Abstract
The copines are a widely distributed class of calcium-dependent, phospholipid-binding proteins of undetermined biological function. Mutation of the Arabidopsis CPN1 (COPINE 1) gene causes a humidity-sensitive lesion mimic phenotype with increased resistance to a bacterial and an oomyceteous pathogen, constitutive pathogenesis-related gene expression, and an accelerated hypersensitive cell death defense response. Here, we show that the disease resistance phenotype of the cpn1-1 mutant was also temperature sensitive, demonstrate increased CPN1 gene transcript accumulation in wild-type plants under low-humidity conditions, and present a detailed analysis of CPN1 gene transcript accumulation in response to bacterial pathogens. In wild-type plants, CPN1 transcript accumulation was rapidly, locally, and transiently induced by both avirulent and virulent strains of Pseudomonas syringae pv tomato bacteria. However, induction of CPN1 transcript accumulation by avirulent bacteria was much faster and stronger than that induced by virulent bacteria. Bacterial induction of CPN1 transcript accumulation was dependent on a functional type III bacterial protein secretion system. In planta expression of the avrRpt2 avirulence gene was sufficient to trigger rapid CPN1 transcript accumulation. CPN1 transcript accumulation was induced by salicylic acid treatment but was not observed during lesion formation in the lesion mimic mutants lsd1 and lsd5. These results are consistent with CPN1 playing a role in plant disease resistance responses, possibly as a suppressor of defense responses including the hypersensitive cell death defense response. The results also suggest that CPN1 may represent a link between plant disease resistance and plant acclimation to low-humidity and low-temperature conditions.
The copines are a class of highly conserved proteins present in organisms ranging from protozoans to complex forms such as mouse (Mus musculus), human (Homo sapiens), and higher plants (Creutz et al., 1998). These proteins are named copine (the French feminine noun meaning “friend”) because of their tight association with lipid membranes (Creutz et al., 1998). The identifying feature of copine proteins is the unique combination of two protein kinase C conserved 2 (C2) domains in the N-terminal region and a von Willebrand A (VWA) domain in the C-terminal region. The C2 domain is a widely distributed protein motif that often has Ca2+-dependent membrane lipid-binding activity (Xu et al., 1997). C2 domain-containing proteins include protein kinase C (Azzi et al., 1992), phospholipase C (Essen et al., 1996), synaptotagmin (Brose et al., 1995), and rabphilin (Wang et al., 2000). The VWA domain is another widely distributed protein motif that is involved in mediating protein-protein interactions in a range of extracellular and intracellular proteins (Whittaker and Hynes, 2002). Although several biochemical studies of copines have revealed that they have a calcium-dependent phospholipid-binding activity (Creutz et al., 1998; Tomsig and Creutz, 2000), no specific biological functions for any copines have been defined.
Previously, we conducted a genetic screen to identify Arabidopsis mutants with increased resistance to virulent Pseudomonas syringae pv tomato (P. s. t.) bacteria (Jambunathan et al., 2001). One of the mutants identified from our screen was the cpn1-1 mutant, a recessive, T-DNA insertion mutant of the CPN1 gene, which encodes a copine-like protein (GenBank accession nos. AY045764 and AY045765). The cpn1-1 mutant exhibits a strict humidity-dependent lesion mimic phenotype: cpn1-1 plants grown under low-humidity (LH) conditions (35%–45% relative humidity [RH]) are small in size and have curled leaves, minute lesions at the leaf margins, dramatically increased resistance to virulent P. s. t. and Peronospora parasitica strains, and display constitutive PR gene expression. In contrast, cpn1-1 plants grown under high humidity (HH; 75%–85% RH) conditions are morphologically indistinguishable from wild-type (WT) plants and no longer exhibit increased resistance to virulent P. s. t. bacteria. However, both LH- and HH-grown cpn1-1 mutant plants have an accelerated hypersensitive cell death defense response (HR) compared with WT plants after avirulent bacterial inoculation. We hypothesized that the CPN1 gene product could act as a mediator of plant acclimation to LH or, alternatively, as a suppressor of defense-related cell death and defense responses (Jambunathan et al., 2001). Hua et al. (2001) isolated mutants with T-DNA insertions at the same locus, characterized the mutants as temperature-sensitive dwarf plants, and named the corresponding gene BON1 (BONZAI 1). The bon1 mutants exhibited a dwarf phenotype under low-temperature (LT) conditions (22°C) but grew in a manner similar to WT plants when grown under high-temperature (HT; 28°C) conditions. Hua et al. (2001) hypothesized that the BON1 gene product could be a regulator of growth homeostasis under LT conditions.
Environmental conditions such as light intensity, day length, RH, and temperature play key roles in the growth and development of most plants. The ability of the plant to acclimate to environmental stress conditions is essential for normal plant development. For example, loss of plant acclimation to LT in mutants such as asculis1, asculis3, and asculis4 in Arabidopsis leads to a defect in leaf expansion and stem elongation (Tsukaya et al., 1993; Akamatsu et al., 1999). One of the major ways that plants respond to the environment is at the level of gene transcription. In many instances, genes involved in controlling the plant response to a particular stress are induced at the transcriptional level. Members of the cold-responsive transcription factor family CBF/DREB1 are induced within 15 min of cold treatment (Gilmour et al., 1998; Liu et al., 1998), and transgenic plants overexpressing CBF/DREB1 transcription factors exhibit accumulation of solutes with cryoprotective activity and increased freezing tolerance (Gilmour et al., 2000). Several genes known to be key regulators of plant defense responses are transcriptionally regulated by pathogen infection. Important plant defense signaling genes such as NDR1 (NON-RACE-SPECIFIC DISEASE RESISTANCE 1; Century et al., 1997), EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1; Falk et al., 1999), EDS5 (Nawrath et al., 2002), and PAD4 (PHYTOALEXIN DEFICIENT 4; Jirage et al., 1999) are rapidly induced by pathogen attack.
These genes are part of a complex signaling network that allows the plant to recognize and protect itself against pathogens and environmental stress. Plant interactions with pathogens may culminate in either disease susceptibility or resistance in the plant. In the case of resistance, the plant is able to recognize quickly the presence of the pathogen and mount appropriate defense responses. In contrast, during pathogenesis, pathogen recognition by the plant is delayed or nonexistent, and the defense responses are slower, less pronounced, and largely ineffective. Some of the early events after pathogen recognition by the plant include an inward flux of Ca2+ and H+ and an outward flux of K+ and Cl– (Hahlbrock et al., 1995; Levine et al., 1996; Yang et al., 1997; Schaller and Oecking, 1999), the activation of a plasma membrane-associated NADPH-oxidase complex leading to production of reactive oxygen intermediates (Bolwell, 1999), and the initiation of the HR (Klement et al., 1964). These early events, in turn, lead to a state of increased disease resistance in the whole plant known as systemic acquired resistance (SAR), which is marked by high levels of PR gene expression and elevated levels of salicylic acid (SA; Malamy et al., 1990; Uknes et al., 1992).
Mutational analysis has lead to the identification of a number of genes that participate in plant defense signaling. A number of Arabidopsis mutants have been identified that lack the ability to express effective defense responses, including ndr1, eds5, and npr1. NDR1, a small membrane-associated protein, is involved in signal transduction of the coiled-coil, nucleotide-binding, Leu-rich repeat class disease resistance proteins (Century et al., 1995, 1997; Aarts et al., 1998). EDS5, a membrane protein with homology to multidrug and toxin extrusion transporters, is important for the accumulation of SA during defense (Nawrath et al., 2002). Downstream of SA signaling, NPR1 is a novel protein with ankyrin repeats that is important in the induction of pathogenesis-related (PR) genes such as PR1, PR2, and PR5 (Cao et al., 1994, 1997).
