Abstract
The tumor suppressor gene FHIT is inactivated by genetic and epigenetic changes in the majority of common human cancers. The human Fhit protein undergoes phosphorylation on tyrosine residue 114 by Src and related kinases both in vitro and in vivo. Src is a key cytoplasmic tyrosine kinase downstream to several growth factor receptors, including those of the EGF receptor family, which are overexpressed and activated in about one-third of human breast and ovarian carcinomas. However, the biological significance of Fhit phosphorylation by Src has remained elusive. In the present study, we demonstrate that FHIT acts as a checkpoint in cell proliferation mediated by activated tyrosine kinase receptors that recruit Src. Activation of EGF receptor family members induced Fhit phosphorylation by Src and the subsequent proteasome degradation of the phosphorylated Fhit protein. Indeed, the use of the Fhit mutant Y114F, which carries a phenylalanine instead of a tyrosine at position 114, unable to be phosphorylated on tyrosine 114 by Src, prevents Fhit degradation. Moreover, Fhit protein reduction is transient and occurs in a specific temporal window. During the signaling pathway of activated tyrosine kinase receptors, the phosphorylation of Fhit induces its degradation and the subsequent reduction in Fhit protein levels allows the transmission of the mitogenic signal; immediately thereafter, Fhit protein levels are restored. Such a scenario would suggest a key role for Fhit in the balance of proliferation/survival/apoptosis signals.
Keywords: Fhit phosphorylation proliferation, src, proteasome inhibition
The tumor suppressor gene FHIT is subjected to genetic or epigenetic changes in the majority of common human cancers (reviewed in ref. 1). This gene, encompassing the constitutive chromosome fragile site FRA3B and located in a region of chromosome 3p14.2, is 1.7 Mb in size and encodes a ubiquitously expressed mRNA of 1.1 kb, for a 147-aa protein of 16.8 kDa (2). The Fhit protein exerts an Ap3A-hydrolase activity that is not required for its tumor suppression function (3). Proof of the tumor suppressor function of FHIT has come from the observed enhanced susceptibility to spontaneous and carcinogen-induced tumors in FHIT knockout mice (4, 5) and from the ability to prevent development of inducible forestomach tumors by oral administration of adenovirus and adeno-associated FHIT viruses (6). Moreover, re-expression of FHIT in FHIT-deficient cancer cell lines induced apoptosis, cell cycle arrest, and suppression of tumorigenity in nude mice (reviewed in ref. 7). However, the mechanism through which Fhit exerts its antitumor effect has remained largely unknown. A recent study showed that the human Fhit protein displays the consensus sequence for targets of phosphorylation by Src tyrosine kinase family members and that it undergoes phosphorylation on tyrosine residue 114 by Src and related kinases both in vitro and in vivo (8). Src is a cytoplasmic tyrosine kinase involved in the regulation of a variety of normal and oncogenic processes. It is commonly activated by growth factors, such as EGF family members, and this activation leads to the formation of a receptor-Src complex (9, 10). EGF receptor (EGFR) family members (including EGFR, HER2, HER3, and HER4) regulate proliferation, survival, motility, and angiogenesis and are often overexpressed in epithelial carcinomas, such as breast and ovarian cancers (11). Tumors overexpressing at least one member of the EGFR family reportedly show elevated expression of WT Src (12), suggesting that members of the two tyrosine kinase families functionally interact to promote cancer development. During mitogenic stimulation with EGF, activated Src controls EGFR turnover by reducing its proteasome degradation, which may in part explain Src/EGFR collaboration in oncogenesis.
Recent studies have attempted to better clarify the relationship between Src and Fhit phosphorylation. Tyr-114 of Fhit is located within a 21-residue loop and appears to be the most highly conserved residue in this segment (13). Crystallographic and biochemical studies revealed that Fhit exists in the cell as a homodimer that binds two molecules of substrate at each active site simultaneously (14), and Tyr-114 of the Fhit dimer can be found in a nonphosphorylated state or phosphorylated on one or both subunits (13). To date, no biological significance for these phosphorylated forms of Fhit is known, despite several contrasting hypotheses.
