Abstract
The Hfq protein was discovered in Escherichia coli in the early seventies as a host factor for the Qβ phage RNA replication. During the last decade, it was shown to be involved in many RNA processing events and remote sequence homology indicated a link to spliceosomal Sm proteins. We report the crystal structure of the E.coli Hfq protein showing that its monomer displays a characteristic Sm-fold and forms a homo-hexamer, in agreement with former biochemical data. Overall, the structure of the E.coli Hfq ring is similar to the one recently described for Staphylococcus aureus. This confirms that bacteria contain a hexameric Sm-like protein which is likely to be an ancient and less specialized form characterized by a relaxed RNA binding specificity. In addition, we identified an Hfq ortholog in the archaeon Methanococcus jannaschii which lacks a classical Sm/Lsm gene. Finally, a detailed structural comparison shows that the Sm-fold is remarkably well conserved in bacteria, Archaea and Eukarya, and represents a universal and modular building unit for oligomeric RNA binding proteins.
INTRODUCTION
The Hfq protein was first described in Escherichia coli as a host factor (HF-I) for the replication of the Qβ phage RNA (1). In 1994, Tsui et al. reported that the inactivation of the hfq gene in E.coli provokes a wide variety of phenotypes (2) and the first cellular role observed for Hfq was its participation in the regulation of rpoS, a gene coding for a stress-induced RNA polymerase σs factor (3,4). During the last 5 years, it has been shown that Hfq is a pleiotropic regulator which controls the expression of many proteins by affecting mRNA translation, stability or polyadenylation (5–8). Small RNAs (sRNA) in particular appear to be targets for Hfq (9). Indeed, several studies have established that Hfq, which has a binding preference for A/U-rich sequences (10), binds to uridine-rich tracks of regulatory sRNAs like OxyS, Spot42 or DsrA (11–13). It has been proposed that the protein acts as an RNA chaperone which may simultaneously recognize the regulatory sRNA and its target, and facilitate their interaction. The ability of Hfq to induce structural changes in the 5′ UTR of ompA RNA and to rescue a folding trap of a splicing defective intron confirms this hypothesis (14).
Sequence analysis recently suggested that Hfq may be related to Sm and Sm-like (or Lsm) proteins (T. Gibson, personal communication) found in eukaryotes and in Archaea (15–17). These proteins form ring-like hetero-heptamers in eukaryotes which are the main components of the spliceosomal small nuclear ribonucleoproteins (snRNPs) (18,19). As such they take part in RNA splicing but also participate in many RNA processing events (reviewed in 20). The function of archaeal Lsm proteins is still unknown but they share with their eukaryotic counterparts the ability to bind uridine-rich sequences at the inner part of doughnut-shaped homo-heptamers (21,22). The evolutionary connection between Sm/Lsm proteins and Hfq was for the first time explicitly described by two groups at the beginning of 2002, also showing by electron microscopy (EM) that Hfq forms a ring-like structure with a 6-fold symmetry (11,12). The hexameric organization was confirmed by the crystal structure of the Hfq protein from Staphylococcus aureus (23). Concomitantly, Sm-based homology models were proposed for the E.coli protein (24,25). The latter protein is by far the best studied member of the Hfq family and constitutes a target of choice for structural investigation. We report here its crystal structure at a resolution of 2.15 Å. As could be anticipated from the sequence analysis and former biochemical data, Hfq forms a hexameric ring very similar to that of the S.aureus protein. This observation reinforces the conclusion that the Hfq family is characterized by a hexameric organization. Finally, the structural relationship with Sm/Lsm proteins is discussed as well as implications for the function of these RNA binding proteins.
MATERIALS AND METHODS
Protein and crystal preparation
The open-reading frames for the native Hfq protein and the mutant truncated after Ser72 were obtained by PCR from E.coli lysate and cloned into a modified pET24d expression vector with an upstream sequence coding for a His6 tag followed by a TEV protease site (pETM11). Over-expression of the proteins was carried out in the E.coli strain BL21(DE3) star (Invitrogen). Cells were grown in TB medium supplemented with 0.1 mg/ml kanamycin and the induction was triggered after 3 h at 37°C by adding 1 mM IPTG. Cells were harvested after 18 h at 18°C and lysed using a French press. After centrifugation (20 000 g, 30 min, 4°C), the supernatant was loaded onto a nickel-nitriliacetic acid bead column (Qiagen) and the elution was carried out as recommended by the manufacturer. After protease cleavage at 16°C overnight (enzyme/substrate ratio: 1/50), the samples were further purified on a Superose 12 column (Pharmacia) and concentrated by ultrafiltration to 10 mg/ml. This protocol led to >98% pure samples for both constructs as judged from Coomassie blue-stained gels (data not shown).