At the other extreme are the lesion mimic mutants, which display spontaneous cell death and often develop SAR. Lesion mimic mutants in Arabidopsis include lsd1 to lsd7, acd1, acd2, acd6, cpr5, and cpr22 (Greenberg and Ausubel, 1993; Dietrich et al., 1994; Greenberg et al., 1994; Weymann et al., 1995; Bowling et al., 1997; Dietrich et al., 1997; Rate et al., 1999; Yoshioka et al., 2001). Lesion formation in many lesion mimic mutants is dependent on environmental factors, such as light, day length, and RH (Dietrich et al., 1994; Chamnongpol et al., 1996; Jambunathan et al., 2001; Yoshioka et al., 2001). At least some of these lesion mimic mutants may represent suppressors of plant defenses, including the HR. However, it is possible that many lesion mimic mutants may represent genes that are not necessarily directly involved in plant defense because perturbations of cellular physiology apparently unrelated to disease resistance can result in cell death and SAR (Mock et al., 1999; Molina et al., 1999). In addition, a number of mutants that exhibit constitutive SAR in the absence of spontaneous cell death have also been identified, such as cpr1, cpr6, and mpk4 (Bowling et al., 1994; Clarke et al., 1998; Petersen et al., 2000).
In this report, we extend our previous results by carrying out additional characterization of the cpn1-1 mutant phenotype and analyzing the expression pattern of the CPN1 gene in WT plants in response to temperature, humidity, and pathogen stimuli. Our results indicate that the cpn1-1 lesion mimic phenotype is dependent on both temperature and humidity and that the expression of the CPN1 gene is induced by LH, LT, and pathogen stimulus. Because pathogen-derived signals appeared to be the most effective inducers of CPN1 gene expression, we performed a comprehensive analysis of bacterial pathogen-induced CPN1 gene expression patterns. These results are consistent with the hypothesis that CPN1 is involved in plant disease resistance responses, possibly as a suppressor of plant defense responses.
RESULTS
The cpn1-1 Lesion Mimic, Increased Disease Resistance, and PR Gene Expression Phenotypes Are Temperature Sensitive
We initially identified the cpn1-1 mutant as a humidity-sensitive lesion mimic mutant (Jambunathan et al., 2001). However, mutants in the BON1 gene, which corresponds to CPN1, were identified as temperature-dependent dwarf mutants that had a mutant phenotype when grown at 22°C or lower (Hua et al., 2001). Therefore, we sought to determine whether or not the cpn1-1 lesion mimic phenotype was temperature sensitive and humidity-sensitive. cpn1-1 plants grown at LT (21°C ± 0.5°C) under HH, short-day (SD) conditions (8 h of light/16 h of darkness) developed minute lesions at the leaf margins after 2 to 3 weeks of growth (Fig. 1, B and C). Columbia-0 (Col-0) ecotype WT plants grown under the same conditions did not display any lesions (Fig. 1A). For the first 1 to 2 weeks of growth under LT, HH conditions, the cpn1-1 plants were indistinguishable from the WT plants. With the onset of lesion development after the 2nd week, cpn1-1 plants appeared somewhat stunted compared with WT plants. The lesions appeared consistently in all cpn1-1 plants in the absence of any pathogen. However, the lesion mimic phenotype of cpn1-1 under LT, HH conditions did not appear to be as strong as the phenotype of cpn1-1 grown in LH conditions. The leaves of LT-, HH-grown cpn1-1 plants did not display epinastic curling as severe as that observed for the leaves of cpn1-1 grown under LH, HT (24.5°C ± 0.5°C) conditions (Jambunathan et al., 2001; data not shown). cpn1-1 plants grown under HH, HT conditions were morphologically indistinguishable from the WT plants, with normal leaves and no lesions or dwarfing evident (Jambunathan et al., 2001; data not shown).
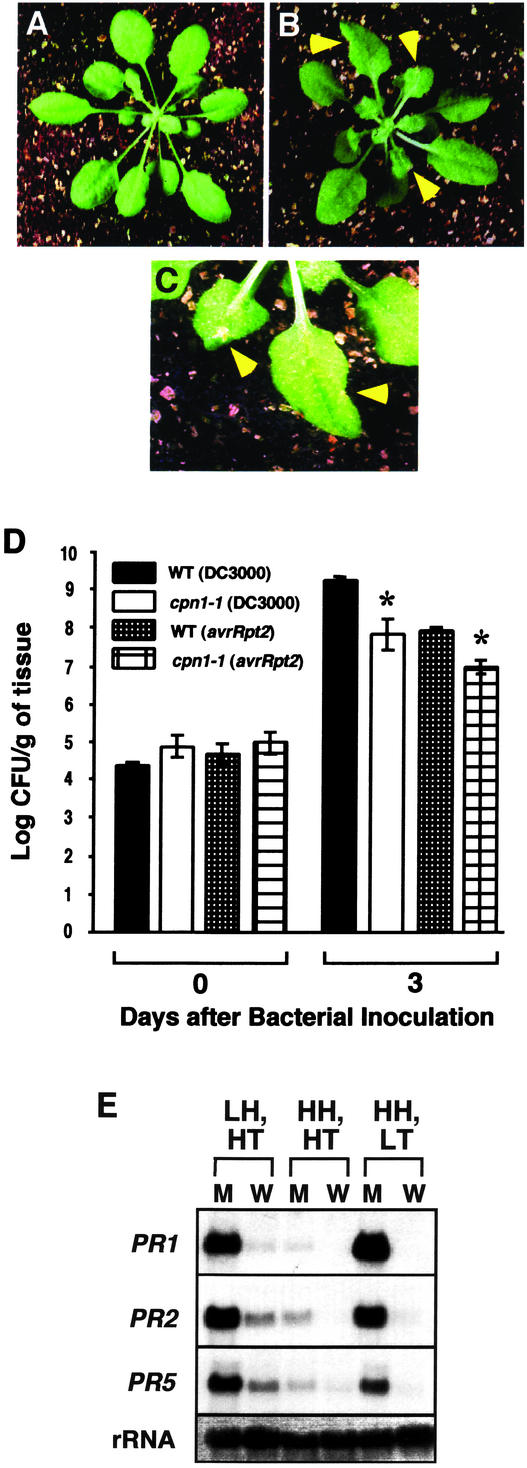
Figure 1.
Temperature-dependent lesion mimic, increased disease resistance, and PR gene expression phenotypes of cpn1-1. A, Five-week-old WT plant grown under LT, HH, SD conditions. B, Five-week-old cpn1-1 mutant plant grown under LT, HH, SD conditions. C, Close-up view of lesions on leaves of a 5-week-old cpn1-1 mutant plant grown under LT, HH, SD conditions. Arrows in B and C indicate lesion locations. D, Growth of virulent P. s. t. DC3000 bacteria and avirulent P. s. t. DC3000 (avrRpt2) bacteria in WT and cpn1-1 plants grown under LT, HH, SD conditions. Plant leaves were infiltrated with a bacterial suspension at a concentration of 1 × 105 colony forming units (cfu) mL–1. Bacterial populations were monitored on d 0 and 3 postinoculation. Bars = se. Asterisks, Significantly lower bacterial populations in cpn1-1 as compared with the corresponding WT plants according to a Student's t test (two-sample Student's t test assuming unequal variances). E, RNA gel-blot analyses of PR1, PR2, and PR5 transcript levels in cpn1-1 mutant (M) and WT (W) plants grown under three different environmental conditions: LH, HT; HH, HT; and HH, LT. rRNA, 28S rRNA stained with methylene blue to show relative amount of RNA in each lane.