Here, we demonstrate that mitogenic stimulation through activation of EGFR family proteins promotes degradation of endogenous and exogenous Fhit protein. FhitY114 phosphorylation by Src is mandatory to observe Fhit protein down-modulation and indeed, treatment of cells with proteasome inhibitors before mitogenic stimulation abolished Fhit protein reduction, suggesting that the phosphorylated form of Fhit undergoes proteasome degradation.
Results
EGF-Dependent EGFR Activation Promotes Decrease in Fhit Protein Steady-State Levels.
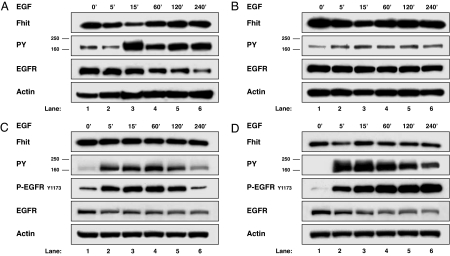
We tested the effect of mitogenic stimulation of EGFR family receptors, known to recruit Src as a key downstream molecule, on Fhit protein steady-state level. Cells of two breast (SkBr-3 and MDA-MB-453) and one ovarian (Igrov1) Fhit-positive cancer cell lines with different levels of expression of EGFR family receptors were stimulated with EGF (20 ng/ml) after starvation for 48 h, and Fhit levels were determined by Western blot analysis. As shown in Fig. 1A and B, Fhit protein levels decreased in the two breast cancer cell lines, reaching the lowest levels within 15 min of EGF stimulation (60% and 22% of Fhit protein reduction in SkBr-3 and MDA-MB-453 cell lines, respectively; Table 1); afterward, the protein amount started to return to the basal level. In both cell lines, the lowest Fhit protein levels coincided with the peak of EGFR family activation, evaluated as global tyrosine-phosphorylated amount of all four EGFR family members migrating at 170–185 kDa (Fig. 1 A and B, lanes 3). In contrast, no reduction in Fhit protein level was observed in the same time frame at EGF stimulation of Igrov1 cells (Fig. 1C). The Igrov1 clone Igrov/S5, which stably overexpresses exogenous HER2 (15), was examined for Fhit steady-state levels. HER2 overexpression increased the content of tyrosine phosphorylation of EGFR family members and the phosphorylation of EGFR detected by using a phospho-EGFR antibody directed against the major EGFR autophosphorylation site Y1173, compared with Igrov1 cells. Moreover, HER2 overexpression caused a decrease in Fhit protein steady-state level in Igrov/S5 compared with the parental cell (33% of Fhit protein reduction) (compare Fig. 1 C and D, lanes 1), and stimulation of these cells with EGF led to a further decrease. The lowest Fhit protein levels (55% of Fhit protein reduction compared with the control) were observed at 5 min of EGF treatment, after EGFR family and EGFR activation (Fig. 1D, lane 2). The total antiphosphotyrosine blots of EGF-stimulated Igrov1 and Igrov/S5 cells are shown in supporting information (SI) Fig. 7. The same behavior has also been observed in other cell lines, such as BT-474, MDA-MB-175, MDA-MB-435, and 293 cells (Table 1). Reduction of endogenous Fhit protein levels in these cell lines varies between 11% and 62%, even though it is rather constant within the same cell model in different experiments as reported in Table 1 for SkBr-3 and BT-474 cells, suggesting that Fhit reduction is a common response of breast and ovarian cancer cell lines to EGF stimulation.
Fig. 1.
Reduction of endogenous Fhit protein steady-state levels through EGF stimulation. Breast cancer cell lines SkBr-3 (A) and MDA-MB-453 (B), ovarian cancer cell line Igrov1 (C), and clone Igrov/S5 stably overexpressing HER2 (D) were starved for 48 h before stimulation with 20 ng/ml of EGF for the indicated times. Western blot analyses of total lysates were carried out by using Fhit polyclonal antibody (Fhit), a mAb to phosphotyrosine (PY), a polyclonal antibody to EGFR (EGFR), a mAb to phospho-EGFR Tyr-1173 (P-EGFRY1173), and mAb to actin (Actin).
Table 1.
