Abstract
Strains of Theiler's murine encephalomyelitis virus (TMEV) are divided into two subgroups, TO and GDVII. TMEV strains show subgroup-specific virus growth and cell tropism and induce subgroup-specific diseases. Using site-directed mutagenesis, we demonstrated that the amino acid at position 57 of the leader protein (L57), which is located at the most N-terminal part of the polyprotein, regulates subgroup-specific virus growth on BHK-21 cells. Further study suggested that L57 may regulate viral RNA encapsidation, although it does not affect the synthesis of viral proteins or the assembly of viral intermediates.
Theiler's murine encephalomyelitis virus (TMEV) belongs to the genus Cardiovirus of the family Picornaviridae and is divided into two subgroups on the basis of their different biological activities (23, 26, 30). GDVII subgroup strains cause acute and fatal polioencephalomyelitis in mice following intracerebral inoculation. In the few surviving mice, no virus persistence or demyelination is observed. In contrast, TO or DA subgroup strains induce a biphasic central nervous system disease. In the acute phase, the virus infects mainly neurons and causes mild polioencephalomyelitis. During the chronic phase, the virus persists in the spinal cord of susceptible strains of mice and causes demyelination. In addition, virus growth differs in vitro between strains of the two subgroups. On baby hamster kidney (BHK-21) cells, GDVII subgroup strains grow well at a titer that is 10-fold higher than that of DA subgroup strains. Furthermore, GDVII subgroup strains produce larger plaques (2.5 mm in diameter), whereas DA subgroup strains produce smaller plaques (less than 0.5 mm) (18).
Leader protein (L) is located at the most N-terminal portion of the polyprotein in members of the Aphthovirus and Cardiovirus genera of the picornaviruses (17). L is known to enable virus growth on specific cell lines because it inhibits alpha/beta interferon (IFN-α/β) transcription (35). Although the capsid proteins have over 94% homology at the amino acid level, the homology of L between TO and GDVII subgroup strains is only 85% (20). The sequence difference between leader proteins of the two subgroups may account for TMEV subgroup-specific biological activities.
There are 11 amino acid (aa) residue differences distributed over the total 76 aa of L between the two subgroup strains (20). One of the amino acid residues is proline, which is known to have an important influence on protein structure; cis-trans isomerization at a proline residue alters the protein structure. In addition, proline has the highest reverse turn probability (9). GDVII L contains eight proline residues, whereas DA L contains seven, since aa position 57 (L57) is a proline in the case of the GDVII strain but is a serine in the case of the DA strain.
We questioned whether the difference in L57 regulates the structure of L and thereby affects TMEV subgroup-specific biological activities. Therefore, by generating a series of mutant viruses, we studied the possibility that L57 is a determinant of subgroup-specific virus growth.
MATERIALS AND METHODS
Cells and virus assay.
BHK-21 cells, a baby hamster kidney-derived fibroblast cell line permissive for TMEV infection, were maintained in Eagle's minimum essential medium (Nissui, Tokyo, Japan) supplemented with 5% calf serum (Invitrogen, Carlsbad, CA). A standard plaque assay was performed at 37°C for 3 days.
Mutant viruses.
A series of mutant cDNAs are shown in Fig. 1. pDALPro and pDALPro/L*-1 were generated by site-directed mutagenesis using a full-length DA infectious cDNA (pDAFL3) (29) and pDAL*-1 (8), respectively, as a template. pDAL*-1 is a mutant construct in which the only difference from pDAFL3 is a change of the AUG at nucleotide (nt) 1079, the initiation codon of the L* protein, to ACG.
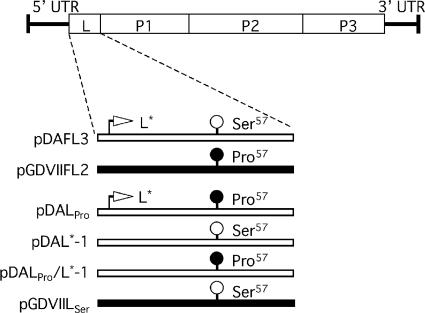
FIG. 1.