Because cpn1-1 mutant plants grown under LH conditions had increased resistance to P. s. t. (Jambunathan et al., 2001), we reasoned that LT-grown cpn1-1 mutant plants might also have increased resistance to P. s. t. bacteria. Growth of virulent P. s. t. strain DC3000 bacteria was monitored in leaves of LT-grown cpn1-1 and WT plants on d 0 and 3 after infiltration with the bacteria. The growth of virulent P. s. t. DC3000 bacteria was reduced by more than 10-fold in LT-grown cpn1-1 mutant plants when compared with LT-grown WT plants (Fig. 1D). In addition, bacterial speck disease symptoms were very weak in LT-grown cpn1-1 plants compared with WT plants (data not shown). The restriction of virulent bacterial growth in LT-grown cpn1-1 was similar to the restriction of the growth of avirulent P. s. t. DC3000 bacteria carrying the avrRpt2 avirulence gene (P. s. t. DC3000 [avrRpt2]) in WT plants (Fig. 1D). This indicates that the level of resistance to virulent P. s. t. bacteria observed in LT-grown cpn1-1 mutant plants was as strong as gene-for-gene resistance in WT plants to avirulent P. s. t. DC3000 (avrRpt2) bacteria mediated by the RPS2 R gene (Whalen et al., 1991; Bent et al., 1994; Mindrinos et al., 1994). The growth of avirulent P. s. t. DC3000 (avrRpt2) bacteria was even more strongly restricted in LT-grown cpn1-1 plants compared with the WT (Fig. 1D). Because the cpn1-1 mutant has a functional RPS2 disease resistance gene (Jambunathan et al., 2001), this indicates that the cpn1-1 mutation has a partially additive effect with the function of RPS2 in mediating resistance to P. s. t. DC3000 (avrRpt2) bacteria.
In previous work (Jambunathan et al., 2001), we observed that LH-grown cpn1-1 plants accumulate high levels of PR gene transcripts. Because LT conditions also triggered the lesion mimic and increased disease resistance phenotypes of cpn1-1, we also examined PR gene transcript accumulation in LT-grown cpn1-1 plants. PR1, PR2, and PR5 gene transcripts accumulated to high levels in uninoculated, LT-grown cpn1-1 plants (Fig. 1E). LT- and LH-grown cpn1-1 plants showed similar patterns of PR1, PR2, and PR5 transcript accumulation. Very low levels of PR1, PR2, and PR5 gene transcript accumulation were seen in HH-, HT-grown cpn1-1 plants. Taken together, these results indicate that the lesion mimic, increased disease resistance, and PR gene expression phenotypes of cpn1-1 are both humidity and temperature sensitive.
Humidity- and Temperature-Dependent CPN1 Transcript Accumulation
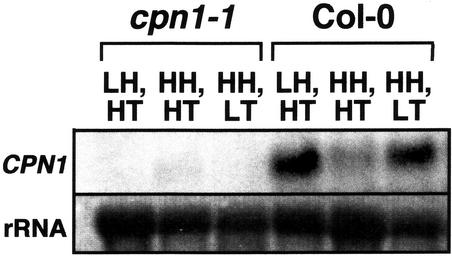
Hua et al. (2001) observed that the BON1 (CPN1) transcript level in WT plants is regulated by temperature conditions. Higher levels of BON1 expression were observed when plants were moved from higher to lower temperature. Because both LH and LT conditions triggered a lesion mimic phenotype in cpn1-1, we tested whether both of these conditions influenced the accumulation of CPN1 transcript in WT plants. WT plants grown under LH, HT or HH, LT conditions showed increased accumulation of CPN1 transcript when compared with WT plants grown under HH, HT conditions (Fig. 2). In cpn1-1 plants, there was no detectable CPN1 transcript under any of the conditions tested. Overall, the CPN1 gene appeared to be expressed at low levels, and the CPN1 transcript was difficult to detect.
Figure 2.
Effects of temperature and humidity on CPN1 transcript accumulation. RNA gel-blot analysis of CPN1 transcript accumulation in WT (Col-0) and cpn1-1 mutant plants grown under three different environmental conditions: LH, HT; HH, HT; and HH, LT. Five-week-old, SD-grown plants were used. rRNA, 28S rRNA stained with methylene blue to show the relative amount of RNA in each lane. For details, see “Materials and Methods.”
The CPN1 Transcript Accumulates in Response to Both Virulent and Avirulent Bacteria
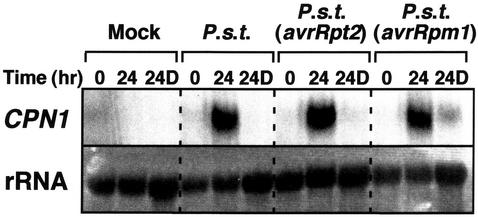
The lesion mimic, accelerated hypersensitive cell death, and increased disease resistance phenotypes of the cpn1-1 mutant suggest that the CPN1 gene may play a role in plant pathogen defense signal transduction, possibly as a repressor of cell death and other defense functions (Jambunathan et al., 2001). Therefore, we speculated that CPN1 gene transcript accumulation might be pathogen regulated. CPN1 transcript accumulation was monitored in LT-, LH-grown WT plants inoculated with virulent and avirulent strains of P. s. t. CPN1 transcript accumulation was monitored both in the inoculated leaves and in distal, uninoculated leaves to test for systemic induction of CPN1 transcript accumulation. In the inoculated leaves, CPN1 transcript accumulation was strongly induced at 24 h after infiltration with virulent P. s. t. DC3000 bacteria and avirulent strains of P. s. t. DC3000 carrying either the avrRpt2 or the avrRpm1 avirulence gene (Fig. 3). In the distal, uninoculated tissues, no induction of CPN1 was detected at 24 h after inoculation with virulent P. s. t. DC3000 bacteria and avirulent P. s. t. DC3000 (avrRpt2) bacteria. However, a very slight induction of CPN1 transcript accumulation in distal tissues was detected 24 h after inoculation with avirulent P. s. t. DC3000 (avrRpm1). Mock inoculation did not induce CPN1 transcript accumulation, indicating that bacteria were required for induction. It should be noted that the induction of CPN1 transcript accumulation by P. s. t. DC3000 bacteria was dramatically higher than that caused by LT and LH because the plants used in these experiments were maintained in LH, LT conditions.
Figure 3.
RNA gel-blot analysis of CPN1 transcript accumulation in WT plants after bacterial inoculation. Five-week-old WT plants were either mock inoculated with 10 mm MgCl2 or inoculated with 1 × 105 cfu mL–1 of one of the following bacterial strains by syringe infiltration of four fully expanded leaves: P. s. t. DC3000, P. s. t. DC3000 (avrRpt2), or P. s. t. DC3000 (avrRpm1). CPN1 transcript accumulation was monitored at 0 and 24 h postinoculation in the inoculated leaves and at 24 h postinoculation in uninoculated, distal leaves. 24D, RNA sample from uninoculated, distal leaves 24 h postinoculation. rRNA, 28S rRNA stained with methylene blue to show relative amount of RNA in each lane.
To determine the timing of CPN1 transcript accumulation in inoculated leaves after bacterial inoculation, a time course experiment was performed. Avirulent P. s. t. DC3000 (avrRpt2) bacteria triggered increased CPN1 transcript accumulation as early as 4 h after inoculation and reached a peak at 6 h (Fig. 4A). The level of CPN1 transcript slowly decreased thereafter, and between 36 and 48 h, it returned to the basal level. The plants used for this experiment were grown under LT, LH conditions; therefore, the induction of CPN1 transcript accumulation by P. s. t. DC3000 (avrRpt2) bacterial inoculation was substantially higher than that induced by LT or LH. Prolonged autoradiographic exposure of the RNA gel blot in this experiment allowed detection of CPN1 transcript in all lanes but resulted in the overexposure of the induced time points. To gauge the rapidity of CPN1 transcript accumulation relative to a known pathogen-inducible gene, we compared the induction time course of CPN1 transcript accumulation with that of PR1. The onset of CPN1 transcript accumulation occurred 5 h earlier than the onset of PR1 expression after inoculation with P. s. t. DC3000 (avrRpt2; Fig. 4A). PR1 transcript accumulation reached a peak at 36 h after inoculation and then decreased but remained elevated until the end of the time course at 72 h. These results indicated that CPN1 transcript accumulated rapidly and transiently after inoculation with avirulent P. s. t. DC3000 (avrRpt2) bacteria.