Maximum Fhit reduction in cell line models upon EGF stimulation
| Cell lines | Fhit protein reduction, % |
|---|---|
| Breast cancer | |
| SKBr-3 | 60 ± 10* |
| BT-474 | 32 ± 5* |
| MDA-MB-453 | 22 |
| MDA-MB-175 | 17 |
| MDA-MB-435 | 62 |
| Ovarian cancer | |
| Igrov 1 | 1 |
| Igrov/S5 | 55 |
| 293 cells cotransfected with FHIT/SRC or EGFR/FHIT | |
| 293 | 12 |
| 293 FHIT/SRC | 31 ± 11† |
| 293 EGFR/FHIT | 31 ± 13† |
*Mean ± SE of three different experiments.
† Mean ± SE of four different experiments.
Fhit Protein Down-Modulation Is Mediated by Src.
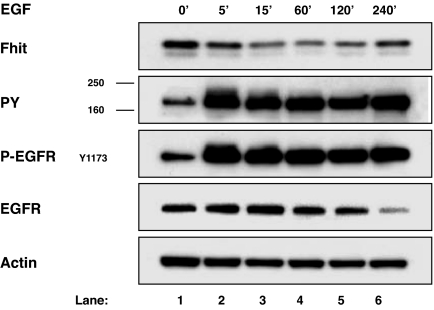
To shed light on the mechanism underlying Fhit protein regulation, human embryonic kidney 293 cells, expressing an endogenous low level of Fhit, were first transiently transfected with expression plasmids for EGFR and FHIT and stimulated with 100 ng/ml of EGF to reproduce what was observed in the endogenous system; also in this model, EGF-dependent Fhit down-modulation occurred, starting at 15 min of EGF stimulation with 39% of reduction (Fig. 2, lane 3). The average maximum Fhit reduction was 31% ± 13 at 15 min (Table 1).
Fig. 2.
Reduction of Fhit protein steady-state levels through EGF stimulation in 293 cells. The 293 cells were transiently transfected with EGFR and FHIT expression plasmids and starved for 48 h before stimulation with 100 ng/ml of EGF for the indicated times. Western blot analyses of total lysates were carried out by using Fhit polyclonal antibody (Fhit), a mAb to phosphotyrosine (PY), a mAb to phospho-EGFR Tyr-1173 (P-EGFRY1173), a polyclonal antibody to EGFR (EGFR), and mAb to actin (Actin).
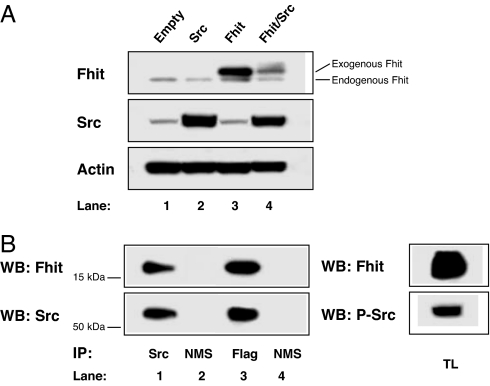
Because it is known that Src, a key downstream modulator of activated cell surface receptors, integrins, and cell-cell adhesion molecules (12, 16), can phosphorylate Fhit at residue Y114 (8), we investigated the role of Src in Fhit down-modulation by using 293 cells transiently transfected for 48 h with a FHIT expression plasmid and cotransfected or not with a SRC expression plasmid. Transfection of SRC plasmid alone led to a slight decrease in the endogenous amount of Fhit in the cells (Fig. 3A, lane 2 vs. lane 1); cotransfection of FHIT and SRC expression plasmids led to a dramatic decrease in exogenous Fhit levels (53% of Fhit protein reduction; Fig. 3A, lane 4 vs. lane 3); the faint slow-migrating band above exogenous Fhit in Fig. 3A, lane 4 is likely to represent phosphorylated Fhit (8, 13). Analysis to determine whether Fhit and Src might interact physically in the FHIT-SRC cotransfected cells revealed that Fhit and Src reciprocally coimmunoprecipitated, indicating that the two molecules are associated in a complex. Indeed, Fhit protein was revealed in the protein complexes immunoprecipitated by using Src mAb (Fig. 3B Upper, lane 1), and Src protein was revealed in the protein complexes immunoprecipitated with Flag mAb recognizing the transfected Fhit protein (Fig. 3B Lower, lane 3). Immunoprecipitation with the normal mouse serum was used as negative control (Fig. 3B, lanes 2 and 4). Src appeared to be tyrosine phosphorylated (Fig. 3B, lane 5) under this experimental condition.