Crystals of Hfq were obtained at 20°C by vapor diffusion in 2 µl hanging drops (protein to reservoir solution ratio: 1/1). Among the four crystal forms we observed in Wizard screens (deCODE genetics), two were hexagonal and diffracted up to a resolution of 2.2 Å after optimization (Table 1). Crystal form A was obtained with the full-length protein and a reservoir containing 1.6 M NH4SO4, 0.1 M Tris–HCl pH 8.0, and form B with the short Hfq form using a reservoir containing 25% PEG 4000, 0.2 M NH4-acetate and 0.2 M Na-acetate pH 4.6. Complete data were collected using synchrotron radiation with crystals flash-frozen in paraffin oil and were processed using HKL (26). Crystal form A appeared to be twinned (twinning ratio: 0.28) and the corresponding data were corrected using Detwin (27).
Table 1. Crystal characterization and refinement statistics.
| Crystal analysis | ||
| Crystal form | A | B |
| Protein | Hfq | Hfq-short |
| Crystal size (mm3) | 0.03 × 0.08 × 0.1 | 0.2 × 0.05 × 0.05 |
| Beamline | ID14-2 (ESRF) | XRD1 (Elettra) |
| Wavelength (Å) | 0.933 | 1.0 |
| Space group | P6 | P61 |
| a, c (Å) | 61.50, 28.25 | 61.35, 166.1 |
| Asymmetric unit | 1 monomer | 1 hexamer |
| Resolution range (Å) | 62–2.25 | 47–2.15 |
| No. of observations | 22 518 | 139 736 |
| No. of unique reflections | 2789 | 19 131 |
| Completeness (%) | 99 (89)a | 99.8 (96.5)a |
| Rmerge (%) | 6.3 (17)a | 9.8 (27)a |
| Structure refinement | ||
| Resolution range (Å) | 20–2.15 | |
| R-factor (%) | 20.8 | |
| Rfree (%) | 26.2 | |
| No. of protein and solvent atoms | 3104, 136 | |
| RMSD from ideal geometry bond distances (Å) and angles (°) | 0.010, 1.61 | |
| Average B-factors: overall, protein and solvent atoms (Å2) | 20.7, 20.4, 29.0 | |
| Ramachandran plotb: residues in core, allowed, generously allowed regions (%) | 93.5, 4.7, 1.8 |
aIn the high resolution shell: 2.30–2.25 Å, 2.21–2.15 Å, respectively.
bStatistics from PROCHECK (44).
Structure determination
The search for molecular replacement (MR) solutions was performed using AMoRe (28). The low solvent content of the two hexagonal crystal forms rendered MR tricky: neither homology models derived from Sm/Lsm structures nor from the S.aureus hexamer gave any significant signal. The procedure will be detailed elsewhere (Sauter,C., Basquin,J. and Suck,D., manuscript in preparation). The search was carried out using the detwinned P6 data (form A) to reduce the problem to one monomer. A poly-ala monomer encompassing residues 7–65 gave a clear solution using data between 3.5 and 10 Å with a correlation factor and R-factor of 45.1 and 42.1%, respectively. After rigid-body and simulated annealing (SA) refinements using CNS (29), a hexamer with the correct sequence was generated applying the 6-fold symmetry. A new search was performed for crystal form B (same resolution range) using this model leading to an outstanding solution (C = 54.8% and R = 47.4%).
The Hfq model was refined with CNS using a maximum likelihood target, a bulk solvent correction and taking into account the non-crystallographic symmetry (NCS). Eight percent of the reflections were randomly selected for Rfree testing. After rigid-body refinement (resolution range: 3–20 Å) the R-factor was 44.4% (Rfree 46.0%), and after SA and B-factor refinement rounds followed by a stepwise increase of the resolution from 3 to 2.15 Å, it dropped to 30.5% (Rfree 32.3%). The model was further inspected in O (30) and water molecules developing sensible hydrogen bonds with protein or solvent atoms were added. NCS constraints were progressively relaxed according to the decrease of the Rfree. The final model consisting of 388 protein residues and 136 water molecules led to an R-factor of 20.7% (Rfree 26.2%). Refinement statistics are given in Table 1. Residues 7–68 are observed in all six subunits and additional residues were modeled at the N-terminus and the C-terminus (from Gly4 in monomers D and E, and up to His71 in F) depending on the local quality of the electron density map. Atomic coordinates and structure factors are accessible at the Protein Data Bank (1HK9).
Sequence and structure comparisons
A BLAST search was carried out in non-redundant data bases at EBI (http://www.ebi.ac.uk/blastall/) and NCBI (http://www.ncbl.nlm.nih.gov/BLAST/) using the E.coli Hfq sequence as a query. Multiple alignments of Hfq and Sm/Lsm sequences were built using CLUSTAL W and manually adjusted with SEAVIEW (31,32). A consensus 2D structure was determined using Jpred (33). A 3D search in Superfamily (http://supfam.mrc-lmb.cam.ac.uk/SUPERFAMILY/) was per formed in parallel to look for structural homologs that clearly identified human B, D1, D2 and D3 Sm proteins (results not shown).
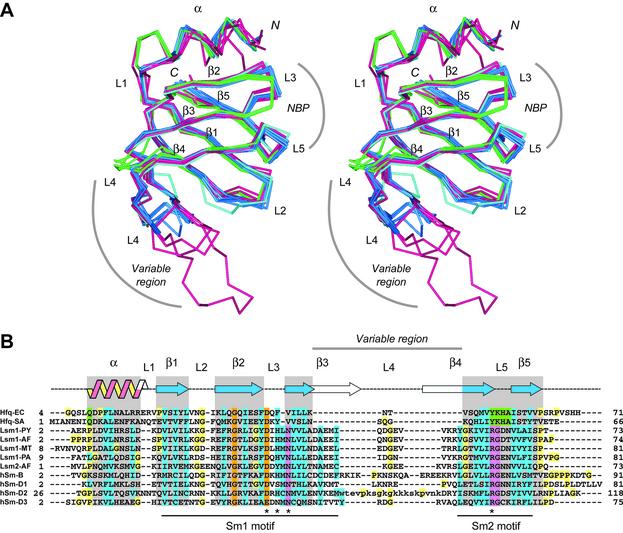
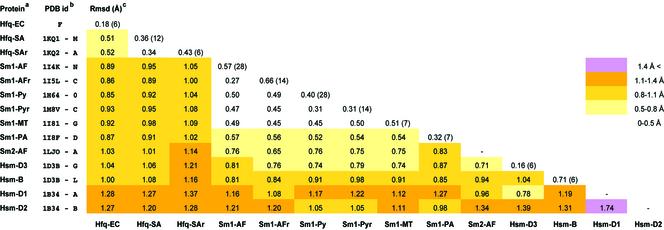
LSQMAN (30) was used to compare the E.coli Hfq monomer with other known Sm/Lsm and Hfq structures (see Fig. 3 for details). When more than one copy of a monomer was present in a PDB entry, the central model, i.e. the model which has the lowest value for root-mean-square (RMS) [root-mean-square deviation (RMSD)] as defined in LSQMAN, was first determined based on the RMSD of main chain atoms (N, Cα, C) of equivalent subunits. Central models were then superimposed using their main chain atoms in the regions of conserved secondary structures (Fig. 3); RMSD values are reported in Table 2.
Figure 3.
Sequence and structure comparison of the Sm-fold in Sm/Lsm and Hfq monomers. (A) This stereoview shows the superposition of central monomers (see Materials and Methods) from each available structure using the following color code: Hfq monomers from E.coli and S.aureus without and with RNA (23), respectively called Hfq-EC, Hfq-SA and Hfq-SAr in (B), are colored in green; archaeal Lsm1 proteins from A.fulgidus and P.abyssi alone and with RNA (21,22,35), Pyrobaculum aerophilum (42) and M.thermoautotrophicum (43), respectively called Lsm1-PY, Lsm1-PYr, Lsm1-AF, Lsm1-Afr, Lsm1-PA, Lsm1-MT, are colored in blue; and Lsm2 from A.fulgidus, or Lsm2-AF, (35) is represented in cyan; human Sm momomers D1, D2, B and D3 (18), respectively called hSm-D1, hSm-D2, hSm-D3 and hSm-B, are shown in magenta. (B) This structure-based sequence alignment is restricted to the regions revealed by crystallographic studies (first and last observed residues are indicated on the left- and right-hand sides of the corresponding sequences). Gray boxes highlight the common backbone regions defining a minimal Sm-fold. These regions were used to superimpose the monomers (A) and to calculate RMSD values (Table 2). They mainly fit to the secondary structure features shown on top [nomenclature according to Kambach et al. (18)]. Overall conserved residues appear in orange, those specific to the Hfq family in green and those characteristic to Archaea and eukaryotes in magenta. Blue boxes indicate conserved patches of aliphatic or aromatic residues. Residues in loops L3 and L5 that form the NBP (A) in the structures of Lsm–RNA complexes are indicated by stars. Residues of the hSm-D2 variable region which are not seen in the structure are indicated by lower case letters. The residues belonging to loop L4 are separated from adjacent β-strands by gaps to highlight the variability of this region. Panel (A) was generated using ViewerLite (Accelrys Inc.).
Table 2. RMSD of conserved Sm-fold in Sm/Lsm/Hfq monomers.
aThe proteins are named as in Figure 3.
bPDB IDs are followed by the name of the central model used to perform the analysis (see Materials and Methods). When several copies of a monomer are present in a given PDB entry, the average RMSD of their main chain atoms is indicated in the diagonal followed by the number of copies between brackets.
cRMSD values are based on 135 common positions of main chain atoms (N, Cα, C).
RESULTS AND DISCUSSION
The Hfq family
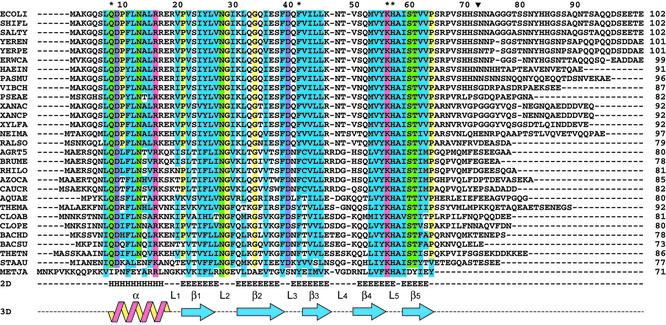
The information provided by microbial sequencing projects has recently led to the identification of Hfq candidates in about half of the 140 complete or nearly complete genomes, a few of them showing gene duplication (25). Figure 1 shows the result of a BLAST search using the E.coli sequence as a query and illustrates the high sequence conservation throughout the Hfq family. Secondary structure prediction suggested to us, as well as to other groups (24,25), a topology very similar to the Sm-fold consisting of an α-helix followed by five β-strands. This hypothesis is now validated by crystallographic data: the Hfq core (residues 7–66 in E.coli) is common to all bacterial proteins and displays a few strictly conserved residues, either important for the structure like Gly29 which allows the bending of β-strand 2, or involved in RNA binding like Gln8, Phe39 or Lys56, His57 in the YKHAI motif (23). Some less conserved residues are characteristic for bacterial phyla (25). The β-hairpin L4 is the most divergent part of the core region and consists of either two (E.coli type) or three residues (S.aureus type). The C-terminal extension following the Hfq core is almost non-existent in Bacillus species, but consists of up to 38 (mainly hydrophilic) amino acids in E.coli and close relatives. No 2D structure is predicted for this variable extension which probably forms a floppy tail, in agreement with circular dichroism analysis (12,24).
Figure 1.
The Hfq family. The organisms corresponding to the sequences are indicated on the left from top to bottom with entry names or access numbers in parenthesis. Proteobacteria: E.coli (HFQ_ECOLI), Shigella flexneri (HFQ_SHIFL), Salmonella typhimurium (HFQ_SALTY), Yersinia enterocolitica (HFQ_YEREN), Yersinia pestis (HFQ_YERPE), Erwinia carotovora (HFQ_ERWCA), Haemophilus influenzae (HFQ_HAEIN), Pasteurella multocida (HFQ_PASMU), Vibrio cholerae (HFQ_VIBCH), Pseudomonas aeruginosa (HFQ_PSEAE), Xanthomonas axonopodis (HFQ_XANAC), Xanthomonas campestris (HFQ_XANCP), Xylella fastidiosa (HFQ_XYLFA), Neisseria meningitidis (HFQ_NEIMA), Ralstonia solanacearum (HFQ_RALSO), Agrobacterium tumefaciens (HFQ_AGRT5), Brucella melitensis (HFQ_BRUME), Rhizobium loti (HFQ_RHILO), Azorhizobium caulinodans (HFQ_AZOCA), Caulobacter crescentus (HFQ_CAUCR). Aquificae: Aquifex aeolicus (HFQ_AQUAE). Thermotogae: Thermotoga maritima (HFQ_THEMA). Firmicutes: Clostridium acetobutylicum (HFQ_CLOAB), Clostridium perfringens (HFQ_CLOPE), Bacillus halodurans (HFQ_BACHD), Bacillus subtilis (HFQ_BACSU), Thermoanaerobacter tengcongensis (HFQ_THETN), S.aureus (Q99UG9). Archaea: M.jannaschii (Q58830). The numbering at the top corresponds to the E.coli sequence and the black arrow to the C-terminus of the short Hfq form. Conserved polar, basic and acidic residues appear in green, pink and violet, respectively, Gly and Pro in yellow, and a star indicates those involved in RNA binding in S.aureus (23). Blue boxes are conserved patches of hydrophobic residues. The 2D structure prediction from Jpred is indicated at the bottom as well as the 2D features seen in 3D structures [nomenclature according to Kambach et al. (18)].
Overall, the Hfq family appears to be a widespread and well conserved class of bacterial factors. Nevertheless, it is not restricted to bacteria, since we identified a potential homolog in Methanococcus jannaschii which presents many characteristics of Hfq proteins, in particular an almost conserved YKHAI motif. Interestingly, this archaeon does not host any Sm/Lsm gene. This suggests that Hfq proteins may be structural and functional Sm/Lsm homologs in organisms lacking the latter genes.
Escherichia coli Hfq forms a compact hexameric core
In our attempts to crystallize the Hfq protein from E.coli, we initially focussed on the wild-type sequence (102 residues) and we obtained tetragonal (data not shown) and hexagonal crystals (Table 1). The poor reproducibility and the extremely low solvent content (18% based on the native monomer sequence) of the latter crystal form strongly suggested that proteolytic degradation of the sample occurred prior to crystallization. To achieve reproducibility, we prepared short Hfq forms based on studies showing that C-terminal deletants are still active (2,34) and on sequences suggesting that the minimal Hfq-fold only requires the first 70 residues of the E.coli monomer (Fig. 1). A construct encompassing amino acids 1–72 readily yielded two new crystal forms: a triclinic one diffracting to 2.9 Å (data not shown) and the hexagonal form B (Table 1) which was used to refine the structure at 2.15 Å resolution. The structure was eventually solved by combining the two hexagonal data sets and using the coordinates of the S.aureus Hfq monomer (see Materials and Methods).
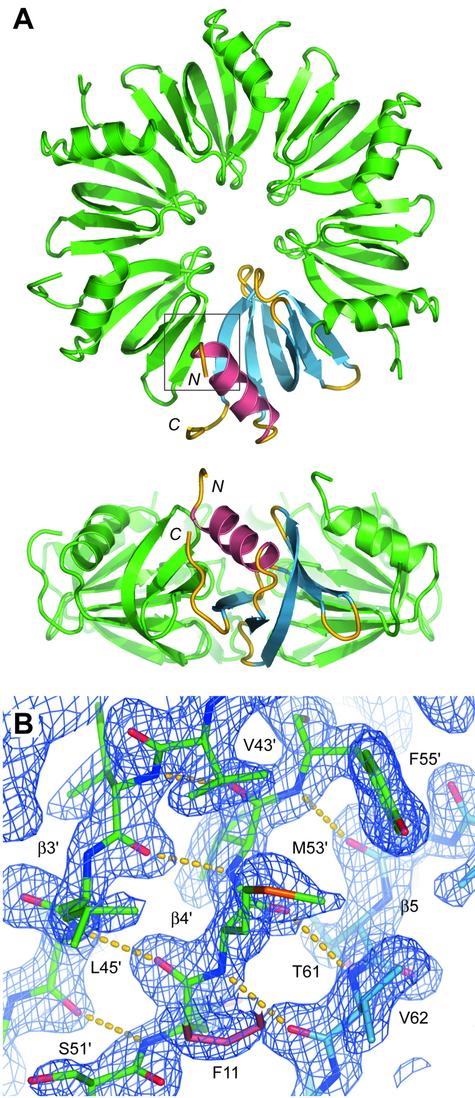
The Hfq protein in E.coli forms a doughtnut-shaped homo-hexamer (Fig. 2). This confirms the oligomeric state described in early biochemical data and recent EM studies. The ring has a diameter of 65 Å, a thickness of 28 Å and the central channel is 11 Å wide at its narrowest point. The diameter is slightly smaller than the 70 Å estimated by EM, but this may be due to the absence of the C-terminal extension in our crystals. Nevertheless, residues 66–71 form a short tail pointing towards the α-helix; this indicates that the C-terminal tail is likely to be located at the top of the compact doughnut and to provide additional possibilities for RNA interaction (see below).
Figure 2.
Structure of the Hfq protein from E.coli. (A) Top and side views of the Hfq hexameric doughnut. Secondary structure elements are highlighted in one monomer with the N-terminal α-helix in pink and the five β-strands in blue. N- and C-termini pointing toward the top of the hexamer are indicated. (B) The dimer interface and H-bond interactions between strands β4′ and β5 of adjacent subunits. The 2Fo–Fc composite omit map (level 1.6σ) is shown in the region indicated by a square in (A). This figure was prepared using PyMol (Delano Scientific, San Carlos, CA).
The overall structure of E.coli Hfq is very similar to its ortholog in S.aureus: their RMSD is 1 Å based on 6 × 57 Cα positions in the ring. This strongly suggests that the hexameric state is a characteristic of the bacterial Hfq family. As in S.aureus Hfq and in Sm/Lsm proteins in general, the oligomer is held together by backbone H-bonds between β-strands 4 and 5 from adjacent monomers (Fig. 2A), reinforced by hydrophobic side chain interactions with the α-helix and neighboring strands 1 and 2. Bacterial subunit interfaces are essentially identical except one interaction, namely the H-bond observed in S.aureus between the side chains of Tyr56 in the YKHAI motif and Tyr63 in β5 (23). The second tyrosine is unique to this bacterium and is predominantly replaced by Val or Ile residues in other Hfq sequences (V62 in E.coli). Thus, in other bacteria the Tyr in the YKHAI motif (Y55 in E.coli) is free to rotate towards the center of the ring and is therefore likely to be involved in RNA binding (see below).
A universal Sm-fold
Sm and Lsm sequences share two sequence motifs, Sm1 and Sm2 (15,16). This Sm hallmark corresponds to hydrophobic patches of residues maintaining the core of the Sm-fold (18) and highly conserved residues involved in RNA binding (21). The link between Hfq and Sm/Lsm families remained unnoticed until recently because at a first look Hfq sequences only contained the Sm1 motif and failed to fit the Sm2 motif. To address the question of the similarity between these proteins, we compared the monomers of four human Sm proteins, five archaeal Lsm proteins and two bacterial Hfq proteins. As shown in Figure 3A, loops and secondary structure elements are conserved in all monomers with some family-specific variability in length. In brief, Hfq proteins are characterized by a two-residue longer α-helix (like hSm-D2), a shorter L3 β-hairpin (three residues instead of four in Sm/Lsm proteins) and a very short ‘variable region’. This region of high sequence variability in Sm/Lsm proteins encompasses the end of strand β3, loop L4 and the start of β4. In Hfq proteins it just consists of a short L4 β-hairpin (two to three residues), a feature shared with Lsm proteins of some Archaea like Halobacterium and Methanobacterium thermoautotrophicum (35). In contrast, this region is generally much longer in Sm/Lsm proteins (14–28 residues), the longest variable region being observed in the structure of hSm-B (Fig. 3A). Surprisingly, the topology of loop L5 is conserved despite its sequence variability. Indeed, L5 clearly introduces a difference in the Sm2 motif between Hfq (YKHA) and Sm/Lsm (RGXX), whereas the Sm1 motif is almost conserved.
A consensus Sm-fold can be defined (gray boxes in Fig. 3B) consisting of 45 common amino acid positions (the variable loops L1–4 were excluded from this analysis) which were used to calculate RMSD values for the 14 known monomer types (Table 2). This analysis performed on 135 main chain atoms reveals that the minimal Sm-fold is remarkably well conserved (average RMSD 0.91 ± 0.28 Å), despite a low sequence conservation. Hfq and Lsm families are homogeneous and present an average RMSD of 0.46 ± 0.10 and 0.55 ± 0.15 Å, respectively. Human proteins are more divergent (RMSD 1.14 ± 0.31 Å) probably as a consequence of structural and functional differences in the hetero-heptameric Sm ring. Hfq monomers display a structure slightly closer to archaeal Lsm (0.85 < RMSD < 1.1Å) than to human Sm monomers (1.0 < RMSD < 1.3 Å).
Oligomerization: hexamer or heptamer
The hetero-heptameric model proposed by Kambach et al. (18) for the human Sm core has been validated by biochemical and EM investigations on snRNPs (36–38) and the rising number of Lsm1 structures already revealed homo-heptamers in four archaeal organisms. On the other hand, considering the structure and sequence conservation characterizing the Sm-fold in the Hfq family, it is almost clear that the hexameric organization is a general feature for Hfq proteins. What drives the preference for hexamer or heptamer formation is not clear yet. Schumacher et al. (23) suggested that a short variable region might constitute a structural switch towards a hexamer but the situation is probably more complicated. For instance, Archaeoglobus fulgidus Lsm2 protein forms a hexamer in the absence of RNA (35) indicating that a long variable region does not necessarily imply a heptameric arrangement. In addition, at least two archaeal Lsm sequences have short variable regions (see above). Since the backbone of the Sm-fold is essentially the same (Table 2), the degree of compaction of the oligomer is probably related to subtle variations of side chain interactions besides the H-bond network of the β-sheet (35). Further structural data, especially of proteins with the archaeal Lsm2 architecture, will help to answer this question.
Independent of the number of monomers, the way the oligomers get assembled may directly affect their mode of interaction with RNA targets. Eukaryotic Sm proteins are found as hetero-dimers or trimers in the cytoplasm and do only form heptamers in the presence of U-rich small nuclear RNAs (UsnRNAs) [for a recent review see Will and Lührmann (20)]. EM data suggest that snRNPs get assembled around the RNA Sm site and the Sm core traps the RNA which seems to be channeled through the central cavity of the ring (37) to build a compact, intricate particle. In contrast, procaryotic Lsm and Hfq proteins generally form stable homo-oligomers. Similarly, eukaryotic Lsm proteins exist, at least in yeast, as stable hetero-heptamers (19). In this situation, the oligomers are likely to operate as a preformed docking unit where RNA targets can bind at one face without going through the central hole.
RNA binding sites in E.coli Hfq
Both Hfq and Sm/Lsm rings present central nucleotide binding pockets (NBPs). The way Sm proteins specifically recognize oligo-U RNA sequences has been exemplified with archaeal Lsm1–RNA complex structures (21,22). Loops 3 and 5 from individual subunits form a NBP consisting of almost universally conserved residues (see Fig. 3): two stacking residues (for instance in Pyrococcus abyssi His37 and Arg63 in L3 and L5, respectively) and an Asn residue (N39) located at the start of strand β3 rendering the cavity specific for a uracil base. The structure of the S.aureus Hfq–AU5G complex (23) revealed a slightly different NBP also able to accommodate an adenine. The base is stacked between Tyr42 residues in L3 from neighboring subunits. Gln8 in the α-helix occupies almost the same position as Asn39 in Sm/Lsm (absent in Hfq) and interacts with the Watson–Crick face of A and U bases. Additional interactions to the base are provided by Lys41 and Lys57, while His58 contacts the RNA backbone buried in the central cavity. An equivalent pocket is potentially present in E.coli, but this situation may be specific for S.aureus. Indeed, it is striking that Tyr55 in the YKHAI motif occupies the same position as Arg63 in Sm/Lsm proteins. As pointed out above, Tyr55 has no H-bond partner in E.coli (unlike in S.aureus where Y55 is H-bonded to Y63). It can therefore be rotated into the central cavity and offer an alternative binding mode similar to the tight base stacking between L3 and L5 observed in Sm/Lsm proteins. This hypothesis still needs to be tested but would account for the strict conservation of residues YKH in loop 5, being all involved in the NBP.
Recent studies on sRNAs show that these riboregulators require Hfq to be fully active and present single-stranded A/U rich sequences. The repetition of identical NBPs on the Hfq ring can be seen as a way of increasing the trapping efficiency of A/U-rich tracks. Furthermore, it is probably essential for the chaperone activity of Hfq by allowing the simultaneous binding of the sRNA and its target RNA, thus facilitating their subsequent interaction. Indeed, ternary complexes of Hfq have been observed with OxyS and its target transcripts rpoS and fhlA, as well as with Spot42 and galK′, and with DsrA and a poly(A) RNA (11–13). These studies also show that Hfq generally recognizes a minimal RNA domain consisting of the A/U track and one or more flanking hairpins. Brescia et al. have proposed a model in which conserved residues at the surface of the ring (R/K16, F/Y41) offer additional interactions to DsrA hairpins (13). Based on sequence conservation in proteobacteria (Fig. 1), we suggest that other positions may be involved in target docking: this is the case for arginines 17 and 19 in the α-helix clustered in the area of the external binding site observed in the Lsm1–PY/U7 complex (22), and for the hydrophilic N-terminal tail located directly above the NBPs at the top of the central cavity. Finally, the long C-terminal tail contains many residues known for their RNA binding capabilities, like His, Tyr, Asn or Asp (39). Although it is not essential for Hfq activity, it may participate in binding of RNA targets, as seen for Sm B, D1 and D3 proteins in yeast (40). This appendix can also provide a platform for other cellular Hfq partners like the ribosome to which the majority of the protein is associated (41). Based on the present structural analysis, site-specific mutagenesis will provide deeper insights concerning the architecture of the NBP and the way E.coli Hfq interacts with its RNA targets.
Evolutionary considerations
The data presented here clearly highlight the conservation of the Sm-fold which represents a universal and modular building unit shared by the Hfq and Sm/Lsm families. Sequence differences in particular in the Sm2 motif suggest a divergent evolution from a common ancestor leading to features specific to bacteria on the one hand, and to Archaea and Eukarya on the other hand. In Hfq hexamers, the NBP is formed by residues belonging to neighboring monomers. This may partly explain the strict conservation of the YKHAI motif in loop 5 which participates in the scaffold of two adjacent NBPs. Hence, Hfq forms a family of RNA binding doughnuts with a homomeric organization and some variations at the N- and C-termini. In Sm/Lsm proteins the U-specific pocket is essentially intra-monomeric. This leaves more room for sequence variations and, thus, for heteromerization as long as RNA binding properties of individual subunits are maintained. To conclude, it appears that bacteria have retained a unique and generalist RNA chaperone involved in many stages of RNA metabolism, whereas a much higher level of complexity has been achieved in eukaryotes hosting several types of heteromers with specialized functions. The Archaea represent an intermediate containing either Hfq or primitive homomeric Sm forms.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge our colleague T. Gibson who triggered our interest for Hfq by pointing out its similarity with Sm/Lsm proteins. We also thank E. Mitchell and colleagues at ID14 beamlines (ESRF, France), K. Djinovic-Carugo and her team at XRD1 beamline (Elettra, Italy) for assistance during data collection. C.S. was the recipient of a Marie Curie Individual Fellowship (IHP Programme, contract number: HPMF-2000-00434).
REFERENCES
- 1.Franze de Fernandez M.T., Hayward,W.S. and August,J.T. (1972) Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem., 247, 824–831. [PubMed] [Google Scholar]
- 2.Tsui H.C., Leung,H.C. and Winkler,M.E. (1994) Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol., 13, 35–49. [DOI] [PubMed] [Google Scholar]
- 3.Brown L. and Elliott,T. (1996) Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol., 178, 3763–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muffler A., Fischer,D. and Hengge-Aronis,R. (1996) The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev., 10, 1143–1151. [DOI] [PubMed] [Google Scholar]
- 5.Zhang A., Altuvia,S., Tiwari,A., Argaman,L., Hengge-Aronis,R. and Storz,G. (1998) The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J., 17, 6061–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vytvytska O., Moll,I., Kaberdin,V.R., von Gabain,A. and Bläsi,U. (2000) Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev., 14, 1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 7.Sledjeski D.D., Whitman,C. and Zhang,A. (2001) Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol., 183, 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hajnsdorf E. and Régnier,P. (2000) Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc. Natl Acad. Sci. USA, 97, 1501–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wassarman K.M., Repoila,F., Rosenow,C., Storz,G. and Gottesman,S. (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev., 15, 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senear A.W. and Steitz,J.A. (1976) Site-specific interaction of Qbeta host factor and ribosomal protein S1 with Qbeta and R17 bacteriophage RNAs. J. Biol. Chem., 251, 1902–1912. [PubMed] [Google Scholar]
- 11.Zhang A., Wassarman,K.M., Ortega,J., Steven,A.C. and Storz,G. (2002) The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell, 9, 11–22. [DOI] [PubMed] [Google Scholar]
- 12.Møller T., Franch,T., Hojrup,P., Keene,D.R., Bachinger,H.P., Brennan,R.G. and Valentin-Hansen,P. (2002) Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell, 9, 23–30. [DOI] [PubMed] [Google Scholar]
- 13.Brescia C.C., Mikulecky,P.J., Feig,A.L. and Sledjeski,D.D. (2003) Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA, 9, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moll I., Leitsch,D., Steinhauser,T. and Bläsi,U. (2003) RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep., 4, 284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermann H., Fabrizio,P., Raker,V.A., Foulaki,K., Hornig,H., Brahms,H. and Lührmann,R. (1995) snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J., 14, 2076–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Séraphin B. (1995) Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J., 14, 2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salgado-Garrido J., Bragado-Nilsson,E., Kandels-Lewis,S. and Séraphin,B. (1999) Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J., 18, 3451–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kambach C., Walke,S., Young,R., Avis,J.M., de la Fortelle,E., Raker,V.A., Lührmann,R., Li,J. and Nagai,K. (1999) Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell, 96, 375–387. [DOI] [PubMed] [Google Scholar]
- 19.Achsel T., Brahms,H., Kastner,B., Bachi,A., Wilm,M. and Lührmann,R. (1999) A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J., 18, 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Will C.L. and Lührmann,R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol., 13, 290–301. [DOI] [PubMed] [Google Scholar]
- 21.Törö I., Thore,S., Mayer,C., Basquin,J., Séraphin,B. and Suck,D. (2001) RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J., 20, 2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thore S., Mayer,C., Sauter,C., Weeks,S. and Suck,D. (2003) Crystal structures of the Pyrococcus abyssi Sm core and its complex with RNA. Common features of RNA binding in Archaea and Eukarya. J. Biol. Chem., 278, 1239–1247. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher M.A., Pearson,R.F., Møller,T., Valentin-Hansen,P. and Brennan,R.G. (2002) Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: a bacterial Sm-like protein. EMBO J., 21, 3546–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arluison V., Derreumaux,P., Allemand,F., Folichon,M., Hajnsdorf,E. and Régnier,P. (2002) Structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol., 320, 705–712. [DOI] [PubMed] [Google Scholar]
- 25.Sun X., Zhulin,I. and Wartell,R.M. (2002) Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res., 30, 3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 27.CC P4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- 29.Brünger A.T., Adams,P.D., Clore,G.M., DeLano,W.L., Gros,P., Grosse-Kunstleve,R.W., Jiang,J.S., Kuszewski,J., Nilges,M., Pannu,N.S., Read,R.J., Rice,L.M., Simonson,T. and Warren,G.L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 30.Kleywegt G.J., Zou,J.Y., Kjeldgaard,M. and Jones,T.A. (2001) Around O. In Rossmann,M.G. and Arnold,E. (eds), International Tables for Crystallography. Volume F. Crystallography of Biological Macromolecules. Kluwer Academic Publishers, Dordrecht, The Netherlands, Vol. F, pp. 353–356, 366–367. [Google Scholar]
- 31.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galtier N., Gouy,M. and Gautier,C. (1996) SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci., 12, 543–548. [DOI] [PubMed] [Google Scholar]
- 33.Cuff J.A., Clamp,M.E., Siddiqui,A.S., Finlay,M. and Barton,G.J. (1998) JPred: a consensus secondary structure prediction server. Bioinformatics, 14, 892–893. [DOI] [PubMed] [Google Scholar]
- 34.Sonnleitner E., Moll,I. and Bläsi,U. (2002) Functional replacement of the Escherichia coli hfq gene by the homologue of Pseudomonas aeruginosa. Microbiology, 148, 883–891. [DOI] [PubMed] [Google Scholar]
- 35.Törö I., Basquin,J., Teo-Dreher,H. and Suck,D. (2002) Archaeal Sm proteins form heptameric and hexameric complexes: crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J. Mol. Biol., 320, 129–142. [DOI] [PubMed] [Google Scholar]
- 36.Raker V.A., Hartmuth,K., Kastner,B. and Lührmann,R. (1999) Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol., 19, 6554–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stark H., Dube,P., Lührmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–542. [DOI] [PubMed] [Google Scholar]
- 38.Walke S., Bragado-Nilsson,E., Séraphin,B. and Nagai,K. (2001) Stoichiometry of the Sm proteins in yeast spliceosomal snRNPs supports the heptamer ring model of the core domain. J. Mol. Biol., 308, 49–58. [DOI] [PubMed] [Google Scholar]
- 39.Jones S., Daley,D.T., Luscombe,N.M., Berman,H.M. and Thornton,J.M. (2001) Protein–RNA interactions: a structural analysis. Nucleic Acids Res., 29, 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang D., Abovich,N. and Rosbash,M. (2001) A biochemical function for the Sm complex. Mol. Cell, 7, 319–329. [DOI] [PubMed] [Google Scholar]
- 41.Kajitani M., Kato,A., Wada,A., Inokuchi,Y. and Ishihama,A. (1994) Regulation of the Escherichia coli hfq gene encoding the host factor for phage Q beta. J. Bacteriol., 176, 531–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mura C., Cascio,D., Sawaya,M.R. and Eisenberg,D.S. (2001) The crystal structure of a heptameric archaeal Sm protein: implications for the eukaryotic snRNP core. Proc. Natl Acad. Sci. USA, 98, 5532–5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins B.M., Harrop,S.J., Kornfeld,G.D., Dawes,I.W., Curmi,P.M. and Mabbutt,B.C. (2001) Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol., 309, 915–923. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]