Figure 4.
Time course of CPN1 transcript accumulation after bacterial inoculation. A, RNA gel-blot analysis of the time course of CPN1 transcript accumulation after inoculation with avirulent P. s. t. DC3000 (avrRpt2) bacteria. Leaves of 5-week-old WT plants were syringe inoculated with 1 × 105 cfu mL–1 of P. s. t. DC3000 (avrRpt2) bacteria, and RNA samples were extracted from inoculated leaves at 0, 2, 4, 6, 9, 12, 24, 36, 48, and 72 h postinoculation. PR1, Same blot reprobed with a PR1 gene-specific probe. Mock, RNA samples from leaves inoculated with 10 mm MgCl2 at 0 and 24 h postinoculation. rRNA, 28S rRNA stained with methylene blue to show relative amounts of RNA in each lane. B, RNA gel-blot analysis of the time course of CPN1 transcript accumulation after inoculation with virulent P. s. t. DC3000 bacteria. This experiment was performed in a manner identical to that described in A.
The time course of CPN1 transcript accumulation in response to inoculation with virulent P. s. t. DC3000 bacteria was also determined (Fig. 4B). The timing of CPN1 transcript accumulation in leaves inoculated with virulent P. s. t. DC3000 was different and slower than that observed after inoculation with avirulent P. s. t. DC3000 (avrRpt2). For these experiments, plants were grown under LH, LT conditions. Induction of CPN1 transcript accumulation was observed at 24 h after inoculation, with peak levels occurring at 36 h postinoculation. By 48 h after inoculation, CPN1 transcript accumulation decreased nearly to the basal level. Severe bacterial speck disease symptoms developed by 72 to 96 h after inoculation. By way of comparison, the timing of PR1 gene transcript accumulation was monitored in leaves inoculated with virulent P. s. t. DC3000. The PR1 gene transcript became detectable at 24 h postinoculation and reached a peak at 36 h after inoculation. The timing of CPN1 gene transcript accumulation after virulent P. s. t. DC3000 inoculation was similar to that of PR1, except that the PR1 transcript remained at elevated levels until the end of the time course at 72 h, whereas CPN1 transcript returned to near basal level at 48 h postinoculation (Fig. 4B).
A Functional Bacterial Type III Protein Secretion System Is Required for Bacterial Induction of CPN1 Transcript Accumulation
Many plant and animal pathogenic bacteria, including P. s. t., use the type III protein secretion pathway to deliver some of their proteins into the host cell during pathogenesis (for review, see Hueck, 1998). In the case of P. syringae, the type III protein secretion system is required both for virulence on compatible host plants and for the elicitation of the HR on incompatible host plants (He et al., 1993; Alfano and Collmer, 1997). The bacterial genes required for pathogenicity in susceptible plants and HR in resistant plants have been defined as hrp (hypersensitive response and pathogenicity) genes (Lindgren et al., 1986). Hrp genes that are highly structurally conserved across bacterial species are called hrc (hypersensitive response and conserved; Bogdanove et al., 1996). A mutation in any of the hrp genes disables the type III protein secretion system of the bacteria and renders them unable to cause disease or elicit an HR.
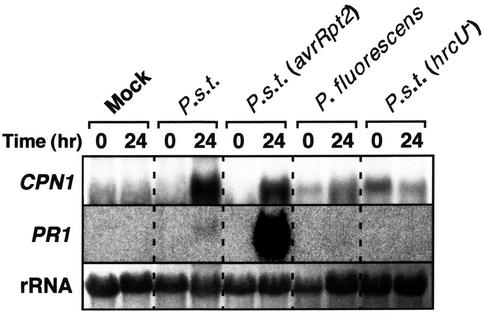
To test whether induction of CPN1 transcript accumulation by inoculation with P. s. t. strains depended on the type III protein secretion system, we monitored CPN1 transcript accumulation in WT plants inoculated with bacterial strains that are nonpathogenic and/or defective in type III secretion (Fig. 5). No induction of CPN1 transcript accumulation above basal levels was observed in leaves of WT plants 24 h after infiltration with an hrcU– mutant strain of P. s. t. DC3000 (P. s. t. DC3000 [hrcU–]). The P. s. t. DC3000 (hrcU–) mutant strain is defective in type III protein secretion and is unable to cause disease in compatible hosts or elicit defense responses, including the HR, in incompatible host plants (Mudgett and Staskawicz, 1999). Also, no induction of CPN1 transcript accumulation above basal levels was observed in leaves of WT plants 24 h after infiltration with nonpathogenic P. fluorescens bacteria. As expected, virulent P. s. t. DC3000 and avirulent P. s. t. DC3000 (avrRpt2) bacteria, which have an intact type III protein secretion system, were able to induce CPN1 transcript accumulation in inoculated leaves at 24 h after infiltration (Fig. 5). For comparison, we also examined the induction of PR1 gene transcript accumulation under the same inoculation conditions. PR1 gene transcript accumulated to high levels in leaves inoculated with avirulent P. s. t. DC3000 (avrRpt2) bacteria and to low levels in leaves inoculated with virulent P. s. t. DC3000 bacteria at 24 h after inoculation. Taken together, these data indicate that CPN1 transcript accumulation was induced specifically by pathogenic bacteria and that this induction required a functional bacterial type III protein secretion system.
Figure 5.
Specific induction of CPN1 transcript accumulation by pathogenic bacteria. RNA gel-blot analysis of CPN1 transcript accumulation after inoculation with pathogenic and nonpathogenic bacterial strains. Leaves of 5-week-old WT plants were either mock inoculated with 10 mm MgCl2 or inoculated with 1 × 105 cfu mL–1 of one of the following bacterial strains by syringe infiltration: virulent P. s. t. DC3000, avirulent P. s. t. DC3000 (avrRpt2), nonpathogenic Pseudomonas fluorescens, or the nonpathogenic mutant P. s. t. DC3000 (hrcU–). RNA samples were extracted from inoculated leaves at 0 and 24 h postinoculation. PR1, Same blot reprobed with a PR1 gene-specific probe. rRNA, 28S rRNA visualized with methylene blue to show relative amounts of RNA in each lane.
In Planta Expression of an Avirulence Gene Is Sufficient to Induce CPN1 Transcript Accumulation
Although CPN1 transcript accumulation occurred in response to both virulent and avirulent bacterial inoculation, the induction was most dramatic with avirulent P. s. t. DC3000 (avrRpt2). This suggests that the stronger and more rapid induction of CPN1 transcript accumulation by P. s. t. DC3000 (avrRpt2) was due to gene-for-gene recognition of the avrRpt2 determinant by the corresponding RPS2 R gene product in the host (Leister et al., 1996). To determine whether RPS2-mediated recognition of AvrRpt2 was sufficient for induction of CPN1 transcript accumulation, we tested whether or not glucocorticoid-inducible expression of avrRpt2 in transgenic plants was sufficient to induce CPN1 transcript accumulation. For this experiment, we used stable transgenic Arabidopsis lines bearing a glucocorticoid-inducible avrRpt2 gene in either the Col-0 WT genetic background having a functional RPS2 gene or in the rps2-101C mutant genetic background, which lacks a functional RPS2 gene (McNellis et al., 1998). Infiltration of leaves of these transgenic plants with dexamethasone (DEX), a strong, synthetic glucocorticoid, induces the expression of the avrRpt2 transgene. In the RPS2 (WT) genetic background, glucocorticoid-induced avrRpt2 expression triggers hypersensitive cell death within 12 to 24 h due to RPS2-mediated gene-for-gene recognition of the avrRpt2-encoded avirulence determinant. In the rps2-101C mutant, no hypersensitive cell death occurs because gene-for-gene recognition of the avrRpt2 avirulence determinant does not take place.