Fig. 3.
Reduction of Fhit protein steady-state levels by Src. (A) The 293 cells were transiently transfected with empty vector (lane 1), SRC (lane 2), FHIT (lane 3), or FHIT and SRC (lane 4). Western blot analyses of total lysate were carried out by using Fhit polyclonal antibody (Fhit), a mAb to Src (Src), and a mAb to actin (Actin). (B) The 293 cells were transiently cotransfected with SRC and FHIT. (Left) Western blot (WB) analysis of the immunoprecipitates of Src (lane 1) or normal mouse serum (NMS) (lane 2) was carried out with Fhit polyclonal antibody (Fhit) (Upper, lanes 1 and 2). Western blot of the immunoprecipitated Fhit (using anti-Flag mab) (lane 3) or NMS (lane 4) was carried out with Src mAb (Src) (Lower, lanes 3 and 4). The membrane was stripped and reprobed with the mAb to Src (Src) (Lower, lanes 1 and 2) and polyclonal antibody to Fhit (Fhit) (Upper, lanes 3 and 4). (Right) Western blot (WB) analysis of the total lysates (TL) was carried out with Fhit polyclonal antibody (Fhit) (Upper) and phospho-Src Tyr-416 polyclonal antibody (P-Src) (Lower). Immunoprecipitates and total lysate Western blot signals were obtained at the same exposure time.
EGFR-Induced Fhit Proteasome-Dependent Degradation.
Fhit protein down-regulation within a few minutes of mitogenic stimulation may be caused by posttranslational control of Fhit expression levels rather than a decrease in Fhit mRNA levels. Indeed, real- time PCR analysis in one endogenous Fhit model (SkBr-3 cells) untreated or treated with 20 ng/ml EGF for 15 min revealed in three independent experiments a mean variation of only 2% in Fhit mRNA amount, which was found to be not statistically significant by Student's t test.
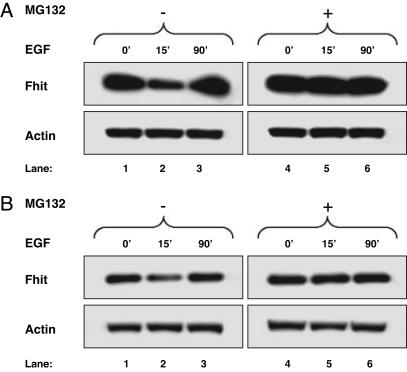
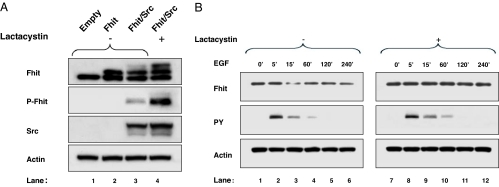
A possible mechanism of protein posttranslational control affecting Fhit is its degradation by the proteasome pathway, with phosphorylation of Fhit serving as the signal that targets this protein for degradation (17). To analyze whether Fhit down-modulation induced by mitogenic stimulation involved the proteasome pathway, we assessed Fhit levels in cells treated with proteasome inhibitors before EGF stimulation. In SkBr-3 and Igrov/S5 cells pretreated with the reversible proteasome inhibitor MG132 followed by stimulation with EGF, Fhit protein levels after 15 or 5 min of EGF stimulation were equal to those in the control (Fig. 4A and B, respectively). In SkBr-3 cells 50% of Fhit reduction was almost completely recovered (93% of recovery); similarly 43% of Fhit reduction in Igrov/S5 was completely recovered. In 293 cells transiently transfected with FHIT and SRC expression vectors and examined after 48 h, a chronic Fhit protein down-regulation was observed, and an irreversible proteasome inhibitor (lactacystin) was used (Fig. 5A). The Fhit protein decrease observed in this experiment was 16% (Fig. 5A, lane 3); the use of lactacystin allowed the visualization that the reversion is complete, considering the accumulation of a slow-migrating extra band (Fig. 5A, lane 4) corresponding to the Fhit phosphorylated form, as revealed by reprobing the membrane with a specific phospho-Fhit (Tyr-114) antibody that detects Tyr-114-phosphorylated Fhit (Fig. 5A, lane 4). Consistently, pretreatment with lactacystin of 293 cells transiently transfected with EGFR and FHIT expression vector and stimulated with 100 ng/ml of EGF completely abrogated 60% of Fhit down-modulation (Fig. 5B, lane 9 vs. lane 3). These data demonstrate that activated EGFR family proteins or Src overexpression causes proteasome degradation of Fhit in cancer cells and 293 cells.
