Abstract
The vascular tissue of higher plants consists of specialized cells that differ from all other cells with respect to their shape and size, their organellar composition, their extracellular matrix, the type of their plasmodesmata, and their physiological functions. Intact and pure vascular tissue can be isolated easily and rapidly from leaf blades of common plantain (Plantago major), a plant that has been used repeatedly for molecular studies of phloem transport. Here, we present a transcriptome analysis based on 5,900 expressed sequence tags (ESTs) and 3,247 independent mRNAs from the Plantago vasculature. The vascular specificity of these ESTs was confirmed by the identification of well-known phloem or xylem marker genes. Moreover, reverse transcription-polymerase chain reaction, macroarray, and northern analyses revealed genes and metabolic pathways that had previously not been described to be vascular specific. Moreover, common plantain transformation was established and used to confirm the vascular specificity of a Plantago promoter-β-glucuronidase construct in transgenic Plantago plants. Eventually, the applicability and usefulness of the obtained data were also demonstrated for other plant species. Reporter gene constructs generated with promoters from Arabidopsis (Arabidopsis thaliana) homologs of newly identified Plantago vascular ESTs revealed vascular specificity of these genes in Arabidopsis as well. The presented vascular ESTs and the newly developed transformation system represent an important tool for future studies of functional genomics in the common plantain vasculature.
Certain cells or tissues are difficult to isolate from Arabidopsis (Arabidopsis thaliana) with classical techniques and, therefore, transcriptome or proteome analyses in these cells or tissues cannot be performed easily. One example of rather inaccessible tissue is the leaf vasculature. In an average Arabidopsis leaf blade, it represents less then 3% of the cell mass and its mechanical isolation is impossible because the strands are built by only few and, typically, tiny cells. The diameter of a companion cell (CC) or a sieve element (SE) can be 3 μm or less.
For several years, the laser microdissection and pressure-catapulting technique (Bonner et al., 1997; Asano et al., 2002; Ivashikina et al., 2003) allows selective isolation of previously inaccessible tissues and amplification and analysis of the included mRNAs. However, there are several critical obstacles on the way from a living tissue to the molecular information, including tissue processing, microdissection itself, and special analytical demands (for review, see Kehr, 2003). Nevertheless, Ivashikina et al. (2003) had successfully applied this technique for the isolation of Arabidopsis vascular mRNA from flower stalks. In fact, the authors confirmed the vascular-specific expression of the AtSUC2 Suc transporter gene (Sauer and Stolz, 1994), of the AKT2 K+-channel gene (Marten et al., 1999), or of AHA3, the gene of the plasma membrane H+-ATPase (DeWitt and Sussman, 1995).
In the same article (Ivashikina et al., 2003), about 700 expressed sequence tags (ESTs) from Arabidopsis CCs were published. However, the corresponding mRNAs were not derived from the laser microdissection and pressure-catapulting technique-generated tissue, but rather from CC protoplasts. These protoplasts had been collected after enzymatic cell wall degradation of leaf tissue from plants expressing green fluorescent protein under the control of the CC-specific AtSUC2 promoter (Truernit and Sauer, 1995; Imlau et al., 1999). However, the lengthy protoplasting procedure (>1 h) resulted in an up-regulation of stress-induced transcripts. This became obvious, for example, from the high levels of AtSUC3 transcripts (AtSUC3 encodes a Suc transporter [Meyer et al., 2004]) that were detected in an mRNA fraction prepared from protoplasted mesophyll cells that were used as controls. It is known that unstressed mesophyll cells do not express AtSUC3 (Meyer et al., 2000, 2004).
Several authors (Zhao et al., 2000; Oh et al., 2003) succeeded in the mechanical isolation of secondary phloem and secondary xylem from Arabidopsis root hypocotyls. This organ produces relatively large amounts of secondary vasculature when senescence is delayed by continuous removal of inflorescences (Zhao et al., 2000, 2005; Beers and Zhao, 2001). Using this material, secondary phloem-specific and secondary xylem-specific mRNAs could be obtained and used for microarray analyses. For several of the predicted phloem-specific or xylem-specific genes, Zhao et al. (2005) confirmed localization via promoter-reporter gene plants.
A different approach was used by Vilaine et al. (2003), who switched to celery (Apium graveolens), a plant that has frequently been used for analyses of phloem mannitol transport (Pharr et al., 1995; Zamski et al., 1996, 2001; Noiraud et al., 2000, 2001). Celery allows simple and rapid isolation of phloem, xylem, and storage parenchyma from its long petioles. cDNA libraries were prepared from petiolar phloem at different developmental stages and 989 ESTs have been published. For other, mostly woody plants, such as pine (Pinus taeda; Allona et al., 1998), poplar (Populus spp.; Sterky et al., 1998), or aspen (Populus spp.; Hertzberg et al., 2001), similar approaches were used for xylem transcriptome analyses.
An entirely different and very elegant approach is the direct isolation of proteins from phloem sap. This technique was used by Balachandran et al. (1997) and by Walz and coworkers (2002, 2004), who isolated phloem sap proteins from cucumber (Cucumis sativus), pumpkin (Cucurbita maxima Duch.), or castor bean (Ricinus communis) and identified phloem proteins after one- and two-dimensional gel electrophoresis and microsequencing or by mass spectrometry (Barnes et al., 2004).
Despite these different approaches, the total number of ESTs, especially from the phloem, is limited. Moreover, many of the plants that were used to collect phloem or vascular EST data (e.g. celery) are difficult or impossible to transform and EST information on these species can, therefore, only be used for other, heterologous plant systems. The largest dataset from a single plant is the collection of 989 ESTs from celery petioles (Vilaine et al., 2003), where 73 of 793 identified independent genes are expressed in the phloem or in the ontogenetically related vascular tissue (phloem and xylem). Considerably larger numbers of ESTs and independent RNAs were published for the cambial tissue of Populus tremula L. × tremuloides Michx. (4,809 ESTs and 2,988 independent RNAs; Sterky et al., 1998) and for the wood-forming tissue of Populus trichocarpa Trichobel (883 ESTs and 731 independent RNAs; Sterky et al., 1998).
In our lab, common plantain (Plantago major) has been used repeatedly for analyses of SE or CC transporters (Gahrtz et al., 1994; Barth et al., 2003; Ramsperger-Gleixner et al., 2004). Like celery, Plantago allows simple isolation of pure vascular tissue in large amounts by extraction of the bundles from the leaf base (images are shown in Gahrtz et al., 1994). Unlike in celery, it cannot be separated into phloem and xylem. However, Plantago vascular tissue is derived from leaf blades, not from petioles, and tissue preparation is very fast (<5 s).
Here, we describe a representative vascular transcript profile for Plantago, based on analysis of 5,900 EST sequences that represent 3,247 different mRNAs. The specificity of the library is confirmed by the identification of ESTs of genes previously identified in Plantago CCs, by the high expression levels of genes known to be vascular specific in other plants, by comparing expression levels in vascular versus nonvascular tissue using northern, reverse transcription (RT)-PCR and macroarray analyses, and in transgenic Plantago. Moreover, tissue specificity of promoters from Arabidopsis homologs of predicted vascular Plantago genes was analyzed and shown to confer vascular specificity in Arabidopsis. This result demonstrates that information obtained in Plantago may be transferred to other plant systems. All common plantain vascular EST sequence data are accessible via the Internet (http://www.plantain.de). Finally, a transformation technique is presented for common plantain. Together, these data provide the basis to perform functional genomics in Plantago, a species that differs significantly in vascular architecture and function from the model plant Arabidopsis.
RESULTS
Characterization of the Plantago Vascular EST Library
Vascular bundles (up to 10 cm and longer) are easily and rapidly pulled out from common plantain leaves and petioles due to the presence of an endodermis that surrounds the entire vascular tissue. The Casparian stripes within this endodermis rupture during extraction of the bundles (Gahrtz et al., 1994), leaving only the tips of the vascular tissue (including the minor veins) in the leaf blades. The extracted bundles show bicollateral anatomy with a central xylem and an abaxial and an adaxial phloem (Gahrtz et al., 1994).
A size-fractionated cDNA library from Plantago vascular tissue was generated in λ-ZAP II (enriched in cDNAs longer than 500 bp; 2 × 106 plaque-forming units in the initial library). Inserts were excised to create a plasmid library in pBluescript SK−. According to the β-galactosidase staining on petri plates with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), almost 100% of the clones in this library contained cDNA inserts; the average length of these inserts was about 750 bp. From this plasmid library, the 5′ ends of 7,680 cDNA inserts were sequenced. A small number of clones were also sequenced from the 3′ end. All of these 3′ sequences had a poly(A+) tail, confirming that the library was correctly oriented.
Of the obtained 5′ sequences, 5,900 were used for further analyses; 40.1% of these sequences (2,347 ESTs) represented singlets and 59.9% (3,553 sequences) were found in two or more copies (maximum 150 copies). Overlapping and redundant sequences were assembled to 900 contigs with an average of 3.8 ESTs per contig, yielding a total of 3,247 independent Plantago vascular mRNAs. These sequences were characterized using BLASTX and BLASTN similarity searches in publicly available data libraries. With a threshold E value of 10−5, significant similarity matches were found for 55% of these sequences.