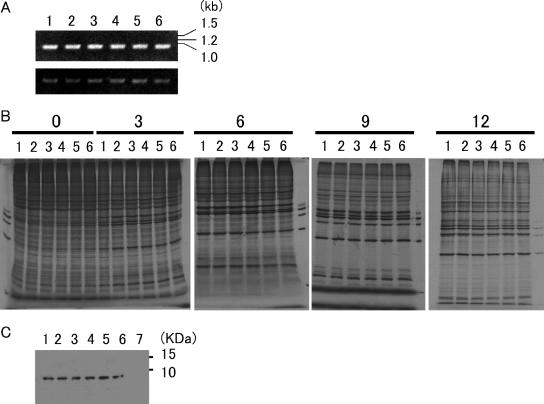
Series of parental and mutant cDNAs. The position of the TMEV coding area is shown at the top. pDAFL3 and pGDVIIFL2 are parental cDNAs derived from DA and GDVII, respectively. DA and GDVII genomes are shown as open and solid bars, respectively. pDAL*-1 has a mutation changing the initiation codon AUG of L* to ACG; therefore, it fails to synthesize L*. Mutant constructs DALPro and DALPro/L*-1 were generated by introducing a mutation at nt 1234, leading to a predicted change in the amino acid residue at position 57 (L57) from the serine of DA to proline. Another mutant construct, GDVIILSer, was generated by introducing a mutation into nt 1236, leading to a predicted change in L57 from the proline of GDVII to serine. UTR, untranslated region.
In the case of pDALPro, the segment spanning from XhoI to MscI sites was amplified by PCR using the following primers, containing a single nucleotide substitution changing serine to proline: primers A (5′-CTC GAG CAA TCA ACC TGA AAC-3′) and B (5′-AGA CAC CTC CGT CCT CGC CAG-3′) and primers C (5′-GTT TCA GGT TGA TTG CTC GAG-3′) and D (5′-AAC ATG CAG AGT AAC GCG AAG-3′) (the nucleotide inducing a single amino acid substitution of serine to proline is underlined). The PCR products were hybridized after purification using a QIAquick gel extraction kit (QIAGEN, United Kingdom). The hybridized DNA was then amplified by PCR with primers A and D. The PCR product and pDAFL3 were digested with XhoI and MscI (TOYOBO, Osaka, Japan), and the digested PCR fragment was replaced with the corresponding fragment in pDAFL3. In the case of pDAL*-1/LPro, the XhoI- and MscI-digested fragment amplified by PCR as described above was replaced with the corresponding fragment in pDAL*-1. This point mutation of L did not change the amino acid sequence of L*, which is synthesized out of frame with the polyprotein only in TO subgroup strains (23, 25, 30).
pGDVIILSer was constructed by using a full-length GDVII infectious cDNA, pGDVIIFL2 (11), as a template. The segment spanning from KpnI to MscI sites was also amplified by PCR using the following primers: primers E (5′-TGA ACC CCT GAA TGG CGA TC-3′) and F (5′-CGG ACA CTA GTA CTC AAT CTC-3′) and primers G (5′-GAG ATT GAG TAC TAG TGT CCG-3′) and H (5′-GAT TGT CAG CAT TGA TCT TGG TG-3′) (the nucleotide inducing a single amino acid substitution of proline to serine is underlined). The PCR product and pGDVIIFL2 were digested with KpnI and MscI (TOYOBO), and the digested PCR fragment was replaced with the corresponding fragment in pGDVIIFL2.
In pDALPro, pDAL*-1/LPro, and pGDVIILSer, PCR-amplified segments and the regions adjacent to the sites of ligation were sequenced completely on both strands. No unexpected mutations were observed. The plasmids were linearized with XbaI (TOYOBO), and viral RNA was synthesized by T7 RNA polymerase (Promega, Madison, WI). BHK-21 cells were then transfected with the synthesized RNA using Lipofectin (Invitrogen) according to the manufacturer's instructions. Virus was purified by a standard plaque purification technique described previously (2). The sizes of plaques before and after the purification were identical. Purified virus was propagated on BHK-21 cells and used in the following experiments.
Kinetics of virus growth.
The kinetics of growth of parental and mutant viruses on BHK-21 cells were examined as previously described (22). A 35-mm-diameter plastic culture dish containing 1 × 106 cells was infected at a multiplicity of infection (MOI) of 10 PFU per cell. At the indicated times, the infected cells with supernatants were harvested, and plaque formation on BHK-21 cells was assayed after freezing and thawing three times.
RT-PCR.