Fig. 4.
Proteasome-dependent degradation of endogenous Fhit. SkBr-3 (A) and Igrov/S5 (B) cells were treated with or without proteasome inhibitor MG132 overnight and stimulated with 20 ng/ml of EGF for the indicated time. Western blot analyses of total lysates were carried out by using Fhit polyclonal antibody (Fhit) and a mAb to actin (Actin).
Fig. 5.
Proteasome-dependent degradation of Fhit in 293 cells. (A) The 293 cells were transiently transfected with empty vector (lane 1), FHIT (lane 2), or FHIT and SRC (lanes 3 and 4). The sample in lane 4 was from cells pretreated with 15 μM lactacystin. (B) The 293 cells were cotransfected with FHIT and EGFR and stimulated with 100 ng/ml of EGF for the indicated time. Cells were pretreated (lanes 7–12) or not (lanes 1–6) with 15 μM lactacystin. Western blot analyses of total lysates were carried out by using Fhit polyclonal antibody (Fhit), a phospho-Fhit polyclonal antibody (P-Fhit), a mAb to phosphotyrosine (PY), a mAb to Src (Src) and a mAb to actin (Actin).
Induction of Fhit Degradation Requires Fhit Phosphorylation by Src.
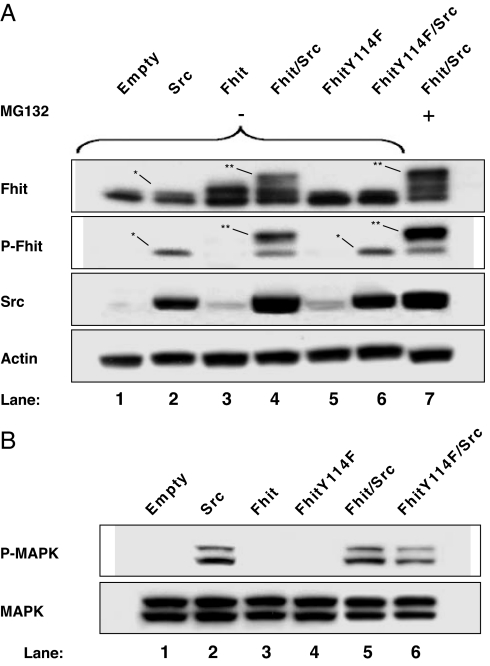
To determine whether Fhit phosphorylation by Src is essential for mediating Fhit protein degradation, we used the FHIT mutant Y114F, which carries a phenylalanine instead of a tryrosine residue at position 114 and is unable to be phosphorylated on this site by Src (8). Analysis of 293 cells transiently transfected with the expression plasmid for the FHIT Y114F mutant revealed no decrease in Fhit protein levels (Fig. 6A, lanes 5 and 6), whereas a decline in Fhit level was observed in 293 cells cotransfected with WT FHIT and SRC (Fig. 6A, lane 4 vs. lane 3). Evaluation of phospho-Fhit in the same samples showed that phosphorylation of exogenous Fhit occurred only in cells double-transfected with WT FHIT and SRC expression plasmids (Fig. 6A, lane 4). The lack of a decrease in FhitY114F levels corresponded to the absence of Fhit phosphorylation (Fig. 6A, lanes 5 and 6). Slight phosphorylation of the endogenous Fhit protein after expression of exogenous Src was observed (Fig. 6A, lane 2). Similarly, phosphorylation of the endogenous Fhit (Fig. 6A, lane 6) was caused by exogenous Src phosphorylation, because it migrated to the same molecular weight position and showed similar band intensity as the phospho-Fhit observed in Fig. 6A, lane 2. Again, abrogation of Fhit protein down-modulation induced by Src in cells pretreated with MG132 was accompanied by a general increase in Fhit protein levels compared with untreated cells. The inhibition of the proteasome activity induces the accumulation of slow-migrating additional bands of Fhit protein (Fig. 6A, lane 7) corresponding to phosphorylated forms of Fhit (Fig. 6A, lane 7 vs. lane 4; see also Fig. 5A). Together, these data indicate that phosphorylation of Fhit on Tyr-114 is required for Fhit protein degradation and the phospho-Fhit is the form committed to proteasome degradation.