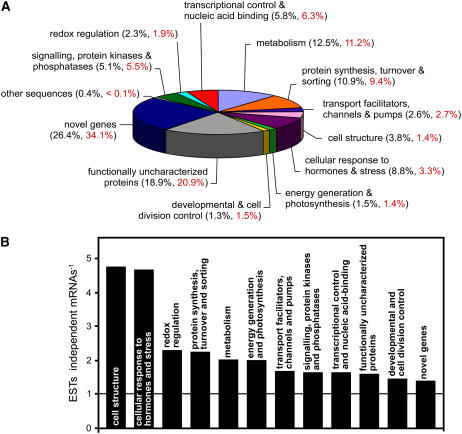
Based on the predicted function of the encoded proteins, mRNAs with similarity to already known sequences were assigned to 11 functional groups. mRNAs for proteins with no functional predictions were divided into two groups that were named functionally uncharacterized proteins, for mRNAs giving similarity matches with functionally uncharacterized proteins in another organism, or novel genes, if no similarity matches were found. In Figure 1A, the percentage of independent mRNAs in each of the 13 groups is presented.
Figure 1.
Plantago vascular ESTs were divided into different groups. A, The obtained sequences were assigned either to one of 10 functional groups or to one of three groups standing for functionally uncharacterized proteins, for novel genes, or for a single mRNA with homology to sequences of viral origin (other sequences). The percentage of ESTs per group (black) and the percentage of independent mRNAs per group (= contigs plus singlets; red) are indicated. B, The EST-to-mRNA ratio was determined for 12 of the 13 groups shown in A. This ratio is a measure for the average strength of expression of the genes in a given group (higher bar = stronger average expression). The ratio is not shown for the other sequences group, which is represented by a single mRNA resulting from 26 ESTs.
Of the 5,900 EST sequences, 26.4% (34.1% of the independent mRNAs) represent novel genes, making this the largest of the 13 groups shown in Figure 1A. The second largest group (18.9% of all ESTs and 20.9% of the independent mRNAs) is the functionally uncharacterized proteins group. The largest groups with known functions are the metabolism group, with 12.5% of all ESTs and the protein synthesis, turnover, and sorting group, with 10.9% of all ESTs. The smallest groups are the developmental and cell division control group, with 1.3% of all ESTs and the energy generation and photosynthesis group, with 1.5% of all ESTs. This distribution is expected because the corresponding genes are known to be either highly expressed (e.g. the protein synthesis, turnover, and sorting genes and many of the metabolism genes) or they are expected to have either low expression levels (developmental and cell division control genes) or expressed mainly in nonvascular tissue (e.g. the photosynthetic genes). The residual groups comprise between 2.6% (transport facilitators, channels, and pumps) and 8.8% (cellular response to hormones and stress) of all ESTs. The relatively high number of ESTs in the cellular response to hormones and stress group does not result from lengthy tissue sampling. The isolation of vascular tissue from Plantago leaves is too short (<5 s) to allow induction of stress-related genes. The other sequences group comprises 26 independent ESTs for a single mRNA with high homology to viral sequences (e.g. Japanese yam mosaic virus genomic RNA [accession no. AB027007] or the leek yellow stripe potyvirus [accession no. AJ307057]).
The Library Is Highly Enriched in Vascular ESTs
Figure 1A presents the percentage of ESTs (black numbers) and the percentage of different mRNAs (red numbers) for each of the 13 groups. The EST-to-mRNA ratios vary from 1.4 in the novel genes group to 4.8 in the cell structure group (Fig. 1B). Higher ratios indicate that one or more of the mRNAs in the respective group are encoded by highly expressed genes. This is expected for the cell structure group with mRNAs of genes, such as actins, tubulins, lamins, or matrix proteins. Interestingly, the mRNAs with the largest number of ESTs in this group encode a Gly/Pro-rich protein (GPRP1; Fig. 2; Table I). Expression of such genes is known to be associated with the vascular system (Keller et al., 1988; Ryser and Keller, 1992; Parsons and Mattoo, 1994; Liu et al., 2003). The increased ratio obtained for the cellular response to hormones and stress group is due to the large number of ESTs for metallothionein (MT)-like proteins (Fig. 2). In fact, mRNAs for three different MT genes (PmMT1, PmMT2, and PmMT3) stand for 225 independent ESTs. MT genes are known to be specifically and highly expressed in the phloem (Vilaine et al., 2003; Barnes et al., 2004) and the high numbers of ESTs for MTs and GRPs are, therefore, the first evidence for the vascular specificity of this EST library.
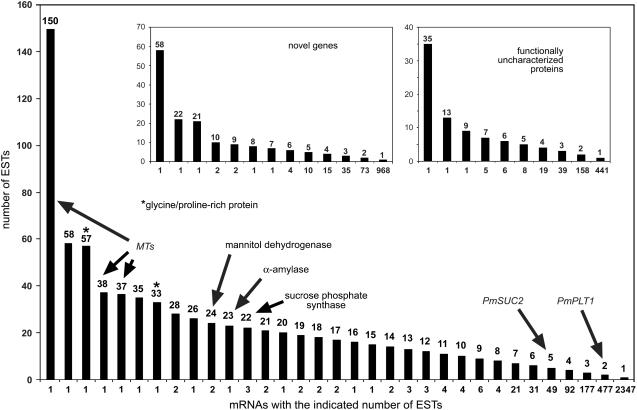
Figure 2.
Number of ESTs identified per individual mRNA. The number of identified and sequenced ESTs per mRNA is shown for all mRNAs (large image) and for the mRNAs of the novel genes and functionally uncharacterized proteins groups (insets). mRNAs for MTs, GRPs (asterisk), three biosynthetic enzymes (mannitol dehydrogenase, α-amylase, Suc phosphate synthase), or for previously characterized CC-specific transporters (PmSUC2, PmPLT1) are indicated.
Table I.
Genes expressed in common plantain vascular tissuea
| ESTs mRNA−1 | Putative Function or Contig Name | Accession No. |
|---|---|---|
| Redox regulation | ||
| 19 | Thioredoxin h (PmTRX1) | AJ844021 |
| 12 | Thioredoxin (PmTRX3) | AJ844023 |
| 7 | Copper/zinc superoxide dismutase (PmCSD1) | AJ844003 |
| 7 | Thioredoxin (PmTRX2) | AJ844022 |
| 6 | Thioredoxin-dependent peroxidase 1 (PmTPX1) | AJ843119 |
| 5 | Glutaredoxin (PmGLX1) | AJ844008 |
| 5 | Glutaredoxin (PmGLX2) | AM111306 |
| 4 | Ascorbate peroxidase (PmAPX1) | AJ843990 |
| 4 | Ferric-chelate reductase (PmFRO1) | AM111307 |
| 4 | 12-Oxophytodienoate reductases (PmOPR1) | AM111308 |
| Transcriptional control and nucleic acid binding | ||
| 17 | Putative DNA-binding protein | AM111309 |
| 11 | Histone H1 (PmH1a) | AM111310 |
| 7 | Poly(A)-binding protein (PmPABP1) | AM111311 |
| 7 | MADS-domain transcription factor (PmSOC1) | AM111312 |
| 7 | Histone H3 (PmH3a) | AM111313 |
| 6 | MYB-transcription factor (PmMYB1) | AM111314 |
| 6 | HMG protein (PmHMG1) | AM111315 |
| 5 | bZIP transcription factor (PmbZIP1) | AM111316 |
| 5 | Putative reverse transcriptase (PmRT1) | AM111317 |
| 5 | Transcription initiation factor IIa, γ-chain (PmTFIIa-γ) | AM111318 |
| Metabolism | ||
| 24 | Mannitol dehydrogenase (PmMTD1) | AJ844011 |
| 23 | α-Amylase (PmAAMY1) | AJ843124 |
| 22 | Suc-phosphate synthase (PmSPS1) | AJ843125 |
| 20 | Malate dehydrogenase (PmMDH1) | AJ843126 |
| 14 | 1-Aminocyclopropane-1-carboxylate oxidase (PmACO1) | AJ843131 |
| 14 | Nucleoside-diphosphate sugar dehydratase (PmNSD1) | AM111319 |
| 11 | Putative nitrilase-associated protein (PmNAP1) | AM111320 |
| 9 | Cinnamoyl alcohol dehydrogenase (PmCAD1) | AM111321 |
| 8 | Glyceraldehyde 3-P dehydrogenase (PmGAPDH1) | AM111322 |
| 7 | β-Glucosidase (PmbGLC1) | AM111323 |
| Protein synthesis, turnover, and sorting | ||
| 24 | Polyubiquitin 1 (PmUBQ1) | AJ844615 |
| 21 | Translation initiation factor 5A-1 (PmelF5A1) | AJ843977 |
| 19 | Asparaginyl endopeptidases (PmENP1) | AJ843978 |
| 13 | Polyubiquitin 2 (PmUBQ2) | AJ844616 |
| 13 | Translation elongation factor-1 (PmEF1α) | AM111324 |
| 12 | Polyubiquitin 3 (PmUBQ3) | AJ844617 |
| 12 | Ubiquitin-conjugating enzyme 1 (PmUBC1) | AJ843120 |
| 9 | Ubiquitin-conjugating enzyme 2 (PmUBC2) | AJ843121 |
| 9 | Ubiquitin-conjugating enzyme 3 (PmUBC3) | AM111325 |
| 8 | Cysteine proteinase 1 (PmCPR1) | AJ844005 |
| Transport facilitators, channels, and pumps | ||
| 7 | Putative transport protein | AJ843130 |
| 5 | Suc transporter (PmSUC2) | X75764 |
| 5 | Amino acid transporter (PmAAP2) | AJ843988 |
| 5 | Vacuolar H+-ATPase, subunit C 1 (PmVATPc1) | AJ843122 |
| 5 | Vacuolar H+-ATPase, subunit C 2 (PmVATPc2) | AM111326 |
| 5 | Tetracycline transporter-like protein 1 (PmTTPL1) | AJ844020 |
| 4 | Aquaporin-like protein (PmAQP1) | AJ843991 |
| 4 | Aquaporin-like protein (PmAQP2) | AJ843992 |
| 3 | Plasma membrane H+-ATPase (PmPMA1) | AJ843127 |
| 3 | Peptide transporter 1 (PmPTR1) | AJ843128 |
| Cell structure | ||
| 57 | Glycine/Pro-rich protein 1 (PmGPRP1) | AJ843997 |
| 33 | Putative structural protein/Gly-rich 1 (PmPSP1) | AJ843998 |
| 28 | Profilin 1 (PmPF1) | AM111327 |
| 18 | Actin-depolymerizing factor 1 (PmADF1) | AM111328 |
| 15 | Expansin 1 (PmEXPA1) | AM111329 |
| 7 | α-Tubulin 1 (PmTUA1) | AM111330 |
| 7 | Jacalin-domain protein 1 (PmJDP1) | AM111331 |
| 5 | Actin-depolymerizing factor 2 (PmADF2) | AM111332 |
| 4 | Tubulin-folding cofactor 1 (PmTFC1) | AM111333 |
| 3 | Formin-homology protein 1 (PmFH1) | AM111334 |
| Cellular response to hormones and stress | ||
| 150 | MT-like protein 1 (PmMT1) | AJ843993 |
| 38 | MT-like protein 2 (PmMT2) | AJ843994 |
| 38 | Abscisic acid and salt-responsive lectin-like protein 1 (PmLLP1) | AJ844009 |
| 37 | MT-like protein 3 (PmMT3) | AJ843995 |
| 22 | Ripening-induced/major latex-like protein (PmMLP1) | AJ844012 |
| 18 | Auxin resistance protein 2 (PmAXR2) | AJ843999 |
| 17 | Cold stress-induced protein (PmSRC1) | AJ844001 |
| 11 | Stress-induced/dehydrin (PmDHN1) | AJ844000 |
| 10 | Copper chaperone (PmCCH1) | AJ844002 |
| 7 | BURP domain-containing protein (PmBDC1) | AJ843872 |
| Energy generation and photosynthesis | ||
| 16 | Rubisco SU 1 (PmRBCS1) | AJ843972 |
| 6 | Mitochondrial ATP synthase, β-chain (PmATP2) | AJ843974 |
| 6 | Rubisco SU 1a (PmRBCS1a) | AJ843975 |
| 5 | Mitochondrial ATP synthase, δ-chain (PmATP5) | AJ843973 |
| 3 | Light-harvesting chlorophyll a/b-binding (PmLHC1) | AJ843976 |
| 3 | Mitochondrial ATP synthase, β-chain (PmATP3) | AM111335 |
| 3 | Cytochrome C-oxidase subunit 5b (PmCOX5b) | AM111336 |
| 3 | Ubiquinol-cytochrome C reductase (PmUCR3) | AM111337 |
| 2 | Cytochrome b5-reductase (PmCBR1) | AM111338 |
| 2 | PSII, 22-kD protein (PmPSBS1) | AM111339 |
| Developmental and cell division control | ||
| 6 | Photoperiodic response/GIGANTEA 1 (PmGI1) | AM111340 |
| 4 | Phototropin/nonphototropic hypocotyl-like 1 (PmNPL1) | AM111342 |
| 4 | Abaxial cell fate/YABBY 1 (PmYAB1) | AM111348 |
| 3 | Fimbriata-associated protein 1 (PmFAP1) | AM111343 |
| 3 | BAG-domain protein 1/cell death associated (PmBAG1) | AM111345 |
| 3 | Exostosin 1/growth-related protein (PmEXT1) | AM111347 |
| 3 | Photoperiodic response/GIGANTEA 2 (PmGI2) | AM111341 |
| 3 | Fimbriata-associated protein 2 (PmFAP2) | AM111344 |
| 2 | Fimbriata-associated protein 3 (PmFAP3) | AM111346 |
| 2 | Embryo-specific protein 3 (PmATS3) | AM111349 |
| Functionally uncharacterized proteins | ||
| 35 | P12.43.C2 | AJ843979 |
| 13 | P12.43.C1 | AJ843980 |
| 9 | P12.24.C1 | AJ843981 |
| 7 | P12.76.C1 | AJ843982 |
| 7 | P12.187.C1 | AJ843983 |
| 7 | P12.351.C1 | AJ843984 |
| 7 | P12.0.CB7 | AJ843985 |
| 7 | P12.117.C1 | AJ844614 |
| 6 | P.12.0.C66 | AJ843986 |
| 6 | P12.515.C1 | AJ843987 |
| Novel genes | ||
| 58 | P12.0.C65 | AJ844013 |
| 22 | P12.0.CB9 | AJ844014 |
| 21 | P12.157.C1 | AJ844015 |
| 10 | P12.0.CB8 | AJ844016 |
| 10 | P12.0.CB21 | AJ844017 |
| 9 | P12.53.C1 | AJ844018 |
| 9 | P12.64.C1 | AJ844019 |
| 8 | P12.73.C1 | AM111350 |
| 7 | P12.683.C1 | AM111351 |
| 6 | P12.0.C130 | AM111352 |
| Other sequences | ||
| 26 | Homology to viral polyprotein | AJ844004 |
For each group, the 10 genes with the highest expression are presented.
Figure 2 shows the distribution of ESTs per mRNA and the number of mRNAs for each EST count. The distribution is shown for all identified mRNAs (large image) and for the mRNAs of the novel genes and functionally uncharacterized proteins groups (Fig. 2, insets). As already mentioned, the large image demonstrates that MTs and GRPs are among the most highly expressed genes. It also shows the positions of the three mRNAs with the highest EST numbers of the metabolism group. These mRNAs encode a mannitol dehydrogenase, an α-amylase, and a Suc phosphate synthase (PmMTD1, PmAAMY1, PmSPS1; for accession numbers, see Table I).
Moreover, two mRNAs are marked that encode the CC-specific transporters PmSUC2 (for Suc; accession no. X75764; Gahrtz et al., 1994) and PmPLT1 (for sorbitol; accession no. AJ532589; Ramsperger-Gleixner et al., 2004). The CC specificity of these proteins has previously been confirmed by immunolocalization (Stadler et al., 1995; Ramsperger-Gleixner et al., 2004). This demonstrates that both mRNAs represented by many ESTs (e.g. MTs or GRPs) and mRNAs represented by only a few ESTs (e.g. PmSUC2 or PmPLT1) stand for vascular-specific or vascular-enriched genes. This is likely to be the case also for the mRNAs from the novel genes or the functionally uncharacterized protein groups (Fig. 2, insets).
In Table I, the mRNAs of the 10 most highly expressed genes from 12 groups plus the single mRNA from the other sequences group are listed together with their gene bank accession numbers. These 121 sequences, plus all other Plantago cDNA sequences, are deposited on a Web site (http://www.plantain.de). In addition to mRNAs from genes expressed in all tissues (e.g. histones, ubiquitins, tubulins, profilins), Table I shows numerous mRNAs that were previously shown to be specifically or preferentially expressed in the vascular tissue. These are, for example, mRNAs for thioredoxins (PmTRX1, PmTRX2, PmTRX3; Ishiwatari et al., 1998), glutaredoxins (PmGLX1, PmGLX2; Balachandran et al., 1997), a malate dehydrogenase (PmMDH1; Barnes et al., 2004; Walz et al., 2004), a β-glucosidase (PmbGLC1; Walz et al., 2004), a copper/zinc superoxide dismutase (PmCSD1; Walz et al., 2002), a cinnamoyl alcohol dehydrogenase (PmCAD1; Sarni et al., 1984), or the mRNA for a gene with high homology to the vascular-specific amino acid transporter AtAAP2 from Arabidopsis (PmAAP2; Kwart et al., 1993).
Validation by Northern and RT-PCR Analyses
For selected genes, the suggested vascular specificity or nonvascular specificity was tested by northern blots. The nonvascular RNA was isolated from the basal regions of leaves (blade and petiole) from which the vascular bundles had been extracted (the quality of these tissues has been demonstrated in figure 3 in Gahrtz et al., 1994). Figure 3A shows that PmMT1 and PmMT2 mRNA levels are very high in vascular and very low in nonvascular tissue. As expected, mRNA levels for Rubisco (PmRBCS1) show an inverse distribution being higher in nonvascular than in vascular tissue (Fig. 3A). The vascular PmRBCS1 signal may result both from low-level PmRBCS1 expression in the vasculature and from some mesophyll contamination in the vascular preparation. Also, the mRNA of the viral polyprotein-like sequence (see Table I) is highly vascular specific. This is in agreement with a report of Cronin et al. (1995), who analyzed the cell-to-cell movement and long-distance translocation of the tobacco (Nicotiana tabacum) edge virus (potyvirus group) and found an accumulation of β-glucuronidase (GUS)-labeled polyprotein fragments in the phloem. However, the band labeled on the northern blot (Fig. 3A) does not show the size expected for a polyprotein mRNA (>10 kb). It is smaller than the 18S ribosomal RNA (1,900 bp) and is, therefore, most likely not of viral origin and rather may represent a virus-derived sequence that has been inserted into the Plantago genome.
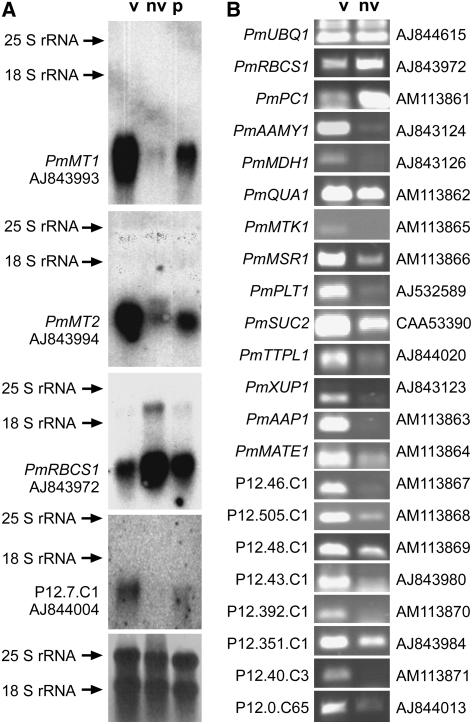
Figure 3.
Validation of vasculature-specific expression by northern blots and RT-PCR. A, Expression of four genes (PmMT1, PmMT2, PmRBCS1, and the contig P12.7.C1 [homologous to viral polyprotein]) in vascular (v) tissue isolated from leaves, in nonvascular (nv) tissue represented by petioles that had their vascular bundles extracted, and in intact petioles (p) was analyzed on northern blots. As expected, PmMT1 and PmMT2 are expressed almost exclusively in the vascular tissue, whereas expression of PmRBCS1 is higher in nonvascular tissue. Expression of P12.7.C1 is also highly specific for the vasculature. The bottom image represents the loading control (methylene blue-stained nylon filter). Gene names and accession numbers are given. B, Expression of 20 genes was analyzed in vascular (v) and nonvascular (nv) tissue. The figure shows the bands obtained by quantitative RT-PCR amplification with total RNA, the names of the respective genes (for novel genes the contig numbers are given), and their accession numbers. PmUBQ1 was used as a control that was expected to have similar expression levels in both tissues. PmPC1 (plastocyanine) was used as a control that was expected to be expressed mainly outside the vasculature. With the exception of PmQUA1, which shows comparable expression levels, all other genes are expressed mainly or exclusively in the vascular tissue.
A larger number of genes was analyzed by RT-PCR (Fig. 3B). Again, the results confirmed the predicted vascular specificity. With the exception of PmUBQ1, which was used as a control gene and has similar expression levels in vascular and nonvascular tissue, and with the exception of PmPC1 (encoding a plastocyanine), which should be expressed more strongly outside the vasculature (similar to PmRBCS1 in Fig. 3A), all other genes showed varying degrees of vascular specificity. Some of them (e.g. THIOMETHYL-RIBOSE KINASE 1 [PmMTK1], POLYOL TRANSPORTER 1 [PmPLT1], XANTHINE/URACILE TRANSPORTER 1 [PmXUP1], AMINO ACID PERMEASE 1 [PmAAP1], two functionally uncharacterized genes [contigs P12.46.C1 and P12.40.C3], and a novel gene [contig P12.7.C65]) were expressed almost exclusively in the vasculature.
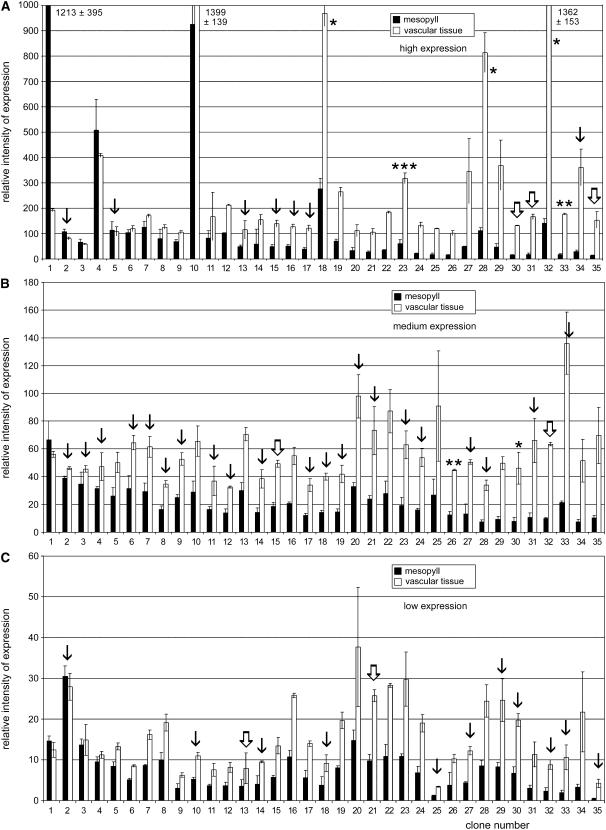
For large-scale expression analyses, macroarrays with several hundred PCR-generated cDNA fragments (typical length: 250–300 bp) were hybridized to radiolabeled cDNA from vascular or nonvascular poly(A+) RNA. Figure 4 shows the results for 105 ESTs sorted by their vascular specificity (from left to right, increasing vascular specificity) and their relative expression intensity (high expression, Fig. 4A; medium expression, Fig. 4B; low expression, Fig. 4C). Genes expressed mainly in nonvascular tissue were identified only among the highly expressed genes (Fig. 4A). For example, the first EST in Figure 4A encodes a BURP domain-containing protein (PmBDC1) and the corresponding gene (accession no. AJ843872) has previously been shown to be expressed strongly and mainly in nonvascular tissue (M. Gahrtz and N. Sauer, unpublished data). Only three of the genes analyzed in Figure 4B (medium expression) and four of the genes analyzed in Figure 4C (low expression) have similar expression levels in vascular and nonvascular tissue. All other ESTs show higher or very high expression in the vasculature. ESTs 18 and 32 in Figure 4A represent the MT genes PmMT2 and PmMT1 that were also analyzed on northern blots (Fig. 3A) and that are listed among the top 10 genes in the cellular response to hormones and stress group (Table I). The relative expression intensities and the vascular specificity that is predicted for these genes from the macroarray in Figure 4A reflects the relative expression intensities predicted by the EST frequency listed in Table I and by the vascular specificity seen in the northern analyses in Figure 3A. This is also true for the PmAQP1 and PmPSP1 genes marked in Figure 4A or for the PmAAMY1 and PmSPS1 genes marked in Figure 4B (see also Table I for frequency of ESTs and/or Fig. 3B for vascular specificity). This demonstrates that the ratios (vascular to nonvascular) obtained by the analyses in Figure 4 are a useful measure for the vascular specificity of these mRNAs.
Figure 4.
Analysis of vascular tissue-specific expression using macroarrays. PCR-derived fragments of 216 different ESTs were spotted in duplicate on nylon filters and hybridized to radiolabeled mRNA isolated from vascular or nonvascular tissue. The relative expression intensities in vascular and nonvascular tissue are presented (±sd; n = 3). A, Genes with high expression levels. Bars marked with one asterisk represent mRNAs of MTs; the bar marked with two asterisks shows a vascular-specific aquaporin (PmAQP1); the bar marked with three asterisks represents a GRP (PmPSP1). B, Genes with medium expression levels. The bar marked with one asterisk represents PmAAMY1, an α-amylase gene; the bar marked with two asterisks shows PmSPS1, the gene of a Suc phosphate synthase. C, Genes with low expression levels. A to C, Thin arrows mark mRNAs from the functionally uncharacterized proteins group; thick and white arrows mark mRNAs from the novel genes group.
PCR-derived fragments from numerous mRNAs of the functionally uncharacterized proteins (Fig. 4, thin arrows) and the novel gene (Fig. 4, thick arrows) groups were included in these macroarrays. The vast majority of the corresponding genes turned out to be expressed preferentially or exclusively in the vascular tissue; some, for example, EST numbers 34 (contig P12.43.C2; accession no.AJ843979) or 35 (contig P12.635.C1; accession no. AM114423) in Figure 4A, even with higher specificity than PmMT1, a phloem marker protein.
Information on the Vasculature Specificity Obtained in Plantago Can Be Transferred to Arabidopsis
An important question is, of course, whether such a set of expression data can be used to predict vascular specificity also for homologous genes from other plants (e.g. from Arabidopsis). Therefore, we isolated promoter sequences by PCR for Arabidopsis genes that showed significant homology to vascular-specific Plantago genes and used these promoters to drive expression of the GUS reporter gene in Arabidopsis.
The first gene we selected was AtMTK (At1g49820; 1,220-bp promoter sequence; Plantago homolog PmMTK1 [see Fig. 3B]). When the PmMTK1 sequence was first isolated from Plantago, BLAST searches found similarity only to a then functionally uncharacterized gene from Arabidopsis. Meanwhile, however, this Arabidopsis homolog was characterized as methylthio-Rib kinase (AtMTK; Sauter et al., 2004). AtMTK is a unique Arabidopsis gene and encodes an enzyme involved in the recycling of Met through the methylthioadenosine (MTA) cycle. The tissue specificity of AtMTK expression had not been studied.
The second gene we selected was PmTTPL1 (see Table I), which is homologous to three Arabidopsis genes (At2g16970, At2g16980, and At2g16990; highest identity values with At2g16970). These three genes encode so far uncharacterized tetracycline transporter-like proteins. We isolated the promoter of At2g16970 by PCR (1,104 bp) and used it to drive expression of the GUS reporter gene in Arabidopsis.
The third gene we selected was PmXUP1, which codes for a putative xanthine/uracile transporter. There are six XUP homologs in Arabidopsis (xanthine/uracile permease-like family: At1g60030, At2g05760, At2g26510, At2g34190, At5g49990, and At5g62890), with At5g62890 sharing the highest degree of homology with PmXUP1. We isolated the promoter of At5g62890 by PCR (1,343 bp) and used it to drive expression of the GUS reporter gene in Arabidopsis.
The fourth gene we selected was a Plantago member of the novel genes group (P12.0.C65; 1,428-bp 5′-flanking sequence; accession no. AM156930). We amplified a P12.0.C65 promoter fragment from Plantago genomic DNA by PCR based on sequence information that had been obtained by thermal asymmetric interlaced (TAIL)-PCR. This fragment was used to drive GUS expression in Arabidopsis. Vascular-specific expression of this gene in Plantago has previously been shown by northern analysis (N. Sauer and M. Gahrtz, unpublished data) and was confirmed by the RT-PCR shown in Figure 3B.
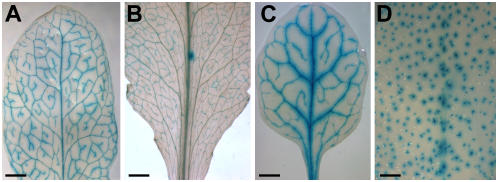
Figure 5 demonstrates that source leaves of AtMTK promoter-GUS plants (seven of 10 analyzed transformants), of At2g16970 promoter-GUS plants (eight of 16 analyzed transformants), and of P12.0.C65 promoter-GUS plants (11 of 24 analyzed transformants) show GUS histochemical staining specifically in their vascular bundles. Only the GUS staining of At5g62890 promoter-GUS plants is not vascular specific and stains the trichomes of these plants with high specificity (11 of 12 analyzed plants). These data show that (1) Plantago-derived information on the vascular specificity of a unique gene is also valid in Arabidopsis (AtMTK); (2) vascular-specific Plantago promoters of genes (P12.0.C65) that do not have homologs in Arabidopsis confer also vascular-specific GUS expression in Arabidopsis; and (3) no prediction can be made on vascular specificity for members of larger Arabidopsis gene families (e.g. for the xanthine/uracile permease family). Nevertheless, the Plantago data suggest that at least one member of the Arabidopsis family may also be vasculature specific.
Figure 5.
Analysis of the vascular specificity of Arabidopsis promoters chosen due to the vascular specificity of the homologous Plantago gene and of a Plantago promoter in Arabidopsis. A, GUS histochemical staining of the leaf of an AtMTK promoter-GUS plant. B, GUS histochemical staining of the leaf of an At2g16970 (tetracycline transporter-like protein) promoter-GUS plant. C, GUS histochemical staining of the leaf of a P12.0.C65 promoter-GUS plant. D, GUS histochemical staining of the leaf of an At5g62890 promoter-GUS plant. Scale bars = 2 mm in A, B, and D; 1 mm in C.
Establishing a Transformation System for Common Plantain
A major prerequisite for analysis of expression patterns and gene functions in a given plant is the availability of a transformation system. So far, transformation had not been transcribed for common plantain. Based on Agrobacterium tumefaciens-mediated transformation of Arabidopsis, we developed an effective and simple system for producing common plantain transgenic plants.
For selection of possible transformants, we used the BASTA resistance gene in the vector pGPTV-bar (Becker et al., 1992). Kanamycin resistance could not be used as a selection marker because Plantago has strong endogenous resistance to this antibiotic (data not shown) that may result from a similar mechanism as has recently been described for Arabidopsis (Mentewab and Stewart, 2005).
In a first approach, we tried to transform calli that had been obtained from leaves or roots of Plantago plants grown under sterile conditions and, in parallel, we tried to set up a regeneration system using untransformed calli. A similar approach has been published recently for Medicago truncatula (Crane et al., 2006). Using media with different phytohormone concentrations, we were able to obtain calli from root and leaf tissue (see “Materials and Methods”). Whereas regeneration of plants from leaf-derived calli was not successful with any of the tested phytohormone combinations, a phytohormone combination could be determined that allowed two-step regeneration of Plantago plants from root-derived calli (shoot-inducing and root-inducing conditions; see “Materials and Methods”). However, when we tried to induce calli from roots that had been incubated or cocultivated with Agrobacterium tumefaciens that harbored a BASTA resistance gene, no BASTA-resistant callus tissue could be obtained (data not shown).
In a second approach, we tried to transform Plantago based on the floral-dip technique described for Arabidopsis (Clough and Bent, 1998). We applied this technique because Plantago plants develop between several hundred and 1,000 seeds. Using the floral-dip technique exactly as published did not yield BASTA-resistant Plantago seedlings. However, eventually a slightly modified technique could be established that yielded significant numbers of transgenic Plantago plants. The main modifications of the Plantago floral-dip technique were the much higher (10-fold) concentrations of the detergent Silwet in the Agrobacterium solution used for dipping and the prolonged application of vacuum (up to 5 min) during the actual dipping step.
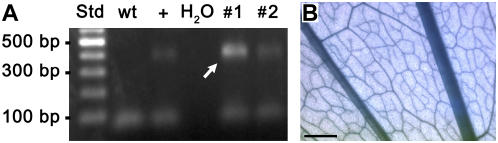
After dipping 24 Plantago plants into a suspension of Agrobacterium that harbored a PmPLT1 promoter-GUS construct, about 2,500 seeds from these plants (1.5 g) were put on soil and kept at 21°C in the growth chamber. Of the germinated seedlings, 32 survived repeated (3-fold) treatment with BASTA. Insertion of the GUS reporter gene into the genomes of these plants was checked by PCR on genomic DNA. In 15 of the analyzed plants, a PCR product could be identified (Fig. 6A) that had the same size as the band obtained in a control reaction. In contrast, PCR with DNA from wild-type plants did not result in a PCR product with GUS-specific primers (Fig. 6A). A GUS assay with fully developed leaves of these 15 plants was performed and GUS histochemical staining was detected in the vascular tissue of 11 plants. GUS staining that was performed repeatedly with wild-type Plantago plants did not result in GUS-positive staining (data not shown).
Figure 6.
GUS histochemical staining of PmPLT1 promoter-GUS plants and identification of the GUS gene in genomic DNA of BASTA-resistant Plantago plants. A, PCR analysis of genomic DNA from two BASTA-resistant Plantago plants (nos. 1 and 2). The used primers amplify a 436-bp fragment of the GUS gene (arrow). The same fragment is seen only in a control with +. It is absent from wild-type (wt) Plantago and from the water (H2O) control. B, GUS histochemical staining of a leaf from a PmPLT1 promoter-GUS Plantago plant. Scale bar = 1 mm.
These data demonstrate that (1) transgenic common plantain can be obtained using a modified floral-dip technique; (2) BASTA resistance is a suitable selection marker for Plantago; and (3) the used PmPLT1 promoter drives expression of the GUS reporter gene in Plantago vascular tissue.
DISCUSSION
This article presents data on the generation, characterization, and application of tools for the analysis of vasculature-specific gene expression in common plantain. More than 3,200 independent mRNAs were identified in an EST project that was based on a cDNA library constructed from vascular mRNA. In contrast to previously described vascular, phloem, or xylem ESTs that were obtained from petioles or hypocotyls, or that had been isolated from cells collected under conditions of mechanical stress, the mRNA used for the presented transcriptome analyses was derived from nonstressed vascular tissue of Plantago leaf blades.
Vascular strands extracted from mature Plantago source leaves are branched and up to 15 cm long. The identified mRNAs are derived from genes expressed in the phloem and xylem of fully developed Plantago vasculature. Because most of the mature xylem vessels are no longer alive, the transcriptionally active cells in this tissue preparation represent mainly cells of the xylem parenchyma, phloem CCs, phloem SEs, and phloem parenchyma cells.
Data presented in this article are useful for future studies of the complex and highly specialized long-distance transport system of higher plants. All of the identified genes are expressed within the vasculature and many of them were shown to be active preferentially or exclusively in this tissue. Due to the nature of the material that was used for the generation of the analyzed EST library (vascular bundles from fully developed source leaves), the obtained sequences represent primarily genes that encode proteins responsible for vascular structure and function (transport and signaling). In contrast, mRNAs from genes that regulate vascular development are expected only in smaller numbers because these are expressed mainly in the vasculature of sink leaves.
Metabolic Pathways and Genes Identified in Plantago Vasculature
A comparison of the highly expressed genes of this Plantago EST library with vasculature marker genes previously identified in Plantago or in other plants underlines the vasculature specificity of this library. MTs (Vilaine et al., 2003; Barnes et al., 2004), thioredoxins (Sasaki et al., 1998; Vilaine et al., 2003), Cys proteinases (Zhao et al., 2005), a Suc transporter (Stadler et al., 1995), and polyol transporters (Ramsperger-Gleixner et al., 2004) represent just a small selection of identified proteins that are known to be highly enriched in the vasculature of higher plants.
Moreover, the vasculature specificity of this library is supported by the identification of vasculature-typical metabolic pathways. For example, lignin biosynthesis is xylem specific and numerous mRNAs for enzymes of this pathway were identified. This includes the mRNAs for a Phe ammonia lyase (PmPAL1, AM159090), for three cinnamoyl-alcohol dehydrogenases (PmCAD1, AM11321; PmCAD2, AM159095; PmCAD3, AM159096),for three caffeoyl-CoA O-methyltransferases (PmCCoAOMT1, AM159088; PmCCoAOMT2, AM159089; PmCCoAOMT3, AM159091), for two 4-coumarate:CoA ligases (Pm4CL1, AM159092; Pm4CL2, AM159093), and for one sinapyl alcohol dehydrogenase (PmSAD1, AM159094).
We also identified almost complete sets of mRNAs for phloem-specific pathways. For example, Walz et al. (2002) had analyzed phloem sap proteins from cucumber and pumpkin plants for antioxidant defense enzymes. Their search was based on the assumption that oxidative stress avoidance in sieve tubes is important to maintain the functionality of the phloem, where reactive oxygen species (ROS), such as superoxide anions and hydrogen peroxide, may be produced by the mitochondria and the endoplasmic reticulum. Moreover, ROS may reach the phloem and also the xylem parenchyma during secondary cell wall formation of nearby tracheary elements (Karlsson et al., 2005). In fact, Walz et al. (2002) identified a copper/zinc-superoxide dismutase and a monodehydroascorbate reductase from cucumber phloem and a cytosolic peroxidase from pumpkin and discussed this as evidence for the presence of a complete antioxidant defense system in the phloem.
We also identified the mRNAs for these three proteins (a Cu/Zn-superoxide dismutase [PmCSD1, AJ844003], an ascorbate peroxidase [PmAPX1, AJ843990], a monodehydroascorbate reductase [PmMDAR1, AM158910]), plus several other mRNAs for proteins potentially involved in ROS detoxification. Examples are a glutathione peroxidase (PmGPX1; AM159087), a thioredoxin-dependent peroxidase (PmTPX1; AJ843119), two glutaredoxins (PmGLX1, AJ844008; PmGLX2, AM111306), and three thioredoxins (PmTRX1, AJ844021; PmTRX2, AJ844022; PmTRX3, AJ844023).
Finally, numerous ESTs were identified for mRNAs encoding proteins of the ethylene biosynthetic pathway. We observed high expression levels of the gene for a 1-aminocyclopropane-1-carboxylate oxidase (PmACO1, AJ843131), the last enzyme in ethylene biosynthesis. Only recently, a regulatory function of ethylene in the induction of phloem defense responses has been postulated for different conifers (Hudgins and Franceschi, 2004) and Walz et al. (2004) had found significant amounts of 1-aminocyclopropane-1-carboxylate oxidase protein in phloem sap proteins from cucumber. In summary, this demonstrates that the Plantago vascular EST library contains mRNAs for complete biosynthetic pathways of the xylem and the phloem.
However, we also identified pathways that were so far not described to be vasculature specific or vasculature typical. For example, we identified several mRNAs for the polyamine biosynthetic pathway. Like ethylene biosynthesis (see above), polyamine biosynthesis starts with S-adenosyl-Met (SAM) as the primary substrate. The identified mRNAs for polyamine synthesis include two different SAM decarboxylases (PmSAMDC1, AM156953; PmSAMDC2, AM159097) that produce the aminopropyl groups for the polyamines spermine and spermidine. Moreover, we identified mRNAs for one spermidine synthase (PmSPDS1, AM158913) and for two different spermine synthases (PmSPMS1, AM158911; PmSPMS2, AM158912) that catalyze the formation of spermine or spermidine from decarboxylated SAM. This suggests that Plantago vasculature is an important site for polyamine biosynthesis.
A side product of ethylene and polyamine biosynthesis is 5′ MTA, which is recycled to Met in the MTA cycle (or Yang cycle; Yung and Yang, 1982). Eventually, the recycled (but also de novo synthesized) Met is used to form new SAM. We found mRNAs for three proteins involved in SAM formation. These are the mRNAs for Met synthase (PmMET1, AM158915), 5′-methylthio-ribose kinase (PmMTK1, AM113865), and SAM synthase (PmSAMS1, AM158914). MTK1 is the only known enzyme of the plant MTA cycle and has been characterized only recently in Arabidopsis, where it is encoded by a single gene, and in rice (Oryza sativa), where two genes were found (Sauter et al., 2004). In the PCR analyses shown in Figure 3B, PmMTK1 seems to be expressed weakly, but with high specificity in the vascular tissue of Plantago. When we used the Arabidopsis AtMTK1 promoter to drive GUS expression in Arabidopsis, this promoter also turned out to be vasculature specific (Fig. 5A). This points toward an extremely high demand for Met recycling in the vascular tissues of Plantago and Arabidopsis. Moreover, this is consistent with the high expression levels of genes encoding enzymes for ethylene and polyamine biosynthesis in the Plantago vasculature.
Finally, we found vasculature-specific expression also for several other genes that were not previously described to be vascular specific. For example, vascular-specific expression has so far only been shown for a β-amylase in Streptanthus tortuosus (Wang et al., 1995), but not for α-amylase, and vascular-specific expression for Suc phosphate synthase has so far not been described in any other plant. The vasculature specificity of both genes has been confirmed by macroarray analyses (Fig. 4) and by quantitative RT-PCR (Fig. 3B).
Our data also show vascular-specific expression for numerous genes encoding so far uncharacterized proteins or for novel genes. Analysis of their specific roles within the vasculature will be a major challenge for the next years.
Transformation of Common Plantain with a PmPLT1 Promoter-GUS Construct
Analyses of promoter activities from Arabidopsis genes that are homologs of identified Plantago vascular genes (Fig. 5) revealed vascular-specific expression for AtMTK1 and for the gene for the tetracycline transporter-like protein At2g16970. This demonstrates that careful predictions on the expression pattern of a gene of interest can be made from data obtained with homologous genes in other plant species. However, for numerous questions, it will be essential to modulate the expression of a gene directly in Plantago.
For example, in contrast to Arabidopsis, which translocates Suc and small amounts of raffinose in its phloem (Haritatos et al., 2000), members of several plant families (e.g. of Rosaceae, Apiaceae, or Plantaginaceae) translocate Suc together with linear polyols. Also common plantain translocates Suc plus the linear polyol sorbitol (Lohaus and Fischer, 2002). Ramsperger-Gleixner et al. (2004) had shown a differential regulation for the genes encoding of the CC-specific Suc transporter PmSUC2 and the CC-specific polyol transporter PmPLT1 during the early stages of phloem development. Only recently, B. Pommerrenig, F.S. Papini-Terzi, and N. Sauer (unpublished data) were able to show that these genes are differentially regulated also in response to increased salinity. In fact, the mRNAs of these CC-specific Suc transporters were identified within the EST project. For the PmPLT1 gene, more than 1,400 bp of 5′-flanking sequences were isolated by TAIL-PCR, cloned in front of the GUS reporter gene, and the resulting construct was used to establish transformation of Plantago plants.
Besides the mere fact that this promoter shows the expected GUS staining in the Plantago vasculature (Fig. 6), the successful and simple transformation of common plantain will provide the basis for further analyses. The physiological roles of the identified proteins and metabolic pathways can now be studied by overexpression of the respective genes or by down-regulation of their mRNA levels using RNAi or antisense RNA constructs.
MATERIALS AND METHODS
Strains
Common plantain (Plantago major) plants were grown in a greenhouse on potting soil. Escherichia coli strain DH5α (Hanahan, 1983) was used for basic cloning steps. E. coli SOLR and XL1-BlueMRF′ (both strains from Stratagene) were used for the construction of the Plantago cDNA library. Arabidopsis (Arabidopsis thaliana) Columbia wild-type plants were transformed with Agrobacterium tumefaciens strain GV3101. Transgenic Arabidopsis plants used for GUS histochemical analyses were grown under short-day conditions (8-h light/16-h dark) in growth chambers.
Isolation of Total and Poly(A+) RNA
Total RNA from pure Plantago major vascular bundles (6 g) was isolated as described (Sauer et al., 1990). Poly(A+) RNA was purified using the Oligotex mRNA purification system (Qiagen).
cDNA Library Construction and Sequencing of ESTs
A directionally cloned (EcoRI/XhoI) cDNA library was generated from poly(A+) RNA using a λ-Uni-Zap XR cDNA synthesis kit (Stratagene), according to the manufacturer's instructions. The primary cDNA library (1.34 × 106 plaque-forming units) was amplified once, mass excised in vivo, and the resulting plasmids (pBluescript II SK−) propagated in the E. coli SOLR host strain (Stratagene). The number of blue colonies (no insert) was negligible; 7,776 white colonies were transferred to 96-well plates and stored at −80°C.
cDNA inserts were PCR amplified with pUC-forward (5′-ACGACGTTGTAAAACGACGGCCAG-3′) and pUC-reverse (5′-TTCACACAGGAAACAGCTATGACC-3′) primers using whole cells as a template. PCR products were treated with exonuclease I and shrimp alkaline phosphatase for 30 min at 37°C and purified using 96-well purification plates (Edge Biosystems). Sequencing of the PCR products (Applied Biosystems 3700 automated DNA-sequencing system) was performed using the T3 forward primer. Vector and adaptor sequences were removed from the raw sequences. Resulting sequences were aligned to identify identical or overlapping ESTs for contig formation. Chimeric clones were identified by the presence of a cloning adaptor within the sequence.
Resulting contig and singlet sequences were compared against the Swissprot protein database using BLASTX (Altschul et al., 1997) with a cutoff E-value of 10−5. All sequences were further compared against identified coding sequences (−introns, −untranslated regions) and against genomic sequences (+introns, +untranslated regions) of Arabidopsis using BLASTN (Altschul et al., 1997). Tentative gene ontologies were assigned based on top BLASTX alignments or BLASTN alignments, respectively.
RT-PCR Analysis and TAIL-PCR
For analysis of the vascular specificity of different Plantago genes, total RNA was extracted from isolated vascular bundles or from nonvascular tissue of Plantago using TrizolR reagent (Invitrogen). cDNA was synthesized from 5 μg of RNA in a total volume of 20 μL using the RevertAid H Minus first-strand cDNA synthesis kit (Fermentas). From these reactions, 0.5 μL were used as PCR templates with gene-specific 20-bp primers.
Promoter sequences were PCR amplified from Arabidopsis genomic DNA using the following primers: MTK-fwd (5′-CAAATCATTTTTATACCTCGATGC-3′) and MTK-rev (5′-GGCTTTTGGTACAAATTTTCAGA-3′) for the amplification of a 1,220-bp promoter fragment of the Arabidopsis AtMTK gene, At5g62890-fwd (5′-CCCGACACTTAGAAATGTGTATCA-3′) and At5g62890-rev (5′-CACAGAGAGAGAGAGAGGGAGAA-3′) for the amplification of a 1,343-bp promoter fragment of the Arabidopsis At5g62890 gene, and At2g16970-fwd (5′-CTCTCTCTAAGCTTTCAAGGGTTATGTGAAATG GTA-3′) and At2g16970-rev (5′-CAAGTCTATATTCCTCCATGGCT-3′) for the amplification of a 1,104-bp promoter fragment of the Arabidopsis At2g16970 gene.
The promoter sequence of the gene encoding the Plantago contig P12.0.C65 was isolated by TAIL-PCR (Liu et al., 1995) using the gene-specific primers P-1 (5′-GAATACTTGTTCAGGATTCATGTG G-3′), P-2 (5′-GGCTGAGTGGTGGTAGCTTT-3′), and P-3 (5′-GACGACCACGTGGGTATGTT-3′), and degenerate primers as described (Liu et al., 1995) and sequenced. Based on the TAIL-PCR-derived sequence, 1,428 bp of 5′-flanking sequence were PCR amplified from the Plantago genomic sequence using the primers P12.0.C65-fwd (5′-GTCTGCATGCTCTAGATGAAACCAGCGCAAAAC-3′) and P12.0.C65-rev (5′-TGGGTATGTTACCATGGTCTTGTT-3′). All promoter fragments were cloned into the XbaI and NcoI sites of the plant transformation vector pAF16 (Stadler et al., 2005).
Northern-Blot and Dot-Blot Analysis
For northern-blot analyses, 10 μg of total RNA were separated on denaturing agarose gels and transferred to nylon membranes as described (Maniatis et al., 1982). Radiolabeled restriction fragments were used as probes (a 366-bp EcoRI/ClaI fragment for PmRBCS1, a 756-bp EcoRI/XhoI fragment for PmMT2, a 600-bp EcoRI fragment for PmMT1, and a 900-bp EcoRI/XhoI fragment for RNA with the viral polyprotein-like sequence). Hybridizations were performed at 42°C in 50% formamide, 5 × SSC (20 × SSC = 3 m NaCl, 0.3 m sodium citrate, pH 7.0), 5 × Denhardt's solution, 0.1% SDS, 50 mm sodium phosphate (pH 8), 0.1% sodium pyrophosphate, and 50 μg mL−1 salmon sperm DNA. Filters were washed for 30 min at 42°C in 2 × SSC, 0.1% SDS, followed by 30-min incubation at 42°C in 0.5 × SSC, 0.1% SDS, and 15-min incubation at 65°C in 0.1 × SSC, 0.1% SDS. Signals were detected in a phosphoimager (BAS 2000; Fuji Photo Film) and quantified using TINA quantification software (Raytest Isotopenmessgeräte GmbH).
For macroarray analyses, 600 ng of PCR products of the cDNA fragments were spotted on nylon membranes and UV cross linked. Duplicates were spotted for each EST. PCR fragments for 216 different ESTs were spotted on the macroarrays used to study vascular specificity. PCR fragments for 108 different ESTs were spotted on those macroarrays that were used to study the salt responsiveness of gene expression. Hybridizations with 32P-labeled cDNAs were performed for 40 h at 42°C in 50% formamide, 5 × SSC, 5 × Denhardt's solution, 0.1% SDS, 50 μg mL−1 salmon sperm DNA, and 20 μg mL−1 polyuridylic acid. Filters were washed as described for the northern-blot analyses. Signals were detected as described for the northern blots.
Isolation of the PmPLT1 Promoter and Construction of the PmPLT1 Promoter-GUS Construct
The PmPLT1 promoter was obtained by two rounds of TAIL-PCR (Liu et al., 1995). The first round with three PmPLT1 cDNA-specific primers (PmPLT1c + 136r, 5′-TTGCTAAAGCATACTTGTTCCTC-3′; PmPLT1c + 103r, 5′-TCTTAGGGAGTGTGTCGAGAG TC-3′; PmPLT1c + 18r, 5′-TGGTGATCAGCAGTCATAGTTGA-3′) and the degenerate primer AD7 [5′-A(A/T)GCANGNC(A/T)GANATA-3′] yielded a 700-bp promoter fragment. The second round with three PmPLT1 promoter-specific primers (PmPLT1pII-P1, 5′-GTATGTTTTACAACGTTGAAGCTG-3′; PmPLT1pII-P2, 5′-GAAGCTGCTTTATAATAGCTGGAAA-3′; PmPLT1pII-P3, 5′-TTCCGCCAATTTTCTTTCTC-3′) and the same degenerate primer yielded 1,475 bp of PmPLT1 promoter sequence. Based on the TAIL-PCR sequences, a 1,475-bp PmPLT1 promoter was PCR amplified (PmPLT1prom-5′, 5′-GTCTGCATGCTCTAGATCAGATTTTGAACATGTCCGTTA-3′ [introduces SphI and XbaI cloning sites at the 5′ end of the promoter] and PmPLT1prom-3′, 5′-GTCTCCATGGTTGAAAACAAGTAGTGTTGGTTTAAT-3′ [introduces a NcoI cloning site at the position of the start ATG]). This fragment was cloned in front of the GUS open reading frame and inserted into the plant transformation vector pAF16 (Stadler et al., 2005) that confers BASTA resistance.
Regeneration of Plantago Plants from Callus Tissue and Transformation of Plantago by Modified Floral-Dip Technique
Callus tissue was obtained from root sections of Plantago plants that were grown on Murashige and Skoog medium (Murashige and Skoog, 1962) under sterile conditions. To this end, sterile root fragments (1–2 cm long) were cultivated at 22°C on Murashige and Skoog medium supplemented with 6-furfurylaminopurine (kinetin; 0.1 mg/L; Sigma-Aldrich) and 2,4-dichlorophenoxy acetic acid (0.5 mg/L; Sigma-Aldrich). For longer propagation, calli were transferred to new medium every 3 to 4 weeks.
Intact Plantago plants were regenerated from root-derived callus tissue in three steps. First, calli were transferred to shoot-inducing medium plates [containing Murashige and Skoog medium with 1-phenyl-3-(1,2,3-thiadiazol-5-yl)-urea (2 mg/L; Sigma-Aldrich)], where first leaves form after 3 weeks. For further growth, shoots were transferred in a second step to plastic boxes (5 cm high) with shoot-inducing medium. In the third and final step, shoots were transferred to boxes with root-inducing medium (Murashige and Skoog medium without hormones), where roots form after 2 to 3 weeks.
Transformation of common plantain plants was performed with Agrobacterium tumefaciens strain GV3101 (Holsters et al., 1980). The used Plantago plants were 12 weeks old and had been grown in the greenhouse. Sucrose (final concentration 75 g/L) and Silwet (Lehle Seeds; final concentration 2 mL/L) were added to a fresh 400-mL overnight culture right before dipping Plantago flowers.
Dipping was performed for 5 min under vacuum. Inflorescences with fully developed female flowers were used for the dipping procedure. Twenty-four and 48 h after the first dipping, dipping was repeated. Between dipping, plants were kept on the lab bench with no extra light. After the last dipping, plants were covered with plastic wrap to maintain sufficiently high humidity for Agrobacterium infection and transferred to the greenhouse (the wrap was removed after 2 d). After about 4 weeks, the dipped inflorescences had developed ripe seeds. At that stage, inflorescences were covered with a paper bag and seeds were harvested. BASTA-resistant offspring were identified by spraying soil-grown seedlings three times with BASTA (250 μL Liberty SL 200 g/L; ArgEvo).
Genomic DNA was isolated (Aitchitt et al., 1993) from leaf tissue of resistant plants and PCR with primers for the GUS gene (GUS-5′ + 996-fwd, 5′-CCCTTACGCTGAAGAGATGC-3′; GUS-3′ + 1,396-rev, 5′-GGCACAGCACATCAAAGAGA-3′) was performed to check for successful integration of the foreign DNA.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Table I.
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. SA 382/15 to N.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Norbert Sauer (nsauer@biologie.uni-erlangen.de).
References
- Aitchitt M, Ainsworth CC, Thangavelu M (1993) A rapid and efficient method for the extraction of total DNA from mature leaves of the date palm (Phoenix dactylifera L.). Plant Mol Biol Rep 11: 317–319 [Google Scholar]
- Allona I, Quinn M, Shoop E, Swope K, St Cyr S, Carlis J, Riedl J, Retzel E, Campbell MM, Sederoff R, et al (1998) Analysis of xylem formation in pine by cDNA sequencing. Proc Natl Acad Sci USA 95: 9693–9698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano T, Masumura T, Kusano H, Kikuchi S, Kurita A, Shimada H, Kadowaki K (2002) Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: toward comprehensive analysis of the genes expressed in the rice phloem. Plant J 32: 401–408 [DOI] [PubMed] [Google Scholar]
- Balachandran S, Xiang Y, Schobert C, Thompson GA, Lucas WJ (1997) Phloem sap proteins from Cucurbita maxima and Ricinus communis have the capacity to traffic cell to cell through plasmodesmata. Proc Natl Acad Sci USA 94: 14150–14155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes A, Bale J, Constantinidou C, Ashton P, Jones A, Pritchard J (2004) Determining protein identity from sieve element sap in Ricinus communis L. by quadrupole time of flight (Q-TOF) mass spectrometry. J Exp Bot 55: 1473–1481 [DOI] [PubMed] [Google Scholar]
- Barth I, Meyer S, Sauer N (2003) PmSUC3: characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15: 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D, Kemper E, Schell J, Mastersen R (1992) New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol Biol 20: 1195–1197 [DOI] [PubMed] [Google Scholar]
- Beers EP, Zhao C (2001) Arabidopsis as model for investigating gene activity and function in vascular tissues. In N Morohoshi, A Komamine, eds, Molecular Breeding of Woody Plants. Elsevier Science, New York, pp 43–52
- Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA (1997) Laser capture microdissection: molecular analysis of tissue. Science 278: 1481–1483 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Crane C, Wright E, Dixon RA, Wang ZY (2006) Transgenic Medicago truncatula plants obtained from Agrobacterium tumefaciens-transformed roots and Agrobacterium rhizogenes-transformed hairy roots. Planta 223: 1344–1354 [DOI] [PubMed] [Google Scholar]
- Cronin S, Verchot J, Haldeman-Cahill R, Schaad MC, Carrington JC (1995) Long-distance movement factor: a transport function of the Potyvirus helper component proteinase. Plant Cell 7: 549–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt ND, Sussman MR (1995) Immunocytological localization of an epitope-tagged plasma membrane proton pump (H+-ATPase) in phloem companion cells. Plant Cell 7: 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz M, Stolz J, Sauer N (1994) A phloem specific sucrose-H+ symporter from Plantago major L. supports the model of apoplastic phloem loading. Plant J 6: 697–706 [DOI] [PubMed] [Google Scholar]
- Hanahan D (1983) Studies on transformation of E. coli with plasmids. J Mol Biol 166: 557–580 [DOI] [PubMed] [Google Scholar]
- Haritatos E, Ayre BG, Turgeon R (2000) Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol 123: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzberg M, Aspeborg H, Schrader J, Andersson A, Erlandsson R, Blomqvist K, Bhalerao R, Uhlen M, Teeri TT, Lundeberg J, et al (2001) A transcriptional roadmap to wood formation. Proc Natl Acad Sci USA 98: 14732–14737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, Silva B, Van Vliet F, Genetello C, De Block M, Dhaese P, Depicker A, Inze D, Engler G, Villarroel R, et al (1980) The functional organization of the nopaline A. tumefaciens plasmid pTiC58. Plasmid 3: 212–230 [DOI] [PubMed] [Google Scholar]
- Hudgins JW, Franceschi VR (2004) Methyl jasmonate-induced ethylene production is responsible for conifer phloem defense responses and reprogramming of stem cambial zone for traumatic resin duct formation. Plant Physiol 135: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwatari Y, Fujiwara T, McFarland KC, Nemoto K, Hayashi H, Chino M, Lucas WJ (1998) Rice phloem thioredoxin has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta 205: 12–22 [DOI] [PubMed] [Google Scholar]
- Ivashikina N, Deeken R, Ache P, Kranz E, Pommerrenig B, Sauer N, Hedrich R (2003) Isolation of AtSUC2 promoter-GFP-marked companion cells for patch-clamp studies and expression profiling. Plant J 36: 931–945 [DOI] [PubMed] [Google Scholar]
- Karlsson M, Melzer M, Prokhorenko I, Johansson T, Wingsle G (2005) Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J Exp Bot 56: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Kehr J (2003) Single cell technology. Curr Opin Plant Biol 6: 617–621 [DOI] [PubMed] [Google Scholar]
- Keller B, Sauer N, Lamb CJ (1988) Glycine-rich cell wall proteins in bean: gene structure and association of the protein with the vascular system. EMBO J 7: 3625–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwart M, Hirner B, Hummel S, Frommer WB (1993) Differential expression of two related amino acid transporters with differing substrate specificity in Arabidopsis thaliana. Plant J 4: 993–1002 [DOI] [PubMed] [Google Scholar]
- Liu Y-G, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Liu ZZ, Wang JL, Huang X, Xu WH, Liu ZM, Fang RX (2003) The promoter of a rice glycine-rich protein gene, Osgrp-2, confers vascular-specific expression in transgenic plants. Planta 216: 824–833 [DOI] [PubMed] [Google Scholar]
- Lohaus G, Fischer K (2002) Intracellular and intercellular transport of nitrogen and carbon. In C Foyer, G Noctor, eds, Advances in Photosynthesis. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 239–263
- Maniatis T, Fritsch EF, Sambrook J (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Marten I, Hoth S, Deeken R, Ache P, Ketchum KA, Hoshi T, Hedrich R (1999) AKT3, a phloem-localized K+ channel, is blocked by protons. Proc Natl Acad Sci USA 96: 7581–7586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentewab A, Stewart CN Jr (2005) Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol 23: 1177–1180 [DOI] [PubMed] [Google Scholar]
- Meyer S, Lauterbach C, Niedermeier M, Barth I, Sjolund RD, Sauer N (2004) Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol 134: 684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Melzer M, Truernit E, Hümmer C, Besenbeck R, Stadler R, Sauer N (2000) AtSUC3, a gene encoding a new Arabidopsis sucrose transporter, is expressed in cells adjacent to the vascular tissue and in a carpel cell layer. Plant J 24: 869–882 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Noiraud N, Delrot S, Lemoine R (2000) The sucrose transporter of celery: identification and expression during salt stress. Plant Physiol 122: 1447–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiraud N, Maurousset L, Lemoine R (2001) Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13: 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Park S, Han KH (2003) Transcriptional regulation of secondary growth in Arabidopsis thaliana. J Exp Bot 54: 2709–2722 [DOI] [PubMed] [Google Scholar]
- Parsons BL, Mattoo AK (1994) A wound-repressible glycine-rich protein transcript is enriched in vascular bundles of tomato fruit and stem. Plant Cell Physiol 35: 27–35 [PubMed] [Google Scholar]
- Pharr DM, Stoop JMH, Williamson JD, Studer-Feusi ME, Massel MO, Conkling MA (1995) The dual role of mannitol as osmoprotectant and photoassimilate in celery. HortScience 30: 1182–1188 [Google Scholar]
- Ramsperger-Gleixner M, Geiger D, Hedrich R, Sauer N (2004) Differential expression of sucrose transporter and polyol transporter genes during maturation of common plantain companion cells. Plant Physiol 134: 147–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser U, Keller B (1992) Ultrastructural localization of a bean glycine-rich protein in unlignified primary walls of protoxylem cells. Plant Cell 4: 773–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarni F, Grand C, Boudet AM (1984) Purification and properties of cinnamoyl-CoA reductase and cinnamyl alcohol dehydrogenase from poplar stems (Populus × euramericana). Eur J Biochem 139: 259–265 [DOI] [PubMed] [Google Scholar]
- Sasaki T, Chino M, Hayashi H, Fujiwara T (1998) Detection of several mRNA species in rice phloem sap. Plant Cell Physiol 39: 895–897 [DOI] [PubMed] [Google Scholar]
- Sauer N, Friedländer K, Gräml-Wicke U (1990) Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J 9: 3045–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer N, Stolz J (1994) SUC1 and SUC2: two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine tagged protein. Plant J 6: 67–77 [DOI] [PubMed] [Google Scholar]
- Sauter M, Cornell KA, Beszteri S, Rzewuski G (2004) Functional analysis of methylthioribose kinase genes in plants. Plant Physiol 136: 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Brandner J, Schulz A, Gahrtz M, Sauer N (1995) Phloem loading by the PmSUC2 sucrose carrier from Plantago major L. occurs into companion cells. Plant Cell 7: 1545–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N (2005) Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J 41: 319–331 [DOI] [PubMed] [Google Scholar]
- Sterky F, Regan S, Karlsson J, Hertzberg M, Rohde A, Holmberg A, Amini B, Bhalerao R, Larsson M, Villarroel R, et al (1998) Gene discovery in the wood-forming tissues of poplar: analysis of 5,692 expressed sequence tags. Proc Natl Acad Sci USA 95: 13330–13335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E, Sauer N (1995) The promoter of the Arabidopsis thaliana sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: evidence for phloem loading and unloading by SUC2. Planta 196: 564–570 [DOI] [PubMed] [Google Scholar]
- Vilaine F, Palaqui JC, Amselem J, Kusiak C, Lemoine R, Dinant S (2003) Towards deciphering the phloem: a transcriptome analysis of the phloem of Apium graveolens. Plant J 36: 67–81 [DOI] [PubMed] [Google Scholar]
- Walz C, Giavalisco P, Schad M, Juenger M, Kehr J (2004) Proteomics of curcurbit phloem exudate reveals a network of defense proteins. Phytochemistry 65: 1795–1804 [DOI] [PubMed] [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J (2002) Evidence for the presence and activity of a complete antioxidant defense system in mature sieve tubes. Plant J 31: 189–197 [DOI] [PubMed] [Google Scholar]
- Wang Q, Monroe J, Sjolund RD (1995) ldentification and characterization of a phloem-specific β-amylase. Plant Physiol 109: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung KH, Yang SF (1982) Methionine synthesis from 5-methylthioribose in apple tissue. Biochem Biophys Res Commun 104: 771–777 [DOI] [PubMed] [Google Scholar]
- Zamski E, Guo WW, Yamamoto YT, Pharr DM, Williamson JD (2001) Analysis of celery (Apium graveolens) mannitol dehydrogenase (Mtd) promoter regulation in Arabidopsis suggests roles for MTD in key environmental and metabolic responses. Plant Mol Biol 47: 621–631 [DOI] [PubMed] [Google Scholar]
- Zamski E, Yamamoto YT, Williamson JD, Conkling MA, Pharr DM (1996) lmmunolocalization of mannitol dehydrogenase in celery plants and cells. Plant Physiol 112: 931–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Craig JC, Petzold HE, Dickerman AW, Beers EP (2005) The xylem and phloem transcriptomes from secondary tissues of the Arabidopsis root-hypocotyl. Plant Physiol 138: 803–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Johnson BJ, Kositsup B, Beers EP (2000) Exploiting secondary growth in Arabidopsis: construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol 123: 1185–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]