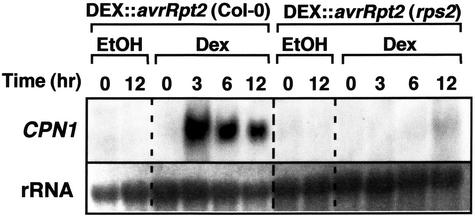
Transgenic plants with the glucocorticoid-inducible avrRpt2 gene in the RPS2 genetic background exhibited strong induction of CPN1 transcript accumulation as early as 3 h after DEX infiltration (Fig. 6). The levels of CPN1 transcript decreased slowly after a peak at 3 until 12 h postinfiltration, at which time the leaves showed near complete collapse due to hypersensitive cell death. Transgenic plants with the glucocorticoid-inducible avrRpt2 transgene in the rps2-101C mutant genetic background did not exhibit induction of CPN1 transcript accumulation after DEX infiltration during the time frame tested (Fig. 6). These results show that in planta expression of the avrRpt2 avirulence gene was sufficient to stimulate CPN1 transcript accumulation and that this effect depended on the presence of a functional RPS2 gene. This result indicates that RPS2-mediated gene-for-gene recognition of the avrRpt2-derived avirulence determinant is sufficient to trigger CPN1 transcript accumulation.
Figure 6.
CPN1 transcript accumulation in response to in planta expression of avrRpt2. RNA gel-blot analysis of CPN1 transcript accumulation in transgenic plants bearing a glucocorticoid-inducible avrRpt2 avirulence gene over time after glucocorticoid treatment. DEX::avrRpt2 (Col-0), Transgenic line bearing the glucocorticoid-inducible avrRpt2 gene in the Col-0 WT genetic background; DEX::avrRpt2 (rps2), glucocorticoid-inducible avrRpt2 gene in the rps2-101C mutant genetic background. Leaves of both plant lines were infiltrated with either 30 μm DEX in 0.1% (v/v) ethanol (DEX) or 0.1% (v/v) ethanol (EtOH) as a control. RNA samples were extracted from DEX-infiltrated leaves at 0, 3, 6, and 12 h postinfiltration and from ethanol-infiltrated leaves at 0 and 12 h postinoculation. rRNA, 28S rRNA visualized by methylene blue staining to show relative amounts of RNA in each lane.
SA Stimulates CPN1 Transcript Accumulation
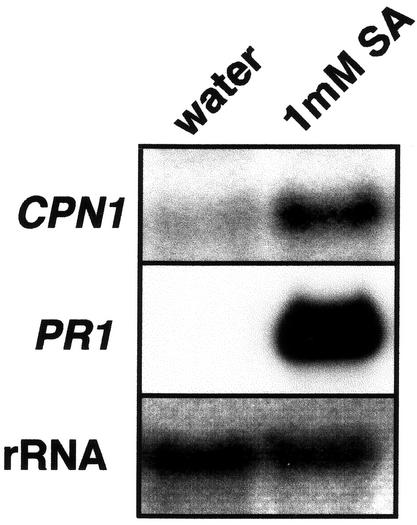
SA is a key chemical inducer of plant defense responses and is required for the development of SAR (Gaffney et al., 1993). Biochemical and genetic data suggest that SA can potentiate defense responses by promoting cell death (Weymann et al., 1995). SA treatment of plants can also induce SAR and the expression of PR genes (Ward et al., 1991). To test whether SA treatment can induce the accumulation of the CPN1 gene transcript, WT Arabidopsis plants were sprayed with 1 mm SA, and the accumulation of CPN1 transcript was monitored 24 h later. SA treatment induced substantial accumulation of CPN1 transcript relative to the water-treated control plants at 24 h after treatment (Fig. 7). High-level accumulation of PR1 transcript was also observed in SA-treated plants but not in the water-treated controls, as expected. These results indicate that accumulation of CPN1 gene transcript can be induced by SA.
Figure 7.
SA induction of CPN1 transcript accumulation. RNA gel-blot analysis of CPN1 gene transcript accumulation in 5-week-old WT plants 24 h after treatment with either 1 mm SA or water. PR1, Same blot reprobed with a PR1 gene-specific probe. rRNA, 28S rRNA visualized with methylene blue to show relative amounts of RNA in each lane.
CPN1 Transcript Accumulation in Other Lesion Mimic Mutants
Conditional lesion mimic mutants such as lsd1 and lsd5 are sensitive to day length conditions. lsd1 has a non-lesion phenotype under permissive, SD conditions. Spreading cell death in the lsd1 mutant can be initiated by shifting the plants from SD to long-day (LD; 16 h of light/8 h of dark) conditions (Dietrich et al., 1994; Jabs et al., 1996). lsd5, another conditional mutant, initiates spreading cell death under SD conditions, but has a non-lesion mimic phenotype under permissive, LD conditions (Dietrich et al., 1994; Morel and Dangl, 1999). We speculated that the day length-induced cell death in lsd1 and lsd5 might trigger CPN1 transcript accumulation. However, no induction of CPN1 transcript accumulation was observed in lsd1 or lsd5 mutant plants grown under permissive light conditions or after the mutants were shifted to nonpermissive (lesion-inducing) conditions (Fig. 8). In contrast, PR1 transcript accumulation was strongly induced in both mutants when grown under conditions that favored lesion formation (Fig. 8). The lsd1 mutant plants showed substantial PR1 transcript accumulation even under permissive, SD conditions.
Figure 8.
RNA gel-blot analysis of CPN1 transcript accumulation in the lsd1 and lsd5 lesion mimic mutants. CPN1 transcript accumulation was monitored in 4-week-old lsd1 plants grown continuously in permissive SD conditions and 48 h after transfer from SD conditions to nonpermissive, LD conditions, at which point spreading lesions were observed. Similarly, CPN1 transcript accumulation was monitored in 3-week-old lsd5 mutant plants grown continuously under permissive LD conditions and 48 h after transfer to nonpermissive, SD conditions that triggered cell death and visible lesion formation. RNA samples from WT plants 0 and 12 h after inoculation with 1 × 105 cfu mL–1 of P. s. t. (avrRpt2) served as a positive control. PR1, Same blot reprobed with a PR1 gene-specific probe. rRNA, 28S rRNA visualized with methylene blue to show relative amounts of RNA in each lane.
DISCUSSION
The results presented herein support both of our current working hypotheses regarding the potential function of the CPN1 gene product: that CPN1 may be a mediator of plant acclimation to LH and LT conditions and a negative regulator of defense-related cell death and other defense responses. The recessive, temperature- and humidity-dependent lesion mimic phenotype of the cpn1-1 mutant implies that the CPN1 gene product is required to prevent damaging effects of or suppress inappropriate responses to LT and LH. The increased accumulation of CPN1 transcript under LH and LT conditions supports the potential role for CPN1 as a mediator of acclimation to LT and LH: Increased levels of the CPN1 protein may be required to deal with the stresses of LT and LH conditions. As a Ca2+-dependent membrane-associated protein, CPN1 may be involved in maintaining cellular homeostasis under LT and LH conditions by regulating some aspect of membrane trafficking (Hua et al., 2001; Jambunathan et al., 2001). It is interesting to note, however, that the LT conditions (22°C) that trigger the cpn1-1 lesion mimic phenotype are considered optimal for the growth of Arabidopsis, and researchers routinely grow their Arabidopsis plants at this temperature.
The rapid, specific, and tightly regulated accumulation of CPN1 gene transcript in response to pathogen signals implies a direct role for CPN1 in plant defense. This finding makes it unlikely that the effects of the cpn1-1 mutation on plant defense responses are simply due to perturbations of plant cell homeostasis unrelated to plant defense signaling. The relatively rapid and high level of CPN1 transcript accumulation after inoculation with avirulent P. s. t. DC3000 (avrRpt2) bacteria as compared with that observed with virulent P. s. t. DC3000 bacteria (Fig. 4) indicated that RPS2-mediated recognition of the avrRpt2 signal triggered the rapid accumulation of CPN1 transcript. It is not unusual for pathogen-induced genes to be induced by both virulent and avirulent pathogens, although induction by avirulent pathogens is generally much stronger and more rapid than that by virulent pathogens, as observed with both CPN1 and PR1 (Fig. 4). Additional evidence for the induction of CPN1 transcript accumulation via gene-for-gene-mediated pathogen recognition came from the induction of CPN1 transcript accumulation by glucocorticoid-inducible expression of avrRpt2 in transgenic plants having a functional RPS2 gene (Fig. 6). The fact that avrRpt2 expression in planta could specifically trigger CPN1 transcript accumulation, in the absence of any pathogen inoculation, and that this induction required the presence of a functional RPS2 disease resistance gene, suggests that CPN1 transcript accumulation is responsive to gene-for-gene-mediated signaling.
The dependence of bacterial induction of CPN1 transcript accumulation on a functional type III protein secretion system also supports a specific role for the CPN1 gene product in plant responses to pathogens (Fig. 5). The lack of CPN1 transcript accumulation after inoculation with P. fluorescens, a nonpathogenic strain related to P. syringae, implies that CPN1 transcript accumulation is specifically triggered by pathogenic bacteria. The lack of CPN1 transcript accumulation after inoculation with the P. s. t. DC3000 (hrcU–) mutant, which is defective in type III protein secretion, indicates that a functional type III protein secretion system is specifically required for induction of CPN1 transcript accumulation by P. s. t. DC3000 bacteria (Mudgett and Staskawicz, 1999). This also supports the conclusion that pathogenic bacteria specifically stimulate CPN1 transcript accumulation because the P. s. t. DC3000 (hrcU–) mutant is nonpathogenic (Mudgett and Staskawicz, 1999).
Our results also suggest that CPN1 may be involved in early steps of plant defense: Induction of CPN1 transcript accumulation was observed within 4 h after inoculation, which was substantially faster than PR1 gene transcript accumulation (Fig. 4). The transient nature of CPN1 transcript accumulation implies that the role of the CPN1 gene product in defense may be restricted to early steps. The local rather than systemic induction of CPN1 transcript accumulation is also consistent with a role for CPN1 as a suppressor of hypersensitive cell death because in that case, CPN1 activity might be needed primarily near the site of infection rather than systemically.
It is interesting to note that although SA treatment could induce CPN1 transcript accumulation in WT plants, the induction of cell death in the lesion mimic mutants lsd1 and lsd5 did not stimulate CPN1 expression, even though, in lsd1 at least, runaway cell death requires SA accumulation (Aviv et al., 2002). This could indicate that CPN1 is not involved in the cell death signaling pathways defined by lsd1 and lsd5, or, perhaps, induction of CPN1 expression in the mutants occurred in a transient manner but was overlooked due to the time intervals used. Alternatively, if CPN1 is a necessary repressor of the HR, then the lack of expression of CPN1 in the lsd1 and lsd5 mutants may represent part of the defect of these mutants, and the lack of CPN1 transcript accumulation might actually contribute to the lesion mimic phenotype of these mutants.
Taken together, the increased disease resistance, lesion mimic, and accelerated HR phenotypes of the cpn1-1 mutant and the gene expression patterns of the CPN1 gene suggest that the CPN1 gene product may function as a negative regulator of plant defense responses, including the HR. The strong, rapid, and specific activation of CPN1 gene transcript accumulation in response to pathogen inoculation implies that plant defense functions could represent a primary role of CPN1. However, the temperature- and humidity-related aspects of the cpn1-1 mutant phenotype and the activation of CPN1 transcript accumulation by these same environmental parameters adds another level of complexity to the biological role of CPN1. Apparently, CPN1 plays a nonredundant role as a suppressor of potentially cell death-inducing effects of LT and LH environmental conditions.
Humidity and temperature play important roles in plant disease development (Agrios, 1997), and overlaps between environmental and pathogen signaling are not unusual in plants. In the Arabidopsis lesion mimic mutants lsd6 and cpr22, HH has been found to suppress the spontaneous lesion phenotype (Weymann et al., 1995; Yoshioka et al., 2001). Plants treated with avirulent pathogen and grown under HH conditions have been found to have a delayed HR with reduced SA levels (Hammond-Kosack et al., 1996). Although the mode of action of HH in modifying plant defense responses is not clear, these observations suggest that HH has the potential to suppress the HR- and SA-dependent defenses in plants. Similarly, a range of factors including LT, LH, hyperosmolarity, wounding, and harpin elicitors have been found to activate ATMPK4 and ATMPK6 rapidly in Arabidopsis (Ichimura et al., 2000; Desikan et al., 2001). In addition, phenotypic analysis of the mpk4 mutant has revealed that ATMPK4 may serve as a negative regulator of SAR (Petersen et al., 2000). Also, the EDS5 gene is activated by UV light and pathogens (Nawrath et al., 2002). These findings implicate a connection between abiotic and biotic stress signaling. This connection could provide a molecular basis for the phenomenon of cross tolerance in plants, in which a plant subjected to one stress, such as UV light or ozone, for example, can become more resistant to pathogens (Yalpani et al., 1994; Sharma et al., 1996; Bowler and Fluhr, 2000).
But what could account for the apparent involvement of CPN1 in plant responses to both biotic and abiotic stimuli? It is possible that the answer could be related to Ca2+. Ca2+ is a ubiquitous second messenger that is involved in plant responses to diverse stimuli, such as drought, touch, cold, heat, and oxidative stress (for review, see Knight, 2000; Reddy, 2001). Ca2+ fluxes are involved in defense signaling in plants (Zimmermann et al., 1997; Grant et al., 2000). Because Ca2+ is such a nonspecific signaling molecule that is involved in many different types of signaling pathways, the specificity of Ca2+ signaling must be accomplished by the timing, duration, and location of Ca2+ fluxes (McAinsh and Hetherington, 1998; Bowler and Fluhr, 2000). It is possible that CPN1, as a Ca2+-dependent membrane-associated protein, is involved in determining the specificity of Ca2+ signaling and preventing inappropriate defense responses to LT and LH conditions. The mechanism of action of the CPN1 protein is unknown, but it has been hypothesized that copines may function by recruiting proteins with which they interact via their VWA domain to a membrane location (Tomsig et al., 2003).
MATERIALS AND METHODS
All experiments described were replicated independently at least two to four times with similar results.
Plant Growth Conditions
All plants were grown in a soil-less potting mix (Scotts Redi-earth Plug and Seedling Mix, E.C. Geiger, Inc., Harleysville, PA) and irrigated with distilled water. Plants grown for cpn1-1 mutant phenotypic analysis and for analysis of humidity and temperature dependence of CPN1 transcript accumulation were grown under SD conditions with a light intensity of 75 to 100 μmol m–2 s–1, whereas temperature and humidity parameters were varied as described in “Results.” Plants used for analysis of bacterial induction of CPN1 gene expression, including WT, cpn1-1, and DEX:: avrRpt2 plants, were grown under LT, LH, SD conditions and 60 to 70 μmol m–2 s–1 light intensity. lsd1 plants were grown under LT, LH, SD conditions and 60 to 70 μmol m–2 s–1 light intensity for 4 weeks and then moved to LT, LH, LD conditions and 75 to 100 μmol m–2 s–1 light intensity. lsd5 mutant plants were grown under LT, LH, LD and 75 to 100 μmol m–2 s–1 light intensity conditions for 3 weeks and then moved to LT, LH, SD conditions and 60 to 70 μmol m–2 s–1 light intensity.
In Planta Bacterial Growth Analyses
These assays were performed by infiltration inoculation as described previously, except that bacterial populations were assayed at 0 and 3 d after inoculation. The bacterial strains used were the same as described previously (Jambunathan et al., 2001).
Bacterial Induction of CPN1 Expression
The Pseudomonas syringae pv tomato (P. s. t.) DC3000 and P. s. t. DC3000 (avrRpt2) bacterial strains were the same as used previously. P. s. t. DC3000 (avrRpm1) bacteria carried the plasmid pVARM (Kunkel et al., 1993). The hrcU– mutant carries a Tn3gus transposon insertion into the hrcU gene (Mudgett and Staskawicz, 1999). All the strains were grown at 28°C on Pseudomonas agar F (Sigma, St. Louis) media supplemented with 100 μg mL–1 rifampicin and 25 μg mL–1 kanamycin (Sigma). The Pseudomonas fluorescens strain was grown at 28°C on Pseudomonas agar F supplemented with 20 μg mL–1 nalidixic acid (Sigma). Leaves of 5-week-old plants were syringe inoculated with bacteria suspended in 10 mm MgCl2 at a concentration of 1 × 106 cfu mL–1. After inoculation, the plants were maintained under SD, LH, LT conditions and 60 to 70 μmol m–2 s–1 light intensity. DEX treatments were performed by syringe infiltration of DEX (Sigma) as described previously (McNellis et al., 1998).
SA Treatments
Five-week-old plants were sprayed to the point of runoff with 1 mm SA (Sigma) in water with 0.025% (v/v) Silwet l-77 surfactant (Lehle Seeds, Round Rock, TX). Control plants were sprayed with water containing 0.025% (v/v) Silwet l-77. The plants were left covered with a dome for 4 h to maintain HH, after which the dome was removed. Tissues were harvested 24 h after treatment.
RNA Isolation and RNA Gel-Blot Analyses
Leaf tissue was collected from treated plants and flash frozen in liquid nitrogen. The permissive condition RNA sample for the lsd1 mutant was obtained from leaf tissue collected from 4-week-old lsd1 mutant plants grown under continuous SD conditions. The nonpermissive condition RNA sample for the lsd1 mutant was obtained from lsd1 mutant plants grown under SD conditions for 4 weeks and then moved to LD conditions for 48 h, at which time spreading lesions were observed. The permissive condition RNA samples for the lsd5 mutant were obtained from leaf tissues collected from 4-week-old lsd5 mutant plants grown under LD conditions. The nonpermissive condition RNA samples for the lsd5 mutant were obtained from lsd5 mutant plants grown under LD conditions for 3 weeks and then moved to SD conditions for 48 h, at which time lesion formation was evident. RNA extractions and RNA gel blotting were performed as described previously, except that 15 μg of total RNA was loaded in each gel lane in all gel-blot experiments (Jambunathan et al., 2001). The PR1, PR2, and PR5 gene probes were the same as described previously (Jambunathan et al., 2001). The CPN1 gene-specific probe consisted of a 738-bp fragment corresponding to amino acids 332 to 578 of the predicted CPN1 protein. This probe allowed the specific detection of CPN1 transcript without hybridization to transcripts from other, highly homologous copine genes. The probe was generated by PCR amplification using a CPN1 cDNA clone plasmid as the template and the following primers: forward primer, 5′-TCTAGAGTACTTGGCATCTGGA-3′; and reverse primer, 5′-GAATTCTCATGGAGGAATCGGTTTCAT-3′; with annealing at 55°C. The primers contain engineered XbaI and EcoRI restriction sites, respectively (in italics); CPN1-homologous sequences are in roman. The PCR-amplified product was cloned into the pCR-Blunt vector using the Zero Blunt PCR cloning kit according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). The cloned product was released using XbaI and EcoRI restriction enzymes (New England Biolabs, Beverly, MA). The released product was resolved on an agarose gel, extracted, and purified using the Qiaex II Gel Extraction Kit (Qiagen, Valencia, CA). About 25 to 50 ng of the cleaned product was used for probe labeling with dCTP 32P using the Redi-prime labeling kit (Amersham-Biotech, Piscataway, NJ) according to the manufacturer's instructions. The labeled probe was cleaned using Performa spin columns (Edge Biosystems, Gaithersburg, MD) according to the manufacturer's instructions. Northern hybridization was performed at 42°C using Ultrahyb hybridization buffer as recommended by the manufacturers (Ambion, Austin, TX). Blot washing and exposure were performed as described previously (McNellis et al., 1998).
Acknowledgments
We thank Jeffery Dangl for the lsd1 and lsd5 seeds; Brian Staskawicz for the ndr1-1, eds5-1, and npr1-2 seeds; Ramesh Raina for the PR1, PR2, and PR5 gene probes; and Brian Staskawicz for all of the P. s. t. strains and the P. fluorescens strain. We thank Seogchan Kang and Ramamurthy Mahalingam for technical assistance. We thank Seogchan Kang, C. Peter Romaine, and four anonymous reviewers for their critical comments on the manuscript. We thank Philip Jensen, Judith Sinn, S. Kang, Tzuu-fen Lee, Jianxin Liu, Justin Dillon, and Andrew Stephenson for many helpful discussions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.022970.
This work was supported by the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service grant program (grant no. 2002–35319–11561 to T.W.M.).
References
- Aarts N, Metz M, Holub E, Staskawicz BJ, Daniels MJ, Parker JE (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 95: 10306–10311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios GN (1997) Plant Pathology, Ed 4. Academic Press, London
- Akamatsu T, Hanzawa Y, Ohtake Y, Takahashi T, Nishitani K, Komeda Y (1999) Expression of endoxyloglucan transferase genes in acaulis mutants of Arabidopsis. Plant Physiol 121: 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfano JR, Collmer A (1997) The type III (hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, avr proteins, and death. J Bacteriol 179: 5655–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv DH, Rustérucci C, Holt IIIBF, Dietrich RA, Parker JE, Dangl JL (2002) Runaway cell death, but not basal disease resistance, in lsd1 is SA- and NIM1/NPR1-dependent. Plant J 29: 381–391 [DOI] [PubMed] [Google Scholar]
- Azzi A, Boscoboinik D, Hensey C (1992) The protein kinase C family. Eur J Biochem 208: 547–557 [DOI] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265: 1856–1860 [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Beer SV, Bonas U, Boucher CA, Collmer A, Coplin DL, Cornelis GR, Huang HC, Hutcheson SW, Panopoulos NJ et al. (1996) Unified nomenclature for broadly conserved hrp genes of pathogenic bacteria. Mol Microbiol 20: 681–683 [DOI] [PubMed] [Google Scholar]
- Bolwell GP (1999) Role of active oxygen species and NO in plant defense responses. Curr Opin Plant Biol 2: 287–294 [DOI] [PubMed] [Google Scholar]
- Bowler C, Fluhr R (2000) The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci 5: 241–246 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X (1997) The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9: 1573–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6: 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brose N, Hofmann K, Hata Y, Südhof TC (1995) Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem 270: 25273–25280 [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Century KS, Holub EB, Staskawicz BJ (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance both to a bacterial and a fungal pathogen. Proc Natl Acad Sci USA 92: 6597–6601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ (1997) NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science 278: 1963–1965 [DOI] [PubMed] [Google Scholar]
- Chamnongpol S, Willekens H, Langebartels C, Van Montagu M, Inzé D, Van Camp W (1996) Transgenic tobacco with a reduced catalase activity develops necrotic lesions and induces pathogenesis-related expression under high light. Plant J 10: 491–503 [Google Scholar]
- Clarke JD, Liu Y, Klessig DF, Dong X (1998) Uncoupling PR gene expression from NPR1 and bacterial resistance: characterization of the dominant Arabidopsis cpr6–1 mutant. Plant Cell 10: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creutz CE, Tomsig JL, Snyder SL, Gautier MC, Skouri F, Beisson J, Cohen J (1998) The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J Biol Chem 273: 1393–1402 [DOI] [PubMed] [Google Scholar]
- Desikan R, Hancock JT, Ichimura K, Shinozaki K, Neill SJ (2001) Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol 126: 1579–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RA, Delaney TP, Uknes SJ, Ward ER, Ryals JA, Dangl JL (1994) Arabidopsis mutants simulating disease resistance responses. Cell 77: 565–577 [DOI] [PubMed] [Google Scholar]
- Dietrich RA, Richberg MH, Schmidt R, Dean C, Dangl JL (1997) A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of cell death. Cell 88: 685–694 [DOI] [PubMed] [Google Scholar]
- Essen LO, Perisic O, Cheung R, Katan M, Williams RL (1996) Crystal structure of a mammalian phosphoinositide-specific phospholipase Cδ. Nature 380: 595–602 [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE (1999) EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proc Natl Acad Sci USA 96: 3292–3297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friederich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Grant M, Brown I, Adams S, Knight M, Ainslie A, Mansfield J (2000) The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J 23: 441–450 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Ausubel FM (1993) Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J 4: 327–341 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM (1994) Programmed cell death in plants: a pathogen-triggered response activated coordinately with multiple defense functions. Cell 77: 551–563 [DOI] [PubMed] [Google Scholar]
- Hahlbrock K, Scheel D, Logemann E, Numberger T, Parniske M, Reinold S, Sacks WR, Schmelzer E (1995) Oligopeptide elicitor-mediated defense gene activation in cultured parsley cells. Proc Natl Acad Sci USA 92: 4150–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Silverman P, Raskin I, Jones JDG (1996) Race-specific elicitors of Cladosporium fulvum induce changes in cell morphology and the synthesis of ethylene and salicylic acid in tomato plants carrying the corresponding Cf disease resistance gene. Plant Physiol 110: 1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He SY, Huang HC, Collmer A (1993) Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the hrp pathway and elicits the hypersensitive response in plants. Cell 73: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev 15: 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24: 655–665 [DOI] [PubMed] [Google Scholar]
- Jabs T, Dietrich RA, Dangl JL (1996) Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Jambunathan N, Siani JM, McNellis TW (2001) A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell 13: 2225–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J (1999) Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc Natl Acad Sci USA 96: 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klement Z, Farkas GL, Lovrekovich L (1964) Hypersensitive reaction induced by phytopathogenic bacteria in the tobacco leaf. Phytopathology 54: 474–477 [Google Scholar]
- Knight H (2000) Calcium signaling during abiotic stress in plants. Int Rev Cytol 195: 269–324 [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ (1993) RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. Plant Cell 5: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Ausubel FM, Katagiri F (1996) Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci 93: 15497–15502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Pennell RI, Alvarez ME, Palmer R, Lamb C (1996) Calcium-mediated apoptosis in a plant hypersensitive disease resistance response. Curr Biol 6: 427–437 [DOI] [PubMed] [Google Scholar]
- Lindgren PB, Peet RC, Panopoulos NJ (1986) Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J Bacteriol 168: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy J, Carr JP, Klessig DF, Raskin I (1990) Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250: 1002–1004 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Hetherington AM (1998) Encoding specificity in Ca2+ signaling systems. Trends Plant Sci 3: 32–36 [Google Scholar]
- McNellis TW, Mudgett MB, Li K, Aoyama T, Horvath D, Chua NH, Staskawicz BJ (1998) Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis induces hypersensitive cell death. Plant J 14: 247–257 [DOI] [PubMed] [Google Scholar]
- Mindrinos M, Katagiri F, Yu GL, Ausubel FM (1994) The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089–1099 [DOI] [PubMed] [Google Scholar]
- Mock HP, Heller W, Molina A, Neubohn B, Sandermann H, Grimm B (1999) Expression of uroporphyrinogen decarboxylase or coproporphyrinogen oxidase antisense RNA in tobacco induces pathogen defense responses conferring increased resistance to tobacco mosaic virus. J Biol Chem 274: 4231–4238 [DOI] [PubMed] [Google Scholar]
- Molina A, Volrath S, Guyer D, Maleck K, Ryals J, Ward E (1999) Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesionmimic phenotype that induces systemic acquired resistance. Plant J 17: 667–678 [DOI] [PubMed] [Google Scholar]
- Morel JB, Dangl JL (1999) Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 151: 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett MB, Staskawicz BJ (1999) Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol Microbiol 32: 927–941 [DOI] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Métraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen M, Brodersen P, Naested H, Andreasson E, Lindhart U, Johansen B, Nielsen HB, Lacy M, Austin MJ, Parker JE et al. (2000) Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103: 1111–1120 [DOI] [PubMed] [Google Scholar]
- Rate DN, Cuenca JV, Bowman GR, Guttman DS, Greenberg JT (1999) The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN (2001) Calcium: silver bullet in signaling. Plant Sci 160: 381–404 [DOI] [PubMed] [Google Scholar]
- Schaller A, Oecking C (1999) Modulation of plasma membrane H+-ATPase activity differentially activates wound and pathogen defense responses in tomato plants. Plant Cell 11: 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma YK, León J, Raskin I, Davis KR (1996) Ozone-induced responses in Arabidopsis thaliana: the role of salicylic acid in the accumulation of defense-related transcripts and induced resistance. Proc Natl Acad Sci USA 92: 5099–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsig JL, Creutz CE (2000) Biochemical characterization of copine: a ubiquitous Ca2+-dependent, phospholipid-binding protein. Biochemistry 39: 16163–16175 [DOI] [PubMed] [Google Scholar]
- Tomsig JL, Snyder SL, Creutz CE (2003) Identification of targets for calcium signaling through the copine family of proteins: characterization of a coiled-coil copine-binding motif. J Biol Chem 278: 10048–10054 [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Naito S, Redei GP, Komeda Y (1993) A new class of mutants in Arabidopsis thaliana, acaulis 1, affecting the development of both inflorescences and leaves. Development 118: 751–764 [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sugita S, Südhof TC (2000) The RIM/NIM family of neuronal C2 domain proteins: interactions with Rab3 and a new class of Src homology 3 domain proteins. J Biol Chem 275: 20033–20044 [DOI] [PubMed] [Google Scholar]
- Ward ER, Uknes SJ, Williams SC, Dincher SS, Wiederhold DL, Alexander DC, Ahl-Goy P, Métraux JP, Ryals JA (1991) Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann K, Hunt M, Uknes S, Neuenschwander U, Lawton K, Steiner HY, Ryals J (1995) Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7: 2013–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO (2002) Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13: 3369–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu RX, Pawelczyk T, Xia TH, Brown SC (1997) NMR structure of a protein kinase C-γ phorbol-binding domain and study of protein-lipid micelle interactions. Biochemistry 36: 10709–10717 [DOI] [PubMed] [Google Scholar]
- Yalpani N, Enyedi AJ, León J, Raskin I (1994) Ultraviolet light and ozone stimulate accumulation of salicylic acid, pathogenesis-related proteins and virus resistance in tobacco. Planta 193: 372–376 [Google Scholar]
- Yang Y, Shah J, Klessig DF (1997) Signal perception and transduction in plant defense responses. Genes Dev 11: 1621–1639 [DOI] [PubMed] [Google Scholar]
- Yoshioka K, Kachroo P, Tsui F, Sharma SB, Shah J, Klessig DF (2001) Environmentally sensitive, SA-dependent defense responses in the cpr22 mutant of Arabidopsis. Plant J 26: 447–459 [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Nürnberger T, Frachisse JM, Wirtz W, Guern J, Hedrich R, Scheel D (1997) Receptor-mediated activation of a plant Ca2+-permeable ion channel involved in pathogen defense. Proc Natl Acad Sci USA 94: 2751–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]