Fig. 6.
Required Fhit phosphorylation by Src for proteasome degradation. The 293 cells were transiently transfected with SRC (A, lane 2; B, lane 2), FHIT (A, lane 3; B, lane 3), FHIT and SRC (A, lanes 4 and 7; B, lane 5), FHITY114F (A, lane 5; B, lane 4), and FHITY114F and SRC (A, lane 6; B, lane 6). The sample in A, lane 7 was from cells pretreated with 15 μM MG132. (A) Western blot analyses of total lysate were carried out by using phospho-Fhit polyclonal antibody (P-Fhit), a mAb to Src (Src), and a mAb to actin (Actin). The membrane with P-Fhit signal was stripped and reprobed with the polyclonal antibody to Fhit (Fhit). ∗ indicates the endogenous phospho-Fhit; ∗∗ indicates the exogenous phospho-Fhit. (B) Western blot analyses of total lysates were carried out by using a mAb to phospho-MAPK (P-MAPK). The membrane was stripped and reprobed with the polyclonal antibody to MAPK (MAPK).
Biological Significance of Fhit Protein Degradation in Mitogenic Signaling.
We tested whether Fhit phosphorylation and its subsequent degradation affects the mitogenic signaling pathway involving Src, by using an early marker of proliferation, MAPK, because Fhit transient down-modulation occurred in a specific temporal window. The 293 cells were transfected with empty vector, FHIT, SRC, FHITY114F, FHIT/SRC, and FHITY114F/SRC expression vectors. As shown in Fig. 6B, MAPK activation in 293 cells cotransfected with SRC and FHITY114F plasmids was decreased 38% in respect to SRC and WT FHIT cotransfectant (lane 5 vs. lane 6).
Discussion
In this study, we demonstrate that FHIT is tightly regulated during cell proliferation and is mediated by tyrosine kinase receptors involving Src. In our analyses to determine how a mitogenic signal can, through Src recruitment, regulate Fhit protein steady-state levels, we chose to examine mitogenic stimulation driven by EGFR family members based on the phosphorylation of Fhit by Src, which is a key downstream molecule of activated EGFR family members. The overexpression/activation of the EGFR family occurred in almost ⅓ of breast and ovarian carcinomas that are known for their aggressive phenotype (18). We previously showed that low-level or absent Fhit protein correlates with a more aggressive phenotype of human breast carcinomas, i.e., highly proliferative and poorly differentiated (19).
Reduction of Fhit protein steady-state levels in human breast and ovarian cancer cell lines overexpressing EGFR-family members, such as EGFR and HER2, and appropriately stimulated with specific ligands appeared to depend on activation of EGFR family members. Indeed, we observed a decrease in Fhit protein levels in Igrov/S5 cells transfected to overexpress HER2, but not in the parental Igrov1 cell line, which expresses a lower level of HER2.
The variability of Fhit protein down-modulation on cells expressing a comparable level of EGFR family members could depend on the ratio and type of interaction between the four receptors of the family, their activation state, and the recruitment of the downstream molecules leading to Fhit phosphorylation, including Src, and not solely on the mere expression level of a single receptor. Clearly, this regulation of the EGFR family pathway varied from line to line and can justify the magnitude of Fhit protein modulation observed in our models. Moreover, Fhit protein down-modulation caused by EGF treatment appeared to be strictly temporally regulated in endogenous and EGF stimulated models. A key finding of our study is that the decrease in Fhit protein levels results from its degradation through proteasomes. Phosphorylation of a protein is a signal that can target proteins to the ubiquitin-proteasome pathway (20, 21). When Fhit degradation occurred, high levels of phospho-Fhit forms were observed, whereas the use of the FHIT mutant Y114F, which cannot be phosphorylated by Src, abolished Fhit proteasome degradation. These data demonstrate that only the phosphorylated Fhit protein is subject to this regulation.
The role of Fhit in proliferation is not defined. Our data clearly indicate that during the signaling of activated-tyrosine kinase receptors Fhit protein is somehow regulated by Src-mediated Fhit phosphorylation. The activated Src molecule physically interacts with Fhit protein, as shown in coimmunoprecipitation experiments, indicating that the Fhit involved in this control is cytosolic and near the plasma membrane. Our hypothesis is that during the signaling pathway of activated tyrosine kinase receptors the phosphorylation of Fhit induces its degradation and the subsequent reduction in Fhit protein levels allows the transmission of the mitogenic signal; immediately thereafter, Fhit protein levels are restored. Such a scenario would suggest a key role for Fhit in the balance of proliferation/survival/apoptosis signals. The reduced MAPK activation in the presence of the mutated unphosphorylated Fhit as compared with that observed in the presence of WT phospho-Fhit may support this hypothesis. The notion that Fhit-induced effects are strictly dose-dependent is consistent with the findings of Cavazzoni et al. (22) who used a hormone-inducible expression system able to modulate Fhit protein expression levels in a nonsmall cell lung cancer line to demonstrate that Fhit protein must accumulate at a sufficient level to facilitate the apoptotic response.
On the other hand, the Fhit-substrate complex is the active form and thus the one interacting with apoptotic pathways (14, 23). It has been shown that the phosphorylated forms of the Fhit-substrate complex decrease the Km in comparison to the unphosphorylated complex, favoring the formation and the half-life of the complex and, in turn, enhancing the tumor suppressor activity of Fhit (13). Moreover, a recent study demonstrated that the Fhit Y114 residue, but not its phosphorylation, is required for the proapoptotic activity of the protein (24). Further studies are needed to clarify whether phosphorylation influences the lifetime of the Fhit–Ap3A complex or modulates the total amount of complexes formed. A better understanding of the impact of Fhit phosphorylation on cell proliferation or cell survival also awaits additional studies. To date, our data suggest that during mitogenic stimulation the cell regulates Fhit protein levels by decreasing them under a threshold necessary to allow its proapoptotic activity; overexpression and activation of the EGFR family might induce a proliferation/survival signal that overcomes the apoptosis control, and the cell uses Fhit degradation as an economical mechanism to transiently diminish this signal.
Materials and Methods
Cell Culture.
The human breast cancer cell SkBr-3, MDA-MB-453, BT-474, MDA-MB-175, MDA-MB-435, the ovarian cancer cell lines Igrov1, and the human embryonic kidney 293 were obtained from the ATCC (Rockville, MD). Cells were routinely maintained in RPMI medium 1640 (Sigma, St. Louis, MO) supplemented with 10% FCS (Sigma) and 2 mM glutamine (Cambrex, East Rutherford, NJ). Cell lines Igrov/S5 were maintained in RPMI medium 1640 supplemented with 10% FCS, 2 mM glutamine, and 200 μg/ml G418, as described (15). Cells were maintained at 37°C in a water-saturated atmosphere of 5% CO2 in air.
Plasmid Construct, Reagents, and Antibodies.
Plasmid constructs of WT PRC-CMV-FHIT and PRC-CMV-FHIT Y114F were kindly provided by Y.P. PcDNAIII-EGFR plasmid is as in ref. 25. Antibodies used were: mouse monoclonal anti-phosphotyrosine (Upstate Biotechnology, Lake Placid, NY); rabbit polyclonal anti-Fhit (Zymed Laboratories, San Francisco, CA); mouse monoclonal anti-Src clone GD11 (Upstate Biotechnology); mouse monoclonal anti-actin clone AC-40 (Sigma); rabbit polyclonal anti-phospho-Fhit-Tyr-114 (Cell Signaling Technology, Danvers, MA); mouse monoclonal anti-Flag M2 (Sigma); rabbit polyclonal anti-p44/42 MAPK (New England Biolabs, Ipswich, MA); mouse monoclonal anti-phospho p44/42 MAPK (Thr-202/Tyr-204) (Cell Signaling Technology); rabbit polyclonal anti-EGFR clone 1005 (Santa Cruz Biotechnology, Santa Cruz, CA); mouse monoclonal anti-EGFR phospho-Tyr-1173 clone 9H2 (NanoTools, Teningen, Germany); and rabbit polyclonal anti-phospho-Src Tyr-416 (Cell Signaling Technology). EGF (recombinant, human) was from Invitrogen (San Diego, CA); MG-132 and Lactacystin were from Biomol International (Plymouth Meeting, PA); and Restore Western blot stripping buffer was from Pierce (Rockford, IL).
Transient Transfection.
The human embryonic kidney 293 cells were grown in RPMI medium 1640 containing 10% FCS. Cells were plated and transfected with different expression plasmids (0.01–1 μg) with Lipofectamine and PLUS, following the manufacturer's protocol (Invitrogen). When necessary, after 18 h of expression, the cells were starved by serum deprivation for 24 h and then stimulated by EGF at 20 or 100 ng/ml for different periods as indicated for each experiment.
Cell Lysis, Western Blot, and Immunoprecipitation.
For the preparation of crude cell lysates, cells were lysed in lysis buffer containing 50 mM Tris·HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100 (Sigma), 10% (vol/vol) glycerol, 2 mM Na-orthovanadate, 10 mM leupeptin, 10 mM aprotinin, 1 mM phenylmethylsulfonyl-fluoride, 100 mM Na-fluoride, and 10 mM Na-pyrophosphate for 20 min at 4°C. Insoluble material was removed by 10 min of centrifugation at 15,500 × g at 4°C. Protein concentrations were determined by using the Comassie technique. Equal amounts of total lysates were loaded and separated on 12% precasted NuPage SDS-BisTris gel (Invitrogen) then transferred to PVDF membranes (Millipore, Billerica, MA). For the immunoprecipitation studies, cells were lysed in lysis buffer containing 0.5% Nonidet P-40. Cell lysates were precleared by incubation with 20 μl of Sepharose protein-A/G (Pierce) and appropriate amounts of normal mouse or rabbit serum for 30 min at 4°C. Then 1.5 mg of lysates per sample was incubated with appropriated antibodies in the presence of 30 μl of Sepharose protein-A/G (Pierce) for 3 h at 4°C. Samples were washed three times in lysis buffer with 0.05% Nonidet P-40 and eluted in 20 μl of sample buffer (Invitrogen). Thirty micrograms of total lysate was loaded on the same gel. Western blots were performed with indicated antibodies and revealed by using a chemiluminescence ECL or ECL-plus Western blotting kit (GE Healthcare, Little Chalfont, UK) according to the manufacturer's instructions.
RNA Extraction and Real-Time PCR.
Total RNA from 293 cells was extracted following the TRIzol manufacturer's instructions (Invitrogen). cDNA was synthesized from 1 μg of total RNA by using Superscipt II RNase H (Invitrogen). Quantitative real-time PCR was performed with a Prism HT 7900 Sequence Detection System (Applied Biosystems, Foster City, CA) with a total volume of 20 μl of reaction mixture containing 1 μl of cDNA. PCR was performed with the ready-to-use FHIT Assay-On-Demand by Applied Biosystems according to the protocol provided by the manufacturer. The endogenous control HPRT was used as reference to normalize the different samples and for the relative quantization of gene expression; the data were analyzed by the relative Ct method (ΔΔCt).
Densitometric Analysis.
The relative level of FHIT protein expression was assessed in comparison with the level of actin in the same sample by Quantity One (Bio-Rad, Hercules, CA).
Supplementary Material
Acknowledgments
We thank C. Ghirelli for cell line preparation; L. Bertola for densitometric analysis; Drs. T. Triulzi and F. Castiglioni for help in real-time PCR data analysis; L. Mameli for manuscript preparation; and G. Bugliesi for artwork preparation. This work was supported by grants from the Associazione and Fondazione Italiana per la Ricerca sul Cancro (to S.M.).
Glossary
Abbreviation
- EGFR
EGF receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605821103/DC1.
References
- 1.Pekarsky Y, Zanesi N, Palamarchuk A, Huebner K, Croce CM. Lancet Oncol. 2002;3:748–754. doi: 10.1016/s1470-2045(02)00931-2. [DOI] [PubMed] [Google Scholar]
- 2.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, Mccue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 3.Siprashvili Z, Sozzi G, Barnes LD, Mccue P, Robinson AK, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L, et al. Proc Natl Acad Sci USA. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fong LY, Fidanza V, Zanesi N, Lock LF, Siracusa LD, Mancini R, Siprashvili Z, Ottey M, