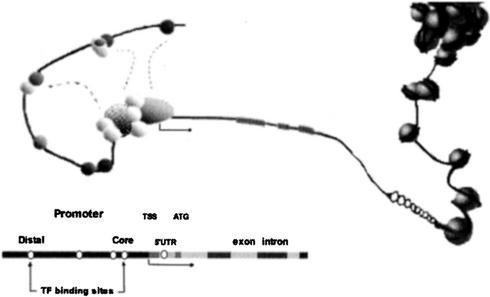
A promoter region, as described above, presents a rather linear view of the promoter. In reality, a supplementary layer of complexity is added by bringing the TFs together on a promoter, by adopting a three-dimensional configuration, enabling the interaction with other parts to activate the basal transcription machinery (Fig. 1; Buratowski; 1997; Berk, 1999; Struhl, 2001). The packaging of DNA into chromatin (Kornberg and Lorch, 2002) limits the accessibility of the DNA template for the transcriptional apparatus and inhibits transcriptional initiation. Therefore, when compared with naked DNA, chromatin is able to repress transcription, which is probably important for the tight regulation of gene activity in vivo (Juo et al., 1996; Marilley and Pasero, 1996; Ioshikhes et al., 1999). Derepression of transcription by partial unfolding of the chromatin structure probably constitutes an important part of gene regulation, and several TFs and transcriptional co-activators have been shown to disrupt or remodel the chromatin structure (Beato and Eisfeld, 1997; Kass et al., 1997; Travers and Drew, 1997; Langst and Becker, 2001; Brower-Toland et al., 2002).
Figure 1.
Graphical, simplified view of the different elements involved in transcription. The pre-initiation complex (PIC) situated at the nucleosome-free TSS is shown containing RNA polymerase II (large gray hatched oval), the TATA box-binding protein (gray sphere), and a number of general TFs (white circles). Gene regulatory proteins upstream or downstream of the TSS that stimulate gene-specific transcription and also contribute to the PIC assembly are shown as small gray circles.
The three-way connection between methylation, gene activity, and chromatin structure has been known for almost two decades. DNA methylation has been shown to repress transcription initiation by interfering directly with the binding of transcriptional activators or indirectly by binding proteins with affinity for methylated DNA (Weber et al., 1990; Razin, 1998; Jones, 1999; Kooter et al., 1999; Ng and Bird, 1999; Meyer, 2000). Proteins, which bind to methylated DNA in a CpG density-controlled manner, have been detected in both mammals and plants. Experiments have indicated that methylation is not a consequence of the transcriptional state but apparently participates actively in the regulation of gene expression (Inamdar et al., 1991; Finnegan et al., 1998b; Pitto et al., 2000). Furthermore, during transcription elongation, RNA polymerase and the DNA template must rotate relative to each other inducing rotary constraints. Scaffold or matrix attachment regions are involved at this level of gene expression by stabilizing the formation of heterochromatin. These repetitive regions enable the formation of Z-DNA dividing the DNA into topological domains, which are delineated by torsionally locked boundaries (Bentin and Nielsen, 2002).
Much attention has been paid to investigate the modular structure of regulatory regions that control the transcription of eukaryotic genes (Dynan, 1989; Johnson and McKnight, 1989; Struhl, 1999; Klingenhoff et al., 2002). The fuzziness of one binding site can be compensated by a higher fitness of the adjacent binding site and enables the positioning of the additional TF thanks to specific protein-protein interactions (Rooney et al., 1995; Struhl, 2001). Thus, promoters can be described as the result of a modular hierarchy, in which the individual CAREs constitute the lowest level; they are then grouped into islands as composite elements, themselves organized in modules that confer the specific expression of a gene. The consequence of this modularity is that each promoter is unique and controls specifically the transcript level of its downstream gene.
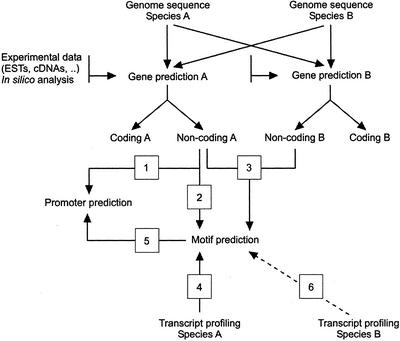
All of these different levels of complexity have great repercussions on the in silico identification of binding sites and promoters. Here, we review current approaches (summarized in Fig. 2) to identify promoters and their regulatory elements.
Figure 2.
Flow chart of the computational approaches to detect promoters and cis-regulatory elements. 1, Promoter prediction through sequence context and structural features, e.g. CpG islands; 2, CARE prediction through statistics on overrepresentation, such as word counting; 3, CARE prediction through comparative genomics (phylogenetic footprinting); 4, CARE prediction through analysis of co-expressed gene clusters, for instance by Gibbs sampling (for details, see text); 5, Promoter prediction through the identification of CAREs; and 6, CARE motif prediction through comparative analysis of expression profiles. These approaches are not described in the text.
CONTENT-BASED FEATURES
Promoter Prediction
Unlike gene prediction (Mathé et al., 2002), prediction of promoters in silico is still in its infancy. One of the main problems is that the promoter is defined functionally and not structurally, which strongly limits the means to model it. Clear and unequivocal descriptions of genomic segments that contain all elements required to activate transcription would be useful but are still unavailable, although the regulatory motifs of some specific genes have been investigated in detail. Therefore, most in silico research on promoters is usually restricted to the so-called intergenic regions of the genome, i.e. between the coding regions of two neighboring genes. The most practical approach is to limit the putative promoter region to an arbitrary number of base pairs upstream of the translation start site of the gene of interest, the location of the TSS being unknown most of the time. However, ideally, this number should be chosen in function of the organism, because the length of intergenic regions may differ considerably (Arabidopsis Genome Initiative, 2000; International Human Genome Sequencing Consortium, 2001; Aparicio et al., 2002). In multicellular organisms, regulatory elements may be found upstream or downstream of the gene, as well as in introns, and may be spread over tens or even hundreds of kilobases (Larkin et al., 1993; Bagga et al., 2000). In such cases, intergenic sequences will contain only a part of the regulatory elements necessary to control transcription.
In 1997, Fickett and Hatzigeorgiou (1997) thoroughly reviewed the existing promoter prediction tools. The programs tested could not reliably identify promoters in a genomic sequence, predicting too many false positives, i.e. on average one false positive per 1 kb. One of the reasons why these programs did not perform better was that they were focused on “search by signal” using only one or two given features, such as the presence of a TATA box or Initiator element, but disregarded structural and more general sequence-based features characteristic for promoter elements (Fickett and Hatzigeorgiou, 1997). Newer approaches do take into account more features; they consider the higher order structure of a promoter DNA sequence important for transcriptional regulation and are based on the concept that they share common content features, although polymerase II promoters are quite different in terms of individual organization.
On the one hand, sequence-based algorithms aim at identifying regulatory regions and promoters based on their sequence composition compared with that of non-promoters. Among others, Scherf et al. (2000) and Bajic et al. (2002) have used this approach, in which the promoter context is described by oligonucleotides (see below; Hutchinson, 1996; Wolfertstetter et al., 1996). On the other hand, promoter regions might be distinguished from non-promoter regions on the basis of specific structural properties. These features are either directly or indirectly correlated with the three-dimensional structure a promoter region should adopt for gene expression in vivo (Baldi et al., 1998; Pedersen et al., 1998, 1999; Zhang, 1998; Fickett and Wasserman, 2000; Hannenhalli and Levy, 2001; Ohler and Niemann, 2001). The three-dimensional structure can depend on characteristic physico-chemical profiles of Z-DNA (Ho et al., 1986) associated with scaffold and matrix attachment regions (Bentin and Nielsen, 2002), stability of duplex DNA (Breslauer et al., 1986; Sugimoto et al., 1996), DNA curvature (Bolshoy et al., 1991), bending and curvature in B-DNA (Goodsell and Dickerson, 1994), DNA bending/stiffness (Sivolob and Khrapunov, 1995), bendability (Brukner et al., 1995a, 1995b), propeller twist (El Hassan and Calladine, 1996), B-DNA twist (Gorin et al., 1995), and protein-induced deformability (Crothers, 1998; Olson et al., 1998). If eukaryotic promoters have such general structural features independently of the genes they control, looking for these should help in identifying promoters in general. We will discuss two prediction tools each representing a different approach to find promoters.
PromoterInspector (Scherf et al., 2000) focuses on the sequence context of a promoter and is based on libraries of IUPAC words. A promoter will be represented by a model that is based on two groups of IUPAC words: one characteristic for promoter sequences and one for non-promoter-related sequences. The IUPAC words that build the model are directly computed from a set of training sequences. New promoter sequences will be assigned to the promoter class when the ratio between the numbers of observed promoter-specific and non-promoter-specific IUPAC words exceeds a certain threshold. Instead of using only one model, the program constructs three different models that differentiate promoters from exons, from introns, and from 3′-untranslated regions (UTRs). A given sequence will be assigned to the class of “promoters” only when all models are in agreement with the decision. The specificity and significance of this program is highly dependent on the given training sets that build up the different models.
McPromoter (Ohler et al., 1999, 2001; Ohler, 2000) is a content-based probabilistic promoter prediction program that uses an integrative approach combining different structural features, such as bendability (Brukner et al., 1995a, 1995b), propeller twist (El Hassan and Calladine, 1996), and CpG content (Antequera and Bird, 1999; Ioshikhes and Zhang, 2000). Here, a promoter is represented as a sequence of consecutive segments represented by joint likelihood of DNA sequence and profiles of physical properties. A profile for a physical property consists of the corresponding values from a chosen parameter, for example the bendability, set along the given DNA sequence. These parameters usually refer to di- and trinucleotides only, so the profiles are generally very noisy and are, therefore, smoothed with a filter. The program tries to divide a given sequence into one region upstream and one downstream from the TSS. A search by signal is used to distinguish the core promoter from the other parts by looking for a TATA and/or Initiator box separated by a spacer of approximately 15 bp.
Although the prediction tools hitherto developed can produce acceptable results for certain species, none of them have been trained and adapted for plants. For example, McPromoter is trained especially to analyze data of fruitfly (Drosophila melanogaster) and has been used in the Genome Annotation Assessment project (Reese et al., 2000; Ohler, 2000; Ohler et al., 2002); however, when applied to plant genomes, it is not as reliable nor as specific. Here, the same rule applies as with gene prediction: Systems have to be trained and tailored for each species separately (Mathé et al., 2002). For the careful training of systems, large amounts of reliable data are needed. Although the availability of large sets of documented promoter sequences is still problematic, we expect this will improve in the near future. An extensive overview of the available programs for the prediction of promoters is given in Table I.
Table I.
Promoter prediction programs
| Programs | Web Sites | References |
|---|---|---|
| McPromoter MM | http://genes.mit.edu/McPromoter.html | Ohler et al. (1999, 2001) |
| PromoterInspector | http://www.gsf.de/biodv/ | Scherf et al. (2000) |
| http://www.genomatix.de/cgi-bin/promoterinspector/promoterinspector.pl | ||
| FunSiteP | http://transfac.gbf.de/programs/funsitep/fsp.html | Kondrakhin et al. (1995) |
| Dragon Promoter Finder | http://sdmc.lit.org.sg/promoter/promoter1_3/DPFV13.htm | Bajic et al. (2002) |
| CONPRO | http://stl.bioinformatics.med.umich.edu/conpro/ | Lui and States (2002) |
| Core-promoter | http://argon.cshl.org/genefinder/CPROMOTER/human.htm | Zhang (1998a, 1998b) |
| WWW PromoterScan | http://bimas.dcrt.nih.gov/molbio/proscan/ | Prestridge (1995) |
| Promoter 2.0 | http://www.cbs.dtu.dk/services/promoter/ | Knudsen (1999) |
| NNPP | http://www.fruitfly.org/seq_tools/promoter.html | Waibel et al. (1989) |
Promoters and CpG Islands
A structural feature that has proven useful in the detection of promoters in the human genome are the so-called CpG islands, i.e. regions that are rich in CpGs, which are important because of their strong link with gene regulation. In general, CpG-rich regions are methylated and are associated with inactive DNA often linked to heterochromatin, gene silencing, and pathogen control (Jeddeloh et al., 1998; Kooter et al., 1999; Wolffe and Matzke, 1999; Meyer, 2000; Bender, 2001; Vaucheret and Fagard, 2001; Richards and Elgin, 2002; Robertson, 2002). In vertebrate genomes, 60% to 90% of all CpGs are normally methylated. Gene-associated CpG islands are mostly not methylated and are usually linked to transcriptionally active DNA (Panstruga et al., 1998; Razin, 1998; Antequera and Bird, 1999; Jones, 1999; Ng and Bird, 1999; Ashikawa, 2001; Li et al., 2001). Prediction programs have been developed to search for the presence of CpG islands in the 5′ region of genes (Ioshikhes and Zhang, 2000; Ohler et al., 2001; Davuluri et al., 2001; Down and Hubbard, 2002; Ponger and Mouchiroud, 2002). However, so far, application of such prediction programs to CpG islands in plants is very limited. A more detailed analysis on CpG and CpNpG islands in Arabidopsis is given below.
Although the functional significance of methylation appears to be similar in humans and plants (Hershkovitz et al., 1990; Weber et al., 1990; Inamdar et al., 1991; Meyer et al., 1994; Sorensen et al., 1996; Rossi et al., 1997; Meza et al., 2002), in plants, DNA methylation is mainly found on the cytosine of the di- and trinucleotide CpG and CpNpG and on nonsymmetrical trinucleotides (Pradhan et al., 1999; Cao et al., 2000; Finnegan and Kovac, 2000; Lindroth et al., 2001; Cao and Jacobsen, 2002). Many plant genomes contain methylated cytosine in asymmetric sequence contexts (CpHpH with H = A, T, or C). Only symmetrical methylation sites have been shown to be maintained through the propagation of cells and methylation of a promoter CpG island has been proposed to play an important role in gene silencing, genomic imprinting, heterochromatin formation, chromatin modification, vernalization, and parent-dependent effects (Finnegan et al., 1998a, 2000; Jeddeloh et al., 1998; Sturaro and Viotti, 2001). CpNpG and CpG islands can occur together in the same promoter region, but their role might be different (Sorensen et al., 1996).
The Landscape of CpG/CpNpG Islands around the TSS in the Arabidopsis Genome
CpG islands are characterized by a locally increased GC percentage (GC%) compared with local averages and by the presence of CpGs (and CpNpGs in plants). The CpG dinucleotide, usually methylated at the fifth position on the cytosine ring, is counter-selected and found much less frequently than expected based on mononucleotide frequencies, for example, 5-fold lower in genomes of vertebrates. This depletion is believed to result from accidental mutations by deamination of 5-methylcytosine to thymine (Sved and Bird, 1990; Duret and Galtier, 2000). In fact, CpG islands are considered evolutionary remnants, because some promoters have somehow been kept free of methylation in the course of evolution, so the deamination process is hampered. Another explanation could be that to function as part of an expression pattern, a selection pressure has to be exerted and, hence, CpG islands stand out in the surrounding regions.
The original pragmatic definition of a CpG island in human sequences considers a GC% higher than 50 and a ratio between observed and expected (o/e) occurrence of CG dinucleotides of 0.6 over a window of 200 bp (Gardiner-Garden and Frommer, 1987). Recently, these parameters have been upscaled to a GC% >55, an o/e CpG >0.65, and a window size of 500 bp, because the previous parameters had been found to overestimate (50-fold) the number of potential CpG islands (Takai and Jones, 2002). In animals, approximately 40% of genes are expected to be associated with CpG islands (Gardiner-Garden and Frommer, 1987; Antequera and Bird, 1999). Actually, this percentage might be too low because a total of 29,000 CpG islands had been estimated after the completion of the human genome sequence (Venter et al., 2001). With the above-mentioned parameters, no CpG islands are discovered in plants (Takai and Jones, 2002; our results). However, DNA methylation occurs in plants, and DNA methylases are even more numerous and diverse in plants than in animals (Finnegan and Kovac, 2000; Cao and Jacobsen, 2002). Therefore, we attempted to define the parameters that could possibly specify CpG and CpNpG islands in Arabidopsis by exploring the compositional landscape around the TSS. To this end, we built a data set of 5,025 gene sequences, designated ARAPROM, by aligning the full-length cDNA sequences generated by Seki et al. (2002) against the genomic sequence (Arabidopsis Genome Initiative, 2000). Generally, these sequences are 2.5 kb long, in which 2 kb represent intergenic sequences upstream from the translation start codon, and 500 bp are taken downstream. Nevertheless, when the upstream neighbor gene lies closer than 2 kb, then only the intergenic sequence is kept, up to the predicted coding boundary of the upstream gene. The genomic sequences in the ARAPROM data set include the promoter region, the 5′-UTR, and the first introns and coding exons of each individual gene.
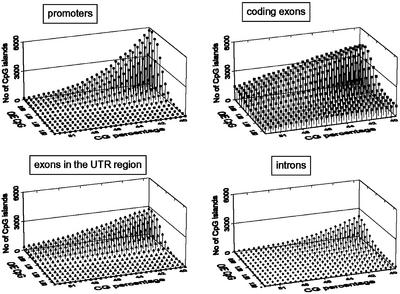
A program in Perl was written that computes the GC content and the o/e ratios of CpG and CpNpG compared with local characteristics over a certain window size. By applying this program to the ARAPROM data set to extract potential CpG/CpNpG islands, we tested the effect of setting the cut-off values for the GC content and the o/e CpG/CpNpG ratios at different levels (39% to 52% with a stepwise increase of 0.5% for the GC content; 0.6% to 2.0% for the o/e CpG and CpNpG ratios with a stepwise increase of 0.1. The results of this analysis with a window size of 200 bp are shown graphically for CpG and CpNpG islands (Figs. 3 and 4, respectively). The first observation is that no CpG island is detected with the cut-off parameters tuned for humans, except for a few in coding exons. Both parameters, CG% and o/e CpG, appear to influence strongly the number of CpG islands detected and, depending on the position in the genome, to affect differently the number of CpGs found. That number found in the “promoter” region sharply increases while the GC% cut-off decreases (Fig. 3). In contrast, for coding exons, the landscape resembles more a plateau, with many CpG islands found already at much higher GC% values. Only at the lowest GC% values, more CpG islands are predicted in the “promoter” region than in the coding exons. In UTR exons, which show a landscape similar to that of coding exons, fewer CpG islands are found and introns, which show a landscape more similar to that of the “promoter” region, show the lowest number of CpG islands.
Figure 3.
CpG island landscape exploration of Arabidopsis gene sequences over a range of CG content and CpG relative frequency. For the various gene elements, on the z axis, the number of CpG islands found in the ARAPROM gene set is plotted against the thresholds defined on the x and y axes, being the CG percentage and the o/e CpG ratio, respectively. The window size was 200 bp. Similar landscapes are obtained for other window sizes (100 and 400 bp) and are available at http://www.psb.rug.ac.be/bioinformatics/.
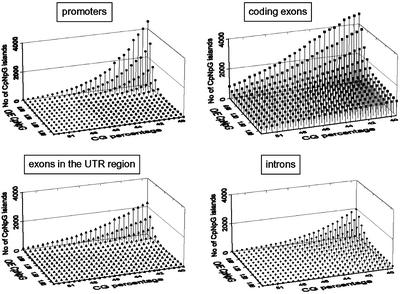
Figure 4.
CpNpG island landscape exploration of Arabidopsis gene sequences over a range of CG content and CpNpG relative frequency. For the various gene elements, on the z axis, the number of CpNpG islands found in the ARAPROM gene set is plotted against the thresholds defined on the x and y axes, being the CG percentage and the o/e CpNpG ratio, respectively. The window size was 200 bp. Similar landscapes are obtained for other window sizes (100 and 400 bp) and are available at http://www.psb.rug.ac.be/bioinformatics/.
Regarding the CpNpG landscape (Fig. 4), the major observation is that the overall number of islands lies well below that of the CpG islands. In addition, the same differences in landscape hold, as observed for CpG islands between promoter (and introns), on the one hand, and coding exons (and UTR exons), on the other hand. Nevertheless, a striking difference is that for CpNpGs, the o/e CpNpG threshold has to be very low for those islands to be detected. In terms of number of genes associated with CpG/CpNpG islands, different parameter settings lead to very different figures (Table II).
Table II.
Percentage of genes (out of 5,025) containing CpG (top) or CpNpG (bottom) islands, for a few different parameter settings
Additional values for other parameter settings can be found at http://www.psb.rug.ac.be/bioinformatics/.
| CG% Threshold | o/e Threshold | With Island in Promoter | With Island in at Least One Exona | With Island in Promoter AND in Exons | With Island in Promoter ONLY | With Island in Exons ONLY |
|---|---|---|---|---|---|---|
| CpG | ||||||
| 39 | 0.6 | 85.75 | 99.42 | 85.73 | 0.02 | 13.69 |
| 42 | 1.6 | 0.82 | 98.95 | 0.82 | 0.00 | 98.13 |
| CpNpG | ||||||
| 39 | 0.6 | 64.98 | 99.38 | 64.84 | 0.14 | 34.47 |
| 45 | 1.6 | 0.02 | 98.73 | 0.02 | 0.00 | 98.71 |
Refer to exons either in coding regions or in the UTR region.
This preliminary in silico analysis shows that prediction of promoter location based on the detection of potential CpG/CpNpG islands in the Arabidopsis genome is not straightforward. Nevertheless, because the landscape of CpG/CpNpG islands differs considerably between promoters and introns on the one side and exons (whether coding or not) on the other, there is some hope that, based on such a classification, more sophisticated approaches can be developed to detect CpG and CpNpG islands in plants.
SIGNAL-BASED FEATURES
Regulatory Elements
As stated in the introduction, CAREs are short, conserved motifs of approximately 5 to 20 nucleotides. Detection of CAREs in the promoter is not self-evident, because such short motifs are statistically expected to occur at random every few hundred base pairs. Therefore, the main problem lies in discriminating “true” from “false” regulatory elements (Blanchette and Sinha, 2001). It is important to distinguish whether unknown or known motifs are looked for. Compared with the detection of unknown motifs, that of known motifs is fairly straightforward and consists of the scanning of the DNA sequence with a given motif, which can be found in specialized databases such as TRANSFAC (Wingender et al., 1996) and TFD (Ghosh, 2000) and in plant-specific databases such as PLACE (Higo et al., 1999) and PlantCARE (Lescot et al., 2002). An overview of the different databases and motif search programs is given in Tables III and IV, respectively. In contrast, the detection of unknown CAREs in large regions of DNA requires the development and use of novel approaches and algorithms. Specifically, local multiple alignment algorithms that identify regulatory motifs have already been developed, which are merely based on statistical properties. Such algorithms search for DNA patterns that are more frequently present in a set of “related” than “unrelated” sequences. Therefore, the successful identification of regulatory DNA patterns depends on the size of the promoter sequence and, to a great extent, on the quality of the set of “related” sequences, i.e. genes that are co-expressed or coregulated and are thus expected to share similar conserved regulatory motifs. Such co-expressed genes are identified based on high-throughput gene expression profiling experiments. Alternatively, instead of coregulated genes, intergenic regions of orthologous sequences can also constitute a valuable data set for motif detection (Duret and Bucher, 1997). When selection pressure tends to conserve DNA patterns in the intergenic regions of homologous genes in related species, such DNA patterns can be expected to be of biological relevance and to reflect a conserved ancestral mode of regulation.
Table III.
Databases of cis-regulatory elements and promoter sequences
| Databases | Description | Web Sites | References |
|---|---|---|---|
| TRANSFAC | Transcription factor database | http://transfac.gbf.de/TRANSFAC/ | Wingender et al. (2000) |
| TFD | Transcription factor database | http://www.tfdg.com/Pages/tfddata.html | Ghosh (2000) |
| TRRD | Transcription regulatory region database | http://www.mgs.bionet.nsc.ru/mgs/dbases/trrd4/ | Kolchanov et al. (2000) |
| PlantCARE | Plant cis-Acting regulatory elements | http://sphinx.rug.ac.be:8080/PlantCARE/ | Lescot et al. (2002) |
| PLACE | Plant cis-acting regulatory elements | http://www.dna.affrc.go.jp/htdocs/PLACE/ | Higo et al. (1999) |
| RegulonDB | Database on transcriptional regulation in Escherichia coli | http://www.cifn.unam.mx/Computational_Genomics/regulondb/ | Salgado et al. (2001) |
| SCPD | Promoter database of Saccharomyces cerevisiae | http://cgsigma.cshl.org/jian | Zhu and Zhang (1999) |
| EPD | Eukaryotic promoter database | http://www.epd.isb-sib.ch/ | Praz et al. (2002) |
Table IV.
Motif search programs
| Programs | Web Sites | References |
|---|---|---|
| PLACE Signal Scan | http://www.dna.affrc.go.jp/htdocs/PLACE/signalup.html | Higo et al. (1999) |
| ScanACE | http://arep.med.harvard.edu/mrnadata/mrnasoft.html | Roth et al. (1998) |
| HMMER | http://bioweb.pasteur.fr/seqanal/interfaces/hmmsearch.htm | |
| Tess | http://www.cbil.upenn.edu/tess/ | |
| PatSearch | http://transfac.gbf.de/cgi-bin/patSearch/patsearch.pl | Pesole et al. (2000) |
| SignalScan | http://bimas.dcrt.nih.gov/molbio/signal/ | |
| http://biosci.cbs.umn.edu/software/software.html | ||
| ftp://molbio.cbs.umn.edu/pub/sigscan/ | Prestridge (1991) | |
| AliBaba2 | http://www.alibaba2.com/ | Grabe (2002) |
| TFBind | http://tfbind.ims.u-tokyo.ac.jp/ | Tsunoda and Takagi (1998) |
| MatInspector | http://www.gsf.de/biodv/matinspector.html | |
| http://www.genomatix.de/matinspector/ | ||
| http://www.genomatix.de/software_services/software/MatInspector/matinspector.html | Werner (2000) |
Regulatory Elements in Coregulated Genes
Co-expressed genes can be identified through transcript profiling techniques, such as microarrays (Brown and Botstein, 1999; Lipshutz et al., 1999; Southern, 2001) and cDNA-AFLP (Vos et al., 1995; Breyne et al., 2002). These high-throughput profiling techniques allow the expression level of hundreds or thousands of genes to be monitored simultaneously under the conditions tested. For each gene, an expression profile is obtained that reflects its dynamic behavior during a time-course experiment or its behavior under distinct conditions. Genes with similar expression profiles are considered “co-expressed”. To identify sets of co-expressed genes from high-throughput expression data, clustering techniques are required (Heyer et al., 1999; Jensen and Knudsen, 2000). In addition to standard cluster algorithms, such as hierarchical clustering, K-means, and self-organizing maps, more advanced algorithms are also being developed, which are specifically fine-tuned for biological applications (for a review, see Moreau et al., 2002).
Because co-expressed genes tend to behave similarly, they are expected to be coregulated. Under the simplifying assumption that this coregulation occurs at the transcriptional level, co-expressed genes should contain similar cis-regulatory elements in their promoter regions. As a consequence, these yet unknown cis-regulatory elements will be statistically overrepresented in the intergenic regions of the co-expressed genes in comparison with their frequent occurrence in a set of unrelated sequences. This overrepresentation constitutes the general principle on which motif detection algorithms is based.
Regulatory Elements in Orthologs
Usually, genes are part of more extensive gene families that have originated through both speciation and duplication events. Homologous genes in distinct species are called orthologs, whereas paralogs refer to homologous genes that are found in the same genome and have been created through gene duplication (Mindell and Meyer, 2001). Regarding promoter analysis and study of regulatory elements, it is important to discriminate between these two types of homologous relationships. True orthologs have usually retained very similar functions in distinct species, whereas this is not necessarily true for paralogs. In many cases, paralogs have only been conserved if they have acquired different or complementary functions. Hughes (1994) and Force et al. (1999) argued that when a gene with multiple functions is duplicated, the duplicates are only redundant for as long as each gene is capable of performing all ancestral roles. When one mutated duplicate is prevented from carrying out one of these ancestral roles, the other duplicate is no longer redundant. According to the “duplication degeneration complementation” model of Force et al. (1999), degenerative mutations preserve rather than destroy duplicated genes, but also change their functions or, at least, restrict them to become more specialized. Duplicated genes can have different expression domains (i.e. the tissue in which both genes are expressed might have changed as well as the time of expression) because of changes in their regulatory elements in the promoter region (Force et al., 1999; Altschmied et al., 2002; Prince and Pickett, 2002). Therefore, promoter regions of true orthologs probably contain similar regulatory motifs, which may no longer be true for paralogs.
Motif Prediction
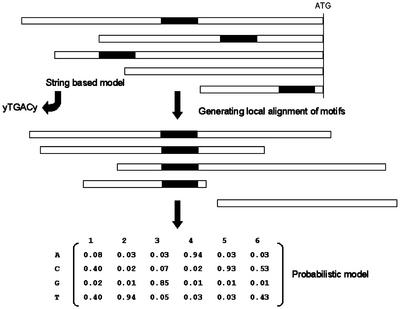
To conceive a general method that can detect regulatory motifs is a great challenge because of both the complexity and flexibility of the regulatory mechanisms (see the introduction). An important distinction between the different approaches used thus far to detect regulatory motifs lies in the representation of the motif, i.e. the TF-binding site. The simplest description for a motif is a string of characters (A, C, G, and T), extended with the 11 IUPAC characters that represent partly unspecified or ambiguous nucleotides, and is used in the string-based approaches, such as word counting. A more sophisticated description is to represent a given motif by describing it in a probabilistic manner in which a certain likelihood is assessed for each nucleotide at a given position in the motif. An example of a probabilistic representation is the position-weight matrix, where each column corresponds to a position in the aligned binding sites and each row to a nucleotide, as shown in Figure 5. The cells of these matrices contain a number indicating the probability to find a given nucleotide at that particular position. Alternatives to describe motifs in a probabilistic manner are the hidden Markov models (Jarmer et al., 2001) or neural networks (Workman and Stormo, 2000). Software tools for motif prediction are listed in Table V.
Figure 5.
Schematic representation of a set of intergenic sequences upstream of the ATG translation initiation site, with a common motif shown as black boxes. On the basis of such a data set, “words” can be counted and statistically evaluated for their overrepresentation. On the other hand, the “putative” motifs can be aligned and frequencies of occurrence of each nucleotide can be calculated for each column within the generated alignment, producing a position weight matrix. See text for details.
Table V.
Motif prediction programs
String-Based Motif Prediction
Counting all of the possible words that may occur across the different promoter sequences is one of the simplest approaches to find CAREs in a set of promoters. Among word-counting methods, enumerative and suffix-tree approaches can be distinguished, the latter being an optimization of the former. Both methods are string based: The DNA sequence is considered as text in which oligonucleotides are represented as words or strings. For a given set of promoter sequences, the frequency of each possible word of a defined length is computed (Hutchinson, 1996; Wolfertstetter et al., 1996; Brāzma et al., 1998; van Helden et al., 1998; Vanet et al., 1999; Bussemaker et al., 2000a, 2000b; Sinha and Tompa, 2000; Hampson et al., 2002). The difference between the two representations is that the enumerative approach will search for each word in the sequence and calculate its frequency, whereas the suffix-tree approach will only look for a certain subset. The suffix tree is used to represent each word together with all of its subwords (or suffixes) so that each word can be reconstructed by going down the tree. For example, when a certain suffix (for instance, ACCT) is not found in the data set, none of the words containing ACCT will be counted anymore (Sagot and Myers, 1998; Marsan and Sagot, 2000). As a consequence, the computational time needed to count words can be highly reduced, which allows the analysis of larger data sets.
Once the frequencies of different words are calculated, the words that are likely to be a “true” regulatory motif have to be differentiated from those that are not. Therefore, in each of these word-counting methods, the number of occurrences of a word needs to be compared with the expected frequency in a set of non-related sequences, represented by a background model, which is used to obtain an expected probability. The simplest way to build a background model is by creating a set of randomly generated sequences, based on the single nucleotide composition of the submitted sequence. More sophisticated ways to generate a background model are based on Markov chain statistics (Schbath et al., 1995; Schbath, 1997, 2000; van Helden et al., 2000a; Thijs et al., 2001), a lexicon (Bussemaker et al., 2000a, 2000b), or by simulations in which words are randomly reassembled to rebuild a set of sequences (Coward, 1999; Marsan and Sagot, 2000). The choice of the background model can be critical. In our experience, representations closest to real biological sequences or a set of well-chosen biological sequences appear to be the most reliable. The statistical methods to evaluate the significance of an observed versus expected frequency and to conclude whether a word is overrepresented or not are, for example, binomial probability (van Helden et al., 1998), composed Poisson law (Robin and Schbath, 2001), z-score (Kleffe and Borodovsky, 1992; van Helden et al., 2000a), and χ2 test (Vanet et al., 1999). Although the latter is a very simple statistical method to evaluate unknown motifs, its merits in previous studies has been proven in looking for regulatory elements in the yeast genome (van Helden et al., 1998; Sinha and Tompa, 2002).
Probabilistic Motif Detection
Probabilistic motif detection aims at constructing a multiple alignment by locally aligning small conserved regions in a set of unaligned sequences. Here, we will focus on the matrix-based approaches to illustrate probabilistic motif detection procedures. All methods start from a random motif model, represented as a weight matrix and altered through a series of iterations by machine-learning algorithms that are aimed at finding the optimal score. The process of optimizing the score for a local alignment already tends to converge toward conserved motifs that occur frequently in the data set. The more advanced algorithms incorporate a background model to compensate for given motifs occurring at high frequencies because of compositions similar to those of the non-conserved parts of the sequence (the “background”). A motif in which the average nucleotide composition differs strongly from the background will be assigned a higher score. Implementations differ from each other in the way the background is represented, in how the score is calculated, and in how the optimization is performed. For motif detection algorithms that describe the motif by a weight matrix, expectation maximization and its stochastic variant, Gibbs sampling, are often used for optimization strategies.
The program CONSENSUS was one of the first algorithms that represented a motif by a weight matrix (Hertz et al., 1990; Hertz and Stormo, 1996, 1999). The algorithm starts with a first sequence from the submitted data set and creates a weight matrix for each possible word of user-specified length. Subsequently, it aligns each possible word from the next sequence with each weight matrix. The obtained alignments are scored for their information content, and those with the highest score are retained for the next iteration. This process is reiterated until all sequences have contributed to the alignment and weight matrix. CONSENSUS was used for example in the identification of CAREs involved in the heat shock response in Caenorhabditis elegans (GuhaThakurta and Stormo, 2001).
The expectation-maximization (EM) method (Stormo, 1988, 1990; Stormo and Hartzell, 1989; Lawrence and Reilly, 1990; Cardon and Stormo, 1992; Bailey and Elkan, 1995) is a two-step iterative procedure that aims at obtaining, for each possible motif position, the likelihood that the motif located at that position corresponds to the current motif model (weight matrix). In the maximization step, the parameters that optimize the likelihood are estimated. Once the motif positions are known, the observed frequencies of the nucleotides at each motif position correspond to the maximum-likelihood estimates of the parameters of the motif model. On the basis of the updated probabilities of all motif positions of the previous step, the model parameters are re-estimated. For motif finding, EM simultaneously computes the alignment positions, the motif weight matrix, and the background model that maximize the likelihood of the sequence. In the original implementation (Lawrence and Reilly, 1990), the “exactly one occurrence” of the motif in each sequence was assumed. This assumption is a problem because in a cluster of co-expressed genes, sequences might be present without a (or the) motif. Because EM-based motif detection algorithms are deterministic, results for particular queries with similar parameter settings and initializations will be identical. A drawback of the method is that results depend strongly on the initial conditions and often converge into local optima. One of the most widely used EM applications is the program MEME (Bailey and Elkan, 1995).
Gibbs sampling-based strategies have originally been developed to detect protein motifs but have been adapted later on to handle DNA sequences (Neuwald et al., 1995; Roth et al., 1998; Hughes et al., 2000; Liu et al., 2001; Thijs et al., 2002a). Gibbs sampling is a stochastic variant of EM (Lawrence et al., 1993; Neuwald et al., 1995). Because of the stochastic nature of the Gibbs sampling approach, an initially detected motif can be replaced by another one that has a higher score, thus allowing escape from local optima. This feature is the reason why the output of a stochastic motif detection algorithm results in different outputs, even with the same input and parameter settings. However, the more pronounced the optimal solution is in a given data set, the more a motif is overrepresented, and the stronger its conservation, the more frequently it will be retrieved over different runs. Statistics on the outcome of multiple runs of a stochastic implementation can facilitate interpretation of the results.
Adaptative quality-based clustering (De Smet et al., 2002) combined with Motif Sampler based on Gibbs sampling (Thijs et al., 2002a) was applied to the data published by Reymond et al. (2000) in which gene expression was studied in response to mechanical wounding in Arabidopsis leaves. After clustering, the four most populated clusters (>3 genes) of co-expressed genes were selected, and the upstream sequences were analyzed with the Motif Sampler to discover common regulatory elements. To avoid the problem of local optima, each data set was submitted 10 times to the Motif Sampler with the same parameters. The output of these 10 runs was compiled taking into account the individual scores of each motif and the order in which they were found. Subsequently, the consensus of the motifs found were compared with regulatory sites described in the PlantCARE database (Lescot et al., 2002). From all of the high-ranking motifs returned by the Motif Sampler, several were similar to known cis-regulatory elements involved in plant defense (methyl jasmonate-, abscisic acid-, or elicitor-responsive elements) or in light responsiveness. Among these elements, a 12-bp motif was found composed of two sites involved in methyl jasmonate responsiveness. These motifs have been described previously in the upstream sequence of the lipoxygenase isoenzyme 1 gene of barley, where they were separated by 15 bp (Thijs et al., 2002a).
Motif Prediction by Phylogenetic Footprinting
The procedure that identifies regulatory elements based on a set of orthologous sequences is named phylogenetic footprinting (Koop, 1995; Duret and Bucher, 1997; Wasserman et al., 2000). Phylogenetic footprinting has proven its usefulness to detect CAREs in the human genome, based on the pairwise comparison between human and mouse (Hardison, 2000; Wasserman et al., 2000; Krivan and Wasserman, 2001; Dermitzakis and Clark, 2002; Jegga et al., 2002). However, producing a reliable data set for phylogenetic footprinting is not self-evident. When the overall degree of conservation in intergenic sequences between two homologs is too high, conserved motifs will not be detected. At the other extreme, when homologs are compared from species that are too distantly related, the intergenic regions may no longer show any similarity (Tompa, 2001). The ideal composition of a data set can only be derived in retrospect, implying that an algorithm suited for phylogenetic footprinting should ideally identify and discard (or counter-weigh) sequences that are too similar and cope with the presence of sequences that do not contain the conserved motif. Furthermore, the phylogenetic distance between organisms should be taken into account in the weighting schemes of the algorithm. As stated before, closely related sequences are less useful for identifying a motif because of their high overall conservation, complicating the search for functionally conserved regions. Alignment algorithms, such as ClustalW (Thompson et al., 1994) and Bayes-Block Aligner (Zhu et al., 1998), have proven useful for phylogenetic footprinting, but the length of the conserved motif is often too small compared with the length of the non-conserved part of the sequence; therefore, multiple sequence alignment will fail.
A promising novel algorithm has recently been published that identifies the most conserved motifs among the input sequences as measured by a parsimony score on the underlying phylogenetic tree (Blanchette et al., 2002; Blanchette and Tompa, 2002). In general, the algorithm selects motifs that are characterized by a minimal number of mismatches and are conserved over long evolutionary distances. Furthermore, the motifs should not have undergone independent losses in multiple branches. In other words, the motif should be present in the sequences of subsequent taxa along a branch. The algorithm, based on dynamic programming, proceeds from the leaves of the phylogenetic tree to its root and seeks for motifs of a user-defined length with a minimum number of mismatches. Moreover, the algorithm allows a higher number of mismatches for those sequences that span a greater evolutionary distance. Motifs that are lost along a branch of the tree are assigned an additional cost because it is assumed that multiple independent losses are unlikely in evolution. To compensate for spurious hits, statistical significance is calculated based on a random set of sequences in which no motifs occur. Phylogenetic footprinting for the detection of CAREs is steadily gaining importance (Koch et al., 2001, 2002; Quiros et al., 2001; Colinas et al., 2002) and will continue to do so when more plant genomic sequences will become available. To give just one example, using phylogenetic footprinting, Tompa (2001) was able to predict several new binding sites in the 5′-UTR of plant genes coding for the small subunit of ribulose-1,5-bisphosphate carboxylase.
Improvements and Fine Tuning of Motif Detection Algorithms
The most obvious reason why motif detection algorithms fail is because of their sensitivity to noise. All parts of a sequence that do not contain the motif constitute noise in the context of motif detection. Moreover, because sets of related sequences are usually based on other predictive tools, for instance clustering, they are expected to contain sequences without any shared motif. A decreasing signal-to-noise ratio exacerbates the identification of statistically overrepresented motifs and increases the chance of finding false positives. Probabilistic motif detection methods have been improved considerably to cope with a large noise level. Current implementations, such as AlignACE (Hughes et al., 2000) and MEME (Bailey and Elkan, 1995), usually take into account that some sequences lack a shared motif and allow the influence of such sequences to be discarded by estimating the motif model parameters. The more advanced implementations derive the optimal number of motif occurrences in each sequence from the data. Modeling the background with a more complex sequence model contributes also considerably to the robustness of the algorithm in the presence of noise (Liu et al., 2001, 2002; Thijs et al., 2001, 2002a). Besides making more robust algorithms that facilitate discrimination between true and false positives, advanced scoring schemes are being developed that assign a statistical significance to the motifs detected, i.e. that describe the probability of observing a motif with a similar score in a set of unrelated sequences.
Because regulatory motifs, in particular in higher eukaryotes, are concentrated in modules, current research is focusing toward adapting motif detection algorithms to retrieve dyads, i.e. motifs spaced by a fixed or variable gap. Within the enumerative statistical methods, Sinha and Tompa (2000) created an algorithm that searches for motifs with a gap of variable size between them. The algorithm developed by van Helden et al. (2000) enables a search for dyads with a fixed number of base pairs between 3 and 20.
Vanet et al. (2000) and Marsan and Sagot (2000) developed approaches that look for two motifs separated by a fixed number of nucleotides by using the suffix-tree method. Cardon and Stormo (1992) adapted their EM-based algorithm to detect dyads with variable gap size. In their Gibbs sampling-based implementation, Liu et al. (2001) have included an extension that allows searching for dyads, whereas the program Co-bind of GuhaThakurta and Stormo (2001) was specifically created to identify two regulatory sites of gap-separated cooperative TFs.
The need for extensive parameter fine tuning complicates nonexpert use of most of the motif detection approaches described above. Novel implementations of motif detection algorithms tackle this problem by estimating the optimal parameter settings themselves, hence, minimizing the number of user-defined parameters. An example of such a user-defined parameter is the motif length. Because the motif length is generally unknown in advance, it is not obvious to choose the parameter setting that results in the true motif. Some algorithms compute the optimal motif length; for instance, Pattern assembly (van Helden et al., 2000b) groups overlapping motifs to build a longer motif consensus. The implementation of AlignAce determines the optimal motif length from the data (Roth et al., 1998; Hughes et al., 2000). Manually generating a suitable data set (see above) that can be used readily for motif detection can be a tedious job. Therefore, some on-line implementations have been developed, such as the INCLUSIve (Thijs et al., 2002b; Engelen et al., 2003) Web site that offers a pipeline to combine microarray preprocessing, the adaptive quality-based clustering, automatic sequence retrieval, and motif detection based on Gibbs sampling (Motif Sampler). A tool similar to INCLUSIve, called expression profiler (Brāzma et al., 1998), is provided by the European Bioinformatics Institute. “Regulatory Sequence Analysis tools” (van Helden et al., 1998) proposes a word counting-based set of tools to analyze a set of intergenic sequences.
CONCLUSIONS
Promoters are very complex structures, defined by many different structural features. The actual regulatory elements are usually very short, which highly complicates their unambiguous identification. As a consequence, the in silico prediction of promoters and regulatory motifs is not straightforward. In addition, our knowledge of transcription regulation in general and organism-specific expression regulation in particular, is still very limited. Especially for plants, solid “intrinsic” genomic data are still needed that can be integrated into existing prediction tools. In this respect, we have started with the analysis of CpG and CpNpG islands, known to be often associated with promoters. Although several implementations for the detection of such “islands” in vertebrates have been described (Ioshikhes and Zhang, 2000), parameter settings used to detect these islands in animals cannot be used to find similar islands in the Arabidopsis genome. Software and parameters have to be adapted to the species under investigation. Moreover, even if a reliable tool were available for the detection of CpG and CpNpG islands associated with plant promoters, it remains to be proven whether these islands would be biologically functional and relevant. In addition to a lack of “intrinsic” genomic data, experimental data on promoters are also scarce, because in general, thorough analysis of even one single promoter is very time consuming. Furthermore, the technology is still missing for exhaustive knowledge of gene expression or for understanding the mechanisms behind it. Therefore, for now and despite its many shortcomings, in silico analysis seems to be a privileged alternative to analyze simultaneously a great number of regulatory elements or promoter regions. Experimental testing of these in silico predictions may be a manner to increase knowledge on promoters more quickly and at a lower cost, especially for plants.
Acknowledgments
We thank two anonymous reviewers for helpful suggestions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.102.017715.
This work was supported by the Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek (grant no. STWW–980396). K.F. is indebted to the Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen for a predoctoral fellowship, K.M. is Research Fellow of the Fund for Scientific Research (Flanders), and P.R. is a Research Director of the Institut National de la Recherche Agronomique (France).
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Altschmied J, Delfgaauw J, Wilde B, Duschl J, Bouneau L, Volff JN, Schartl M (2002) Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 161: 259–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F, Bird A (1999) CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol 9: R661–R667 [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A et al. (2002) Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 23: 1301–1310 [DOI] [PubMed] [Google Scholar]
- Ashikawa I (2001) Gene-associated CpG islands in plants as revealed by analyses of genomic sequences. Plant J 26: 617–625 [DOI] [PubMed] [Google Scholar]
- Bagga R, Michalowski S, Sabnis R, Griffith JD, Emerson BM (2000) HMG I/Y regulates long range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res 28: 2541–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic V, Seah S, Chong A, Zhang G, Koh J, Brusic V (2002) Dragon Promoter Finder: recognition of vertebrate RNA polymerase II promoters. Bioinformatics 18: 198–199 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1995) The value of prior knowledge in discovering motifs with MEME. Proc Int Conf Intell Syst Mol Biol 3: 21–29 [PubMed] [Google Scholar]
- Baldi P, Chauvin Y, Brunak S, Gorodkin J, Pedersen AG (1998) Computational applications of DNA structural scales. Proc Int Conf Intell Syst Mol Biol 6: 35–42 [PubMed] [Google Scholar]
- Barton MC, Madani N, Emerson BM (1997) Distal enhancer regulation by promoter derepression in topologically constrained DNA in vitro. Proc Natl Acad Sci USA 94: 7257–7262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M, Eisfeld K (1997) Transcription factor access to chromatin. Nucleic Acids Res 25: 3559–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J (2001) A vicious cycle: RNA silencing and DNA methylation in plants. Cell 106: 129–132 [DOI] [PubMed] [Google Scholar]
- Bentin T, Nielsen PE (2002) In vitro transcription of a torsionally constrained template. Nucleic Acids Res 30: 803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk AJ (1999) Activation of RNA polymerase II transcription. Curr Opin Cell Biol 11: 330–335 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Sinha S (2001) Separating real motifs from their artifacts. Bioinformatics 17: 30–38 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Schwikowski B, Tompa M (2002) Algorithms for phylogenetic footprinting. J Comput Biol 9: 211–223 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Tompa M (2002) Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res 12: 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolshoy A, McNamara P, Harrington RE, Trifonov EN (1991) Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc Natl Acad Sci USA 88: 2312–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brāzma A, Jonassen I, Vilo J, Ukkonen E (1998) Predicting gene regulatory elements in silico on a genomic scale. Genome Res 8: 1202–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslauer KJ, Frank R, Blocker H, Marky LA (1986) Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA 83: 3746–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyne P, Dreesen R, Vandepoele K, De Veylder L, Van Breusegem F, Callewaert L, Rombauts S, Raes J, Cannoot B, Engler G et al. (2002) Transcriptome analysis during cell division in plants. Proc Natl Acad Sci USA 99: 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower-Toland BD, Smith CL, Yeh RC, Lis JT, Peterson CL, Wang MD (2002) Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci USA 99: 1960–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Botstein D (1999) Exploring the new world of the genome with DNA microarrays. Nat Genet 21: 33–37 [DOI] [PubMed] [Google Scholar]
- Brukner I, Sanchez R, Suck D, Pongor S (1995a) Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J 14: 1812–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brukner I, Sanchez R, Suck D, Pongor S (1995b) Trinucleotide models for DNA bending propensity: comparison of models based on DNaseI digestion and nucleosome packaging data. J Biomol Struct Dyn 13: 309–317 [DOI] [PubMed] [Google Scholar]
- Buratowski S (1997) Snapshots of RNA polymerase II transcription initiation. Curr Opin Cell Biol 12: 320–325 [DOI] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED (2000a) Building a dictionary for genomes: identification of presumptive regulatory sites by statistical analysis. Proc Natl Acad Sci USA 97: 10096–10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED (2000b) Regulatory element detection using a probabilistic segmentation model. Proc Int Conf Intell Syst Mol Biol 8: 67–74 [PubMed] [Google Scholar]
- Cao X, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99: 16491–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Springer NM, Muszynski MG, Phillips RL, Kaeppler S, Jacobsen SE (2000) Conserved plant genes with similarity to mammalian de novo DNA methyltransferases. Proc Natl Acad Sci USA 97: 4979–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Stormo GD (1992) Expectation maximization algorithm for identifying protein-binding sites with variable lengths from unaligned DNA fragments. J Mol Biol 223: 159–170 [DOI] [PubMed] [Google Scholar]
- Colinas J, Birnbaum K, Benfey PN (2002) Using cauliflower to find conserved non-coding regions in Arabidopsis. Plant Physiol 129: 451–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward E (1999) Shufflet: shuffling sequences while conserving the k-let counts. Bioinformatics 15: 1058–1059 [DOI] [PubMed] [Google Scholar]
- Crothers DM (1998) DNA curvature and deformation in protein-DNA complexes: a step in the right direction. Proc Natl Acad Sci USA 95: 15163–15165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Grosse I, Zhang MQ (2001) Computational identification of promoters and first exons in the human genome. Nat Genet 29: 412–417 [DOI] [PubMed] [Google Scholar]
- de Boer GJ, Testerink C, Pielage G, Nijkamp HJ, Stuitje AR (1999) Sequences surrounding the transcription initiation site of the Arabidopsis enoyl-acyl carrier protein reductase gene control seed expression in transgenic tobacco. Plant Mol Biol 39: 1197–1207 [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG (2002) Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol 19: 1114–1121 [DOI] [PubMed] [Google Scholar]
- De Smet F, Mathys J, Marchal K, Thijs G, De Moor B, Moreau Y (2002) Adaptive quality-based clustering of gene expression profiles. Bioinformatics 18: 735–746 [DOI] [PubMed] [Google Scholar]
- Dorsett D (1999) Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr Opin Genet Dev 9: 505–514 [DOI] [PubMed] [Google Scholar]
- Down TA, Hubbard TJ (2002) Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res 12: 458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Bucher P (1997) Searching for regulatory elements in human noncoding sequences. Curr Opin Struct Biol 7: 399–406 [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N (2000) The covariation between TpA deficiency, CpG deficiency, and G+C content of human isochores is due to a mathematical artifact. Mol Biol Evol 17: 1620–1625 [DOI] [PubMed] [Google Scholar]
- Dynan WS (1989) Modularity in promoters and enhancers. Cell 58: 1–4 [DOI] [PubMed] [Google Scholar]
- El Hassan MA, Calladine CR (1996) Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. J Mol Biol 259: 95–103 [DOI] [PubMed] [Google Scholar]
- Engelen K, Coessens B, Marchal K, De Moor B (2003) MARAN: normalizing micro-array data. Bioinformatics 19: 893–894 [DOI] [PubMed] [Google Scholar]
- Featherstone M (2002) Coactivators in transcription initiation: here are your orders. Curr Opin Genet Dev 12: 149–155 [DOI] [PubMed] [Google Scholar]
- Fessele S, Maier H, Zischek C, Nelson PJ, Werner T (2002) Regulatory context is a crucial part of gene function. Trends Genet 18: 60–63 [DOI] [PubMed] [Google Scholar]
- Fickett JW, Hatzigeorgiou AG (1997) Eukaryotic promoter recognition. Genome Res 7: 861–878 [DOI] [PubMed] [Google Scholar]
- Fickett JW, Wasserman WW (2000) Discovery and modeling of transcriptional regulatory regions. Curr Opin Biotechnol 11: 19–24 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Kovac K, Peacock WJ, Dennis ES (1998a) DNA methylation and the promotion of flowering by vernalization. Proc Natl Acad Sci USA 95: 5824–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Genger RK, Peacock WJ, Dennis ES (1998b) DNA methylation in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 223–247 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Kovac KA (2000) Plant DNA methyltransferases. Plant Mol Biol 43: 189–201 [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (2000) DNA methylation, a key regulator of plant development and other processes. Curr Opin Genet Dev 10: 217–223 [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan Y-l, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol Biol 196: 261–282 [DOI] [PubMed] [Google Scholar]
- Ghosh D (2000) Object-oriented transcription factors database (ooTFD). Nucleic Acids Res 28: 308–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel M, Jimenez B, Herrera-Estrella L (1996) The first intron of the Arabidopsis thaliana gene coding for elongation factor 1 β contains an enhancer-like element. Gene 170: 201–206 [DOI] [PubMed] [Google Scholar]
- Goodsell DS, Dickerson RE (1994) Bending and curvature calculations in B-DNA. Nucleic Acids Res 22: 5497–5503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin AA, Zhurkin VB, Olson WK (1995) B-DNA twisting correlates with base-pair morphology. J Mol Biol 247: 34–48 [DOI] [PubMed] [Google Scholar]
- Grabe N (2002) AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol 2: S1–1 [PubMed] [Google Scholar]
- GuhaThakurta D, Stormo GD (2001) Identifying target sites for cooperatively binding factors. Bioinformatics 17: 608–621 [DOI] [PubMed] [Google Scholar]
- Hampson S, Kibler D, Baldi P (2002) Distribution patterns of overrepresented k-mers in noncoding yeast DNA. Bioinformatics 18: 513–528 [DOI] [PubMed] [Google Scholar]
- Hannenhalli S, Levy S (2001) Promoter prediction in the human genome. Bioinformatics 17: S90–S96 [DOI] [PubMed] [Google Scholar]
- Hardison RC (2000) Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet 16: 369–372 [DOI] [PubMed] [Google Scholar]
- Hershkovitz M, Gruenbaum Y, Renbaum P, Razin A, Loyter A (1990) Effect of CpG methylation on gene expression in transfected plant protoplasts. Gene 94: 189–193 [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Hartzell GW III, Stormo GD (1990) Identification of consensus patterns in unaligned DNA sequences known to be functionally related. Comput Appl Biosci 6: 81–92 [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD (1996) Escherichia coli promoter sequences: analysis and prediction. Methods Enzymol 273: 30–42 [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD (1999) Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15: 563–577 [DOI] [PubMed] [Google Scholar]
- Heyer LJ, Kruglyak S, Yooseph S (1999) Exploring expression data: identification and analysis of coexpressed genes. Genome Res 9: 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PS, Ellison MJ, Quigley GJ, Rich A (1986) A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. EMBO J 5: 2737–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond B 256: 119–124 [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM (2000) Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296: 1205–1214 [DOI] [PubMed] [Google Scholar]
- Hutchinson GB (1996) The prediction of vertebrate promoter regions using differential hexamer frequency analysis. Comput Appl Biosci 12: 391–398 [DOI] [PubMed] [Google Scholar]
- Inamdar NM, Ehrlich KC, Ehrlich M (1991) CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol Biol 17: 111–123 [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Trifonov EN, Zhang MQ (1999) Periodical distribution of transcription factor sites in promoter regions and connection with chromatin structure. Proc Natl Acad Sci USA 96: 2891–2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioshikhes IP, Zhang MQ (2000) Large-scale human promoter mapping using CpG islands. Nat Genet 26: 61–63 [DOI] [PubMed] [Google Scholar]
- Jarmer H, Larsen TS, Krogh A, Saxild HH, Brunak S, Knudsen S (2001) Sigma A recognition sites in the Bacillus subtilis genome. Microbiology 147: 2417–2424 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Bender J, Richards EJ (1998) The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev 12: 1714–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegga AG, Sherwood SP, Carman JW, Pinski AT, Phillips JL, Pestian JP, Aronow BJ (2002) Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res 12: 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Knudsen S (2000) Automatic discovery of regulatory patterns in promoter regions based on whole cell expression data and functional annotation. Bioinformatics 16: 326–333 [DOI] [PubMed] [Google Scholar]
- Johnson PF, McKnight SL (1989) Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem 58: 799–839 [DOI] [PubMed] [Google Scholar]
- Jones PA (1999) The DNA methylation paradox. Trends Genet 15: 34–37 [DOI] [PubMed] [Google Scholar]
- Juo ZS, Chiu TK, Leiberman PM, Baikalov I, Berk AJ, Dickerson RE (1996) How proteins recognize the TATA box. J Mol Biol 261: 239–254 [DOI] [PubMed] [Google Scholar]
- Kass SU, Landsberger N, Wolffe AP (1997) DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol 7: 157–165 [DOI] [PubMed] [Google Scholar]
- Kleffe J, Borodovsky M (1992) First and second moment of counts of words in random texts generated by Markov chains. Comput Appl Biosci 8: 433–441 [DOI] [PubMed] [Google Scholar]
- Klingenhoff A, Frech K, Werner T (2002) Regulatory modules shared within gene classes as well as across gene classes can be detected by the same in silico. In Silico Biol 2: S17–26 [PubMed] [Google Scholar]
- Koch MA, Haubold B, Mitchell-Olds T (2002) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol Biol Evol 17: 1483–1498 [DOI] [PubMed] [Google Scholar]
- Koch MA, Weisshaar B, Kroymann J, Haubold B, Mitchell-Olds T (2001) Comparative genomics and regulatory evolution: conservation and function of the Chs and Apetala3 promoters. Mol Biol Evol 18: 1882–1891 [DOI] [PubMed] [Google Scholar]
- Kolchanov NA, Podkolodnaya OA, Ananko EA, Ignatieva EV, Stepanenko IL, Kel-Margoulis OV, Kel AE, Merkulova TI, Goryachkovskaya TN, Busygina TV (2000) Transcription regulatory regions database (TRRD): its status in 2000. Nucleic Acids Res 28: 298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrakhin YV, Kel AE, Kolchanov NA, Romashchenko AG, Milanesi L (1995) Eukaryotic promoter recognition by binding sites for transcription factors. Comput Appl Biosci 11: 477–488 [DOI] [PubMed] [Google Scholar]
- Koop BF (1995) Human and rodent DNA sequence comparisons: a mosaic model of genomic evolution. Trends Genet 11: 367–371 [DOI] [PubMed] [Google Scholar]
- Kooter JM, Matzke MA, Meyer P (1999) Listening to the silent genes: transgene silencing, gene regulation and pathogen control. Trends Plant Sci 4: 340–347 [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y (2002) Chromatin and transcription: Where do we go from here? Curr Opin Genet Dev 12: 249–251 [DOI] [PubMed] [Google Scholar]
- Krivan W, Wasserman WW (2001) A predictive model for regulatory sequences directing liver-specific transcription. Genome Res 11: 1559–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001) Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Oppenheimer DG, Pollock S, Marks MD (1993) Arabidopsis GLABROUS1 gene requires downstream sequences for function. Plant Cell 5: 1739–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC (1993) Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262: 208–214 [DOI] [PubMed] [Google Scholar]
- Lawrence CE, Reilly AA (1990) An expectation maximization (EM) algorithm for the identification and characterization of common sites in unaligned biopolymer sequences. Proteins 7: 41–51 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Chandrasekharan MB, Wolffe AP, Hall TC (2001) Chromatin structure and phaseolin gene regulation. Plant Mol Biol 46: 121–129 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ (1999) High density synthetic oligonucleotide arrays. Nat Genet 21: 20–24 [DOI] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20: 835–839 [DOI] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138 [PubMed]
- Marilley M, Pasero P (1996) Common DNA structural features exhibited by eukaryotic ribosomal gene promoters. Nucleic Acids Res 24: 2204–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsan L, Sagot MF (2000) Algorithms for extracting structured motifs using a suffix tree with an application to promoter and regulatory site consensus identification. J Comput Biol 7: 345–362 [DOI] [PubMed] [Google Scholar]
- Mathé C, Sagot MF, Schiex T, Rouzé P (2002) Current methods of gene prediction, their strengths and weaknesses. Nucleic Acids Res 30: 4103–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P (2000) Transcriptional transgene silencing and chromatin components. Plant Mol Biol 43: 221–234 [DOI] [PubMed] [Google Scholar]
- Meyer P, Niedenhof I, ten Lohuis M (1994) Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrida. EMBO J 13: 2084–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meza TJ, Enerly E, Boru B, Larsen F, Mandal A, Aalen RB, Jakobsen KS (2002) A human CpG island randomly inserted into a plant genome is protected from methylation. Transgenic Res 11: 133–142 [DOI] [PubMed] [Google Scholar]
- Mindell DP, Meyer A (2001) Homology evolving. Trends Ecol Evol 16: 434–440 [Google Scholar]
- Moreau Y, De Smet F, Thijs G, Marchal K, De Moor B (2002) Functional bioinformatics of microarray data: from expression to regulation. IEEE Proc 30: 1722–1743 [Google Scholar]
- Neuwald AF, Liu JS, Lawrence CE (1995) Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci 4: 1618–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Bird A (1999) DNA methylation and chromatin modification. Curr Opin Genet Dev 9: 158–163 [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Burley SK (1997) RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci USA 94: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Hoffman A, Roeder RG, Burley SK (1996) Crystal structure of a human TATA box-binding protein/TATA element complex. Proc Natl Acad Sci USA 93: 4862–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U (2000) Promoter prediction on a genomic scale: the Adh experience. Genome Res 10: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohler U, Harbeck S, Niemann H, Noth E, Reese MG (1999) Interpolated Markov chains for eukaryotic promoter recognition. Bioinformatics 15: 362–369 [DOI] [PubMed] [Google Scholar]
- Ohler U, Niemann H (2001) Identification and analysis of eukaryotic promoters: recent computational approaches. Trends Genet 17: 56–60 [DOI] [PubMed] [Google Scholar]
- Ohler U, Niemann H, Liao GC, Rubin GM (2001) Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics 17: S199–S206 [DOI] [PubMed] [Google Scholar]
- Ohler U, Liao GC, Niemann H, Rubin GM (2000) Computational analysis of core promoters in the Drosophila genome. Genome Biol 3: 0087.1–0087.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki M, Kamakaka RT (2002) Blockers and barriers to transcription: competing activities. Curr Opin Cell Biol 14: 299–304 [DOI] [PubMed] [Google Scholar]
- Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB (1998) DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci USA 95: 11163–11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panstruga R, Buschges R, Piffanelli P, Schulze-Lefert P (1998) A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res 26: 1056–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen AG, Baldi P, Chauvin Y, Brunak S (1998) DNA structure in human RNA polymerase II promoters. J Mol Biol 281: 663–673 [DOI] [PubMed] [Google Scholar]
- Pedersen AG, Baldi P, Chauvin Y, Brunak S (1999) The biology of eukaryotic promoter prediction: a review. Comput Chem 23: 191–207 [DOI] [PubMed] [Google Scholar]
- Pesole G, Liuni S, D'Souza M (2000) PatSearch: a pattern matcher software that finds functional elements in nucleotide and protein sequences and assesses their statistical significance. Bioinformatics 16: 439–450 [DOI] [PubMed] [Google Scholar]
- Pitto L, Cernilogar F, Evangelista M, Lombardi L, Miarelli C, Rocchi P (2000) Characterization of carrot nuclear proteins that exhibit specific binding affinity towards conventional and nonconventional DNA methylation. Plant Mol Biol 44: 659–673 [DOI] [PubMed] [Google Scholar]
- Ponger L, Mouchiroud D (2002) CpGProD: identifying CpG islands associated with transcription start sites in large genomic mammalian sequences. Bioinformatics 18: 631–633 [DOI] [PubMed] [Google Scholar]
- Pradhan S, Urwin NA, Jenkins GI, Adams RL (1999) Effect of CWG methylation on expression of plant genes. Biochem J 341: 473–476 [PMC free article] [PubMed] [Google Scholar]
- Praz V, Perier R, Bonnard C, Bucher P (2002) The Eukaryotic Promoter Database, EPD: new entry types and links to gene expression data. Nucleic Acids Res 30: 322–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS (1991) SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. CABIOS 7: 203–206 [DOI] [PubMed] [Google Scholar]
- Prestridge DS (1995) Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol 249: 923–932 [DOI] [PubMed] [Google Scholar]
- Prince VE, Pickett FB (2002) Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet 3: 827–837 [DOI] [PubMed] [Google Scholar]
- Quiros CF, Grellet F, Sadowski J, Suzuki T, Li G, Wroblewski T (2001) Arabidopsis and Brassica comparative genomics: sequence, structure and gene content in the ABI1-Rps2-Ck1 chromosomal segment and related regions. Genetics 157: 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A (1998) CpG methylation, chromatin structure and gene silencing: a three-way connection. EMBO J 17: 4905–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese MG, Hartzell G, Harris NL, Ohler U, Abril JF, Lewis SE (2000) Genome annotation assessment in Drosophila melanogaster. Genome Res 10: 483–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE (2000) Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12: 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC (2002) Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108: 489–500 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang CZ, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR et al. (2000) Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110 [DOI] [PubMed] [Google Scholar]
- Robertson KD (2002) DNA methylation and chromatin: unraveling the tangled web. Oncogene 21: 5361–5379 [DOI] [PubMed] [Google Scholar]
- Robin S, Schbath S (2001) Numerical comparison of several approximations of the word count distribution in random sequences. J Comput Biol 8: 349–359 [DOI] [PubMed] [Google Scholar]
- Rooney JW, Sun YL, Glimcher LH, Hoey T (1995) Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol 15: 6299–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V, Motto M, Pellegrini L (1997) Analysis of the methylation pattern of the maize opaque-2 (O2) promoter and in vitro binding studies indicate that the O2 B-Zip protein and other endosperm factors can bind to methylated target sequences. J Biol Chem 272: 13758–13765 [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM (1998) Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol 16: 939–945 [DOI] [PubMed] [Google Scholar]
- Sagot MF, Myers EW (1998) Identifying satellites and periodic repetitions in biological sequences. J Comput Biol 5: 539–553 [DOI] [PubMed] [Google Scholar]
- Salgado H, Santos-Zavaleta A, Gama-Castro S, Millan-Zarate D, Diaz-Peredo E, Sanchez-Solano F, Perez-Rueda E, Bonavides-Martinez C, Collado-Vides J (2001) RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res 29: 72–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schbath S (1997) An efficient statistic to detect over- and under-represented words in DNA sequences. J Comp Biol 4: 189–192 [DOI] [PubMed] [Google Scholar]
- Schbath S (2000) An overview on the distribution of word counts in Markov chains. J Comput Biol 7: 193–201 [DOI] [PubMed] [Google Scholar]
- Schbath S, Prum B, de Turckheim E (1995) Exceptional motifs in different Markov chain models for a statistical analysis of DNA sequences. J Comput Biol 2: 417–437 [DOI] [PubMed] [Google Scholar]
- Scherf M, Klingenhoff A, Werner T (2000) Highly specific localization of promoter regions in large genomic sequences by PromoterInspector: a novel context analysis approach. J Mol Biol 297: 599–606 [DOI] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sinha S, Tompa M (2000) A statistical method for finding transcription factor binding sites. Proc Int Conf Intell Syst Mol Biol 8: 344–354 [PubMed] [Google Scholar]
- Sinha S, Tompa M (2002) Discovery of novel transcription factor binding sites by statistical overrepresentation. Nucleic Acids Res 30: 5549–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivolob AV, Khrapunov SN (1995) Translational positioning of nucleosomes on DNA: the role of sequence-dependent isotropic DNA bending stiffness. J Mol Biol 247: 918–931 [DOI] [PubMed] [Google Scholar]
- Sorensen MB, Muller M, Skerritt J, Simpson D (1996) Hordein promoter methylation and transcriptional activity in wild-type and mutant barley endosperm. Mol Gen Genet 250: 750–760 [DOI] [PubMed] [Google Scholar]
- Southern EM (2001) DNA microarrays: history and overview. Methods Mol Biol 170: 1–15 [DOI] [PubMed] [Google Scholar]
- Stormo GD (1988) Computer methods for analyzing sequence recognition of nucleic acids. Annu Rev Biophys Biophys Chem 17: 241–263 [DOI] [PubMed] [Google Scholar]
- Stormo GD (1990) Consensus patterns in DNA. Methods Enzymol 183: 211–221 [DOI] [PubMed] [Google Scholar]
- Stormo GD, Hartzell GW, 3rd (1989) Identifying protein-binding sites from unaligned DNA fragments. Proc Natl Acad Sci USA 86: 1183–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1–4 [DOI] [PubMed] [Google Scholar]
- Struhl K (2001) Gene regulation: a paradigm for precision. Science 293: 1054–1055 [DOI] [PubMed] [Google Scholar]
- Sturaro M, Viotti A (2001) Methylation of the Opaque2 box in zein genes is parent-dependent and affects O2 DNA binding activity in vitro. Plant Mol Biol 46: 549–560 [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Nakano S, Yoneyama M, Honda K (1996) Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Res 24: 4501–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J, Bird A (1990) The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci USA 87: 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA (2002) Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA 99: 3740–3745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouzé P, Moreau Y (2001) A higher order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics 17: 1113–1122 [DOI] [PubMed] [Google Scholar]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouzé P, Moreau Y (2002a) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Comput Biol 9: 447–464 [DOI] [PubMed] [Google Scholar]
- Thijs G, Moreau Y, De Smet F, Mathys J, Lescot M, Rombauts S, Rouzé P, De Moor B, Marchal K, Déhais P et al. (2002b) INCLUSive: INtegrated Clustering, Upstream sequence retrieval and motif Sampling. Bioinformatics 18: 331–332 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjian R, Maniatis T (1994) Transcriptional activation: a complex puzzle with a few easy pieces. Cell 77: 5–8 [DOI] [PubMed] [Google Scholar]
- Tompa M (2001) Identifying functional elements by comparative DNA sequence analysis. Genome Res 11: 1143–1144 [DOI] [PubMed] [Google Scholar]
- Travers A, Drew H (1997) DNA recognition and nucleosome organization. Biopolymers 44: 423–433 [DOI] [PubMed] [Google Scholar]
- Tsunoda T, Takagi T (1998) Estimating transcription factor bindability on DNA. Bioinformatics 15: 622–630 [DOI] [PubMed] [Google Scholar]
- Vanet A, Marsan L, Labigne A, Sagot MF (2000) Inferring regulatory elements from a whole genome. An analysis of Helicobacter pylori σ80 family of promoter signals. J Mol Biol 297: 335–353 [DOI] [PubMed] [Google Scholar]
- Vanet A, Marsan L, Sagot MF (1999) Promoter sequences and algorithmical methods for identifying them. Res Microbiol 150: 779–799 [DOI] [PubMed] [Google Scholar]
- van Helden J, Andre B, Collado-Vides J (1998) Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J Mol Biol 281: 827–842 [DOI] [PubMed] [Google Scholar]
- van Helden J, del Olmo M, Perez-Ortin JE (2000a) Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res 28: 1000–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J, Rios AF, Collado-Vides J (2000b) Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res 28: 1808–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H, Fagard M (2001) Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet 17: 29–35 [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA et al (2001) The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23: 4407–4414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waibel AH, Hanazawa T, Hinton GE, Shikano K, Lang KJ (1989) Phoneme recognition using time-delay neural networks. IEEE Trans Acoustic Speech Signal Process 37: 328–339 [Google Scholar]
- Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE (2000) Human-mouse genome comparisons to locate regulatory sites. Nat Genet 26: 225–228 [DOI] [PubMed] [Google Scholar]
- Weber H, Ziechmann C, Graessmann A (1990) In vitro DNA methylation inhibits gene expression in transgenic tobacco. EMBO J 9: 4409–4415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T (2000) Computer-assisted analysis of transcription control regions: Matinspector and other programs. Methods Mol Biol 132: 337–349 [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28: 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24: 238–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfertstetter F, Frech K, Herrmann G, Werner T (1996) Identification of functional elements in unaligned nucleic acid sequences by a novel tuple search algorithm. Comput Appl Biosci 12: 71–80 [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286: 481–486 [DOI] [PubMed] [Google Scholar]
- Workman CT, Stormo GD (2000) ANN-Spec: a method for discovering transcription factor binding sites with improved specificity. Pac Symp Biocomput 467–478 [DOI] [PubMed]
- Zhang MQ (1998) Identification of human gene core promoters in silico. Genome Res 8: 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SH, Lawton MA, Hunter T, Lamb CJ (1994) atpk1, a novel ribosomal protein kinase gene from Arabidopsis: I. Isolation, characterization, and expression. J Biol Chem 269: 17586–17592 [PubMed] [Google Scholar]
- Zhu J, Liu JS, Lawrence CE (1998) Bayesian adaptive sequence alignment algorithms. Bioinformatics 14: 25–39 [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang MQ (1999) SCPD: a promoter database of the yeast Saccharomyces cerevisiae. Bioinformatics 15: 607–611 [DOI] [PubMed] [Google Scholar]