BHK cells were infected with parental and mutant viruses at an MOI of 10 PFU per cell. Total RNA was isolated from the infected cells at the indicated times (0, 3, 6, 9, and 12 h) after infection using TRIzol reagent (Invitrogen) according to the manufacturer's instruction. Viral RNA synthesis and mRNA expression of mouse β-actin were analyzed by semiquantitative reverse transcription (RT)-PCR. cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen). The 1/20 volume of each RT mixture was subjected to the PCR (94°C for 10 s, 60°C for 20 s, and 72°C for 60 s) using the following primers: 5′-TCAACTCTGACATCCTCACTCTCG-3′ and 5′-GACGTCCGTGAACCTTGTAGTCAA-3′ for TMEV RNA and 5′-ATGGATGACGATATCGCT-3′ and 5′-ATGAGGTAGTCTGTCAGGT-3′ for mouse β-actin as a control. The number of cycles was determined to be 32, since the preliminary data showed that the cycle number was sufficient before the plateau of amplification.
Pulse-labeling experiment for viral and cellular proteins.
BHK-21 cells were infected with virus at an MOI of 10 PFU/cell. The infected cells were incubated in methionine- and cysteine-free medium (Sigma-Aldrich, St. Louis, MO) for 30 min prior to the indicated times, followed by incubation for 1 h in medium containing 75 μCi of l-[35S]methionine and l-[35S]cysteine (GE Healthcare Bio-Science, Piscataway, NJ). The radiolabeled cells were scraped, washed twice with ice-cold phosphate-buffered saline (PBS), and lysed in lysis buffer (5 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% sodium deoxycholate, 1 mM EDTA, 1% NP-40,) for 30 min on ice. After centrifugation at 15,000 rpm for 10 min at 4°C, the radiolabeled proteins in the supernatant were separated by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis (PAGE) and analyzed by autoradiography. The analysis was performed in triplicate.
Determination of viral assembly intermediates by sucrose gradient.
Subconfluent BHK-21 cells (3 × 106 cells in a 25-cm2 flask) were infected with viruses at an MOI of 10 PFU per cell and incubated for 3 h. The infected cells were incubated in methionine- and cysteine-free medium for 30 min prior to the addition of 100 μCi of l-[35S]methionine and l-[35S]cysteine. Following radiolabeling for 4 h, the cells were lysed in 400 μl of lysis buffer as described above. The supernatants were layered onto 5 to 20% or 15 to 30% linear sucrose gradients in lysis buffer (11 ml). Gradients were centrifuged in an SW41 rotor (Beckman, Fullerton, CA) at 35,000 rpm for 16 h or 18,000 rpm for 10 h. Fractions (0.3 ml each) were collected by pumping from the bottom of the sucrose gradient. An aliquot (0.1 ml) of each fraction was counted using a scintillation counter.
Western blotting.
The proteins extracted from 5 × 104 infected cells were separated on a Tris-Tricine gel and transferred onto a polyvinylidene difluoride membrane (Millipore, Billerica, MA). The membrane was blocked with 5% skim milk in Tris-buffered saline-Tween (50 mM Tris-HCl [pH 7.4], 0.45% sodium chloride, and 0.05% Tween 20) for 1 h and incubated at room temperature for 2 h with polyclonal rabbit anti-L antibody (see below), followed by incubation with horseradish peroxidase-conjugated donkey anti-rabbit immunoglobulin G antibody (Jackson Immunoresearch Laboratory, West Grove, PA) for 1 h. Signals were detected using ECL Western blotting detection reagents (GE Healthcare Bio-Science) according to the manufacturer's instructions. In order to generate anti-L antibody, L of the DA strain was expressed as a fusion protein with maltose-binding protein in a bacterial expression plasmid, pMALc2X (New England Bio Labs, Beverly, MA). The L fusion protein was used to immunize a rabbit. The anti-L antibody detected the L of both GDVII and DA strains.
Immunoprecipitation.
Lysis buffer (0.2 ml) was added to a part of each fraction (0.2 ml) of the sucrose gradients. The fractions were then incubated for 1 h at 4°C with protein G (GE Healthcare Bio-Science), which was incubated with the rabbit preimmune sera. The supernatants were separated by centrifugation and then incubated for 3 h at 4°C with protein G, which was incubated with polyclonal rabbit anti-GDVII antibody. The anti-GDVII antibody was raised against GDVII particles purified on a cesium chloride gradient and then dialyzed in PBS. This antibody detected DA as well as GDVII viral capsid proteins. The immunoprecipitates were washed twice with ice-cold PBS, and the proteins were then separated by SDS-15% PAGE. The gels were fixed and subjected to autoradiography.
RESULTS
Growth kinetics and plaque formation of parental and mutant viruses.
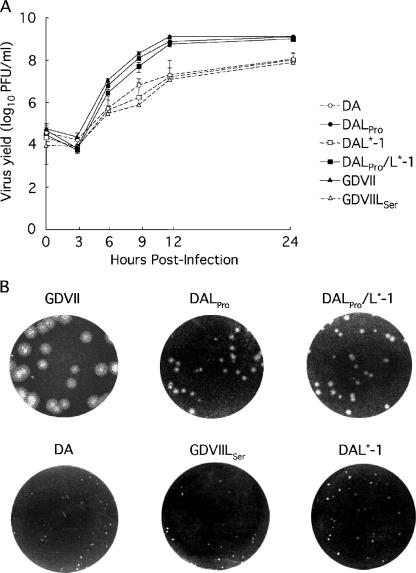
To examine the effect(s) of L57 (proline or serine) on virus growth on BHK-21 cells, growth kinetics were first evaluated (Fig. 2A). DALPro, DALPro/L*-1, and GDVII viruses, which contain proline at L57, showed very similar patterns of growth. The titers of these viruses ranged from 2.5 to 9.3 × 106 PFU/ml at 6 h postinfection (p.i.) and gradually increased thereafter, reaching a peak of 7.0 to 8.8 × 108 PFU/ml at 24 h p.i. On the other hand, the yield of viruses containing serine at L57 was lower; the titer of these viruses ranged from 2.8 to 4.9 × 105 PFU/ml at 6 h p.i. and reached a peak of only 6.0 to 8.4 × 107 PFU/ml at 24 h p.i. Despite a varying genome background, the overall yield of the viruses containing proline at L57 was approximately 10-fold higher than that of the viruses containing serine at the corresponding position. In addition, the size of the plaques correlated with virus growth (Fig. 2B). The plaque size of GDVII, DALPro, and DALPro/L*-1 viruses, in which L57 is proline, was 2 to 4 mm in diameter. In contrast, the size of plaques of DA, DAL*-1, and GDVIILSer viruses, in which L57 is serine, was 0.5 to 1 mm in diameter.
FIG. 2.
(A) Kinetics of parental and mutant virus growth on BHK-21. Cells were infected with each virus at an MOI of 10 PFU per cell. At the indicated times, the infected cells with the supernatants were harvested, and plaque formation on BHK-21 cells was assayed after freezing and thawing three times. The growth curves of DALPro (•), DALPro/L*-1 (▪), and GDVII (▴) viruses, in which L57 is proline, were drawn with solid lines and closed symbols. Those of DA (○), DAL*-1 (□), and GDVIILSer (▵) viruses, in which L57 is serine, were drawn with dotted liens and open symbols. (B) Plaque sizes of parental and mutant viruses on BHK-21. Results were obtained with a 10−6 or 10−7 dilution of parental and mutant viruses.
In summary, the growth of the DA strain is enhanced by the replacement of serine with proline at L57, and the growth of the GDVII strain is suppressed by the replacement of proline with serine. Therefore, the overall data suggest that L57 regulates virus growth on BHK-21 cells.
Viral RNA replication and viral protein synthesis.
To address which step of virus multiplication is influenced by L57, we examined viral RNA replication and protein synthesis of parental and mutant viruses on BHK-21 cells. We first evaluated viral RNA replication at 0, 3, 6, 9, and 12 h p.i. by semiquantitative PCR. No significant differences in the amplified bands were observed among all six viruses throughout the time course. The representative data at 6 h p.i. are shown in Fig. 3A.
FIG. 3.
Viral replication and protein synthesis in infected BHK-21 cells. (A) TMEV genomic RNA (upper panel) and β-actin mRNA (lower panel). BHK-21 cells were infected with DA, DALPro, DAL*-1, DALPro/L*-1, GDVII, or GDVIILSer virus at an MOI of 10 PFU per cell. At 6 h p.i., the infected cells were harvested, and total RNA was extracted from cells infected with parental and mutant viruses. Reverse-transcribed cDNA was subjected to RT-PCR as described in the text. (B) TMEV protein synthesis. BHK-21 cells were also infected with those viruses at an MOI of 10 PFU per cell. The number at the top indicates hours p.i. The cells were labeled with [35S]methionine for 1 h from the indicated time as described in the text. (C) L synthesis. Lysates from BHK-21 cells 7.5 h p.i. were subjected to Western blotting using anti-L antibody as described in the text. In A, B, and C, lane 1 shows DA-infected BHK-21 cells, lane 2 shows DAL*-1-infected cells, lane 3 shows DALPro-infected cells, lane 4 shows DALPro/L*-1-infected cells, lane 5 shows GDVII-infected cells, and lane 6 shows GDVIILSer-infected cells. Lane 7 in C shows noninfected BHK-21 cells as a negative control.
We next evaluated virus protein synthesis at 0. 3, 6, 9, and 12 h p.i. in a pulse-labeling experiment (Fig. 3B). At 3 h p.i., viral proteins started to be prominent, although cellular proteins were still synthesized. At 6 h p.i., host protein shutoff appeared, and in parallel, viral proteins became prominent. This tendency was observed up to 12 h p.i. There were no significant differences in the amounts of viral proteins synthesized by all six viruses that had either proline or serine at L57. The similarity in kinetics suggested that there were no significant differences in viral protein processing among the samples. To further confirm the processing of L, the amounts of L were examined by Western blotting using rabbit anti-L antibody at 7.5 h p.i. L was detected in equal amounts among all the viruses (Fig. 3C). In summary, the data demonstrate that L57 does not affect viral RNA replication and the synthesis and processing of viral proteins.
Virion assembly.
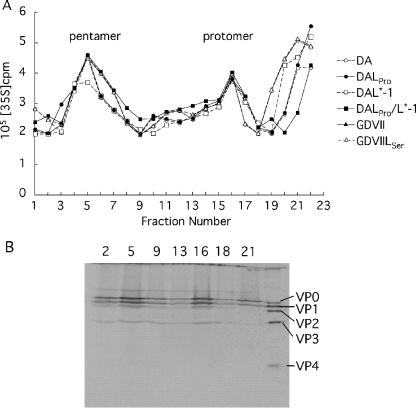
The efficiency of virion assembly may account for the regulation of virus growth by L57. Therefore, we examined the mature virion and assembly intermediates by sedimentation of infected BHK-21 cell lysates on sucrose density gradients. The fractionation from 5 to 20% sucrose gradients identified two peaks: one peak corresponded to the 5S protomer (fraction 16), and the other peak corresponded to the 14S pentamer (fractions 3 to 9). The patterns of the sedimentation profiles of the protomers and pentamers of six viruses were very similar (Fig. 4A).
FIG. 4.
Sucrose gradient sedimentation of capsid precursors synthesized during infection. (A) Virus assembly intermediates, the 5S protomer, and the 14S pentamer were detected in lysates of BHK-21 cells (3 × 106 cells) infected with DA, DALPro, DAL*-1, DALPro/L*-1, GDVIILSer, or GDVII virus at an MOI of 10 PFU per cell. The infected cells were radiolabeled as described in the text. Lysates were fractionated on 5 to 20% gradients. (B) Confirmation of viral assembly intermediates by immunoprecipitation. Representative data for DALPro-infected cells are shown. The numbers at the top indicate the fraction numbers. The fractions were immunoprecipitated and then analyzed by SDS-PAGE.
To confirm whether these fractions contain assembly intermediates, immunoprecipitation using anti-GDVII antibody followed by SDS-PAGE was performed. VP0, VP1, and VP3 were clearly detected in the corresponding fractions (Fig. 4B).
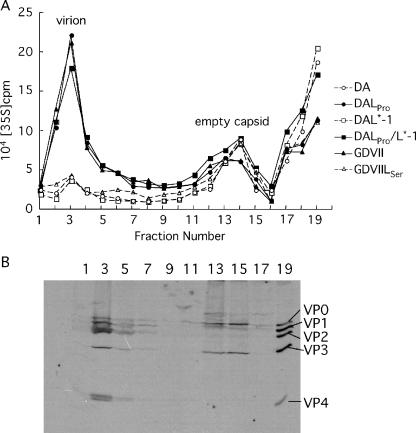
Since no significant differences were observed in the assembly of the protomer and pentamer among all the viruses, virions and empty capsids in the lysates of infected cells were examined (Fig. 5). The fractions from 15 to 30% sucrose gradients showed two peaks corresponding to empty capsids (fractions 17 to 19) and mature virions (fraction 3) (Fig. 5A). No significant differences were detected in the 75S empty capsid peaks among all the viruses. However, the counts of the mature virion peaks of DALPro, DALPro/L*-1, and GDVII viruses were approximately threefold higher than those of GDVIILSer, DAL*-1, and DA viruses. Fraction 3 was shown to contain mature virions, since VP2 and VP4 were detected by immunoprecipitation (Fig. 5B). Immunoprecipitation of fractions 17 and 19 also detected VP0, the precursor protein of VP2 and VP4, suggesting that fractions 17 to 19 contain empty capsids (Fig. 5B).
FIG. 5.
Analysis of virion assembly by sucrose gradient sedimentation. (A) The 75S empty capsid and 150S virion were detected in lysates of BHK-21 cells (3 × 106 cells) infected with parental and mutant viruses at an MOI of 10 PFU per cell. The infected cells were radiolabeled as described in the text. Lysates were fractionated on 15 to 30% gradients. (B) Confirmation of empty capsids and virions by immunoprecipitation. Representative data for DALPro-infected cells are shown. The numbers at the top indicate the fraction numbers. The fractions were immunoprecipitated and then analyzed by SDS-PAGE.
In summary, the amount of mature virions of DA was enhanced by the replacement of serine with proline at L57, and the amount of mature virions of GDVII was suppressed by the replacement of proline with serine at L57. These data suggest that L57 may regulate the efficiency of viral RNA encapsidation.
DISCUSSION
Using a series of mutant viruses, we demonstrated a correlation between virus growth on BHK-21 cells and L57 (proline or serine). GDVIILSer, in which proline is replaced with serine at L57, showed the same pattern of virus growth as DA. On the other hand, DALPro and DALPro/L*-1, in which proline is replaced with serine, showed the same pattern of enhanced virus growth as GDVII. However, the plaque sizes of DALPro and DALPro/L*-1 were not as large as that of GDVII (Fig. 2B). The size of the plaque reflects not only the growth of virus but also the cell-to-cell extension of virus. A sequence(s) other than that of L57 may affect the extension of virus. In addition, we examined the in vivo phenotypes of those mutants. DALPro and DALPro/L*-1 caused acute encephalomyelitis and mainly infected neurons. However, mice showed no apparent clinical signs in the acute stage and survived. On the other hand, GDVIISer caused acute fatal encephalomyelitis with severe neuronal damage, like the parental GDVII strain (data not shown). This indicates that L57 does not regulate in vivo phenotypes of TMEV. Further studies on in vivo phenotypes of those mutant viruses will be important and are now in progress.
TMEV L plays a role in host cell-restricted infection. L-deleted DA mutant virus cannot grow efficiently in L929 cells (16), which can produce IFN. L-deleted DA mutant virus growth is relatively unaffected in BHK-21 cells (6), which have a defect in IFN synthesis. The L of the DA strain inhibits the transcription of IFN-α/β by interfering in the nuclear localization of the transcription factor IFN regulatory factor 3 (10). The zinc-binding motif of L, Cys3-His5-Cys11-Cys14, which is located 44 aa upstream of L57, has the function of inhibiting IFN-α/β transcription (35). In this study, it was demonstrated that L regulates subgroup-specific virus growth on BHK-21 cells. Thus, the L of TMEV works as a multifunctional regulator.
In the present study, the growth pattern on BHK-21 cells was not changed in viruses that expressed or failed to express L*, suggesting that L* does not influence the virus growth on BHK-21 cells. These results show that L* is not important for TMEV subgroup-specific virus growth on BHK-21 cells, although L* is an essential protein for virus growth on macrophages (13, 24, 32) and plays a role in virus persistence and demyelination (8, 23, 30).
L57, whether it is proline or serine, did not affect viral RNA replication, the synthesis of viral proteins, or the assembly of viral intermediates; however, the peak of 150S mature virions was regulated with L57. The pulse-labeling experiment showed that the amounts of VP0, VP2, and VP4 were the same among all the viruses (Fig. 3B), suggesting that the cleavage of VP0 into VP2 and VP4, which is required for virus assembly, was normally processed. It was, however, previously reported that neither virions nor empty capsids were detected in L929 cells infected with L-deleted GDVII mutant virus (3). Those investigators speculated that L may be responsible for the assembly. Since L is present in our mutant viruses, amino acid residue(s) other than L57 may be involved in the formation of empty capsids.
In picornaviruses, it is thought that RNA encapsidation and the replication process of positive-strand RNA are linked to each other (21, 33). The newly synthesized progeny RNA is encapsidated into the capsid proteins as soon as RNA is synthesized. In the case of Aichi virus, which belongs to the genus Kobuvirus of the family Picornaviridae, L is involved in viral RNA replication and encapsidation, since the accumulation of empty capsids was observed in the case of an L-deleted mutant virus (31). In this study, the peaks of empty capsids are almost similar among all the viruses. In addition, no accumulation of empty capsids was observed in the viruses containing serine at L57. Therefore, TMEV L plays a role in viral RNA encapsidation distinct from that of Aichi virus L.
The action of L on viral RNA encapsidation also differs from that of poliovirus 2C protein (PV2C), which is responsible for the binding of newly synthesized viral RNA to the vesicular membranes and for the organization of the replication complex, thereby affecting the efficiency of viral RNA encapsidation (4, 5, 34). The membrane- and RNA-binding domains present within PV2C (28) were not found in TMEV L. Therefore, L of TMEV differs from PV2C in its role in viral RNA encapsidation. Further studies are required in order to clarify the mechanism(s) of L-regulated TMEV RNA encapsidation.
Proline has unique conformational and structural properties. Since L57 of GDVII is a proline residue that has the highest reverse turn probability, GDVII L may be structurally distinct from DA L (9). In addition, cis-trans isomerization at a proline residue alters the protein structure. This structural change plays an important role in protein-protein interactions, ligand-receptor binding, and interconversion of the open and closed states of the ion channel (7, 15, 19). A structural difference in L may alter the utilization and/or interference of cellular and viral proteins, resulting in the change of viral RNA encapsidation efficiency.
Another potentially important reason for the influence of L57 might relate to the phosphorylation of the serine. Phosphorylation of serine can regulate protein-protein interactions as well as kinase, phosphatase, and protease activities (1, 12, 14). Therefore, the phosphorylation of serine at position 57 may foster the binding of L to some host protein(s), resulting in a decrease in TMEV RNA encapsidation.
Finally, previous works (16, 27) reported that the entire substitution of DAL for GDVIIL does not give any differences in in vitro and in vivo phenotypes. As described above, a single amino acid change of L (either proline or serine) may change the structure and property of L or may be compensated for by another divergent amino acid residue of L. This could lead to the alteration of phenotypes of mutants in the present study.
The identification of binding partners of L may provide new insights into RNA encapsidation and may also lead to an increase in our understanding of the pathogenesis of TMEV-induced diseases.
Acknowledgments
We thank M. Higashino, K. Asakura, and S. Saito for technical assistance.
This study was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture; a grant from the Neuroimmunological Research Committee of the Ministry of Health, Labor, and Welfare; a grant for promoted research from Kanazawa Medical University (S2005-12), and a grant for project research from the High-Technology Center of Kanazawa Medical University (2004-7).
Footnotes
Published ahead of print on 27 September 2006.
REFERENCES
- 1.Amaravadi, R., and C. B. Thompson. 2005. The survival kinases Akt and Pim as potential pharmacological targets. J. Clin. Investig. 115:2618-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asakura, K., H. Murayama, T. Himeda, and Y. Ohara. 2002. Epitope-tagged L* protein of Theiler's murine encephalomyelitis virus is expressed in the central nervous system in the acute phase of infection. J. Virol. 76:13049-13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badshah, C., M. A. Calenoff, and K. Rundell. 2000. The leader polypeptide of Theiler's murine encephalomyelitis virus is required for the assembly of virions in mouse L cells. J. Virol. 74:875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calenoff, M. A., C. S. Badshah, M. C. Dal Canto, H. L. Lipton, and M. K. Rundell. 1995. The leader polypeptide of Theiler's virus is essential for neurovirulence but not for virus growth in BHK cells. J. Virol. 69:5544-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, A. M., F. C. Nucifora, Jr., E. T. Fung, D. J. Livingston, R. A. Aldape, C. A. Ross, and S. H. Snyder. 1997. FKBP12 binds the inositol 1,4,5-trisphosphate receptor at leucine-proline (1400-1401) and anchors calcineurin to this FK506-like domain. J. Biol. Chem. 272:27582-27588. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H.-H., W.-P. Kong, L. Zhang, P. L. Ward, and R. P. Roos. 1995. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat. Med. 1:927-931. [DOI] [PubMed] [Google Scholar]
- 9.Creighton, T. E. 1984. Proteins: structural and molecular principles. W. H. Freeman and Co., New York, N.Y.
- 10.Delhaye, S., V. van Pesch, and T. Michiels. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78:4357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, J. L., S. Stein, L. Rosenstein, T. Bodwell, M. Routbort, B. L. Semler, and R. P. Roos. 1990. Neurovirulence determinants of genetically engineered Theiler viruses. Proc. Natl. Acad. Sci. USA 87:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazina, E. V., J. E. Fielding, B. Lin, and D. A. Anderson. 2000. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses. J. Virol. 74:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Himeda, T., Y. Ohara, K. Asakura, Y. Kontani, M. Murakami, H. Suzuki, and M. Sawada. 2005. A lentiviral expression system demonstrates that L* protein of Theiler's murine encephalomyelitis virus (TMEV) is essential for virus growth in a murine macrophage-like cell line. Virus Res. 108:23-28. [DOI] [PubMed] [Google Scholar]
- 14.Hiscott, J., H. Kwon, and P. Genin. 2001. Hostile takeovers: viral appropriation of the NF-κB pathway. J. Clin. Investig. 107:143-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard, B. R., F. F. Vajdos, S. Li, I. Sundquist, and C. P. Hill. 2003. Structural insights into the catalytic mechanism of cyclophilin A. Nat. Struct. Biol. 10:475-481. [DOI] [PubMed] [Google Scholar]
- 16.Kong, W. P., G. D. Ghadge, and R. P. Roos. 1994. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc. Natl. Acad. Sci. USA 91:1796-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leong, L. E. C., C. T. Cornell, and B. L. Semler. 2002. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-197. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 18.Lipton, H. L. 1980. Persistent Theiler's murine encephalomyelitis virus infection in mice depends on plaque size. J. Gen. Virol. 46:169-177. [DOI] [PubMed] [Google Scholar]
- 19.Lummis, S. C. R., D. L. Beene, L. W. Lee, H. A. Lester, R. W. Broadhurst, and D. A. Dougherty. 2005. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature 438:248-252. [DOI] [PubMed] [Google Scholar]
- 20.Michiels, T., N. Jarousse, and M. Brahic. 1995. Analysis of the leader and capsid coding regions of persistent and neurovirulent strains of Theiler's virus. Virology 214:550-558. [DOI] [PubMed] [Google Scholar]
- 21.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Obuchi, M., Y. Ohara, T. Takegami, T. Murayama, H. Takada, and H. Iizuka. 1997. Theiler's murine encephalomyelitis virus subgroup strain-specific infection in a murine macrophage-like cell line. J. Virol. 71:729-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obuchi, M., and Y. Ohara. 1998. Theiler's murine encephalomyelitis virus and mechanisms of its persistence. Neuropathology 18:13-18. [Google Scholar]
- 24.Obuchi, M., J. Yamamoto, T. Odagiri, M. N. Uddin, H. Iizuka, and Y. Ohara. 2000. L* protein of Theiler's murine encephalomyelitis virus is required for virus growth in a murine macrophage-like cell line. J. Virol. 74:4898-4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obuchi, M., T. Odagiri, K. Asakura, and Y. Ohara. 2001. Association of L* protein of Theiler's murine encephalomyelitis virus with microtubules in infected cells. Virology 289:95-102. [DOI] [PubMed] [Google Scholar]
- 26.Oleszak, E. L., J. R. Chang, H. Friedman, C. D. Katsetos, and C. D. Platsoucas. 2004. Theiler's virus infection: a model for multiple sclerosis. Clin. Microbiol. Rev. 17:174-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paul, S., and T. Michiel. 2006. Cardiovirus leader proteins are functionally interchangeable and have evolved to adapt to virus replication fitness. J. Gen. Virol. 87:1237-1246. [DOI] [PubMed] [Google Scholar]
- 28.Rodríguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 29.Roos, R. P., S. Stein, Y. Ohara, J. Fu, and B. L. Semler. 1989. Infectious cDNA clones of the DA strain of Theiler's murine encephalomyelitis virus. J. Virol. 63:5492-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roos, R. P. 2002. Pathogenesis of Theiler's murine encephalomyelitis virus-induced disease, p. 427-435. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 31.Sasaki, J., S. Nagashima, and K. Taniguchi. 2003. Aichi virus leader protein is involved in viral RNA replication and encapsidation. J. Virol. 77:10799-10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takata, H., M. Obuchi, J. Yamamoto, T. Odagiri, R. P. Roos, H. Iizuka, and Y. Ohara. 1998. L* protein of the DA strain of Theiler's murine encephalomyelitis virus is important for virus growth in a murine macrophage-like cell line. J. Virol. 72:4950-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Troxler, M., D. Egger, T. Pfister, and K. Bienz. 1992. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology 191:687-697. [DOI] [PubMed] [Google Scholar]
- 34.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon protection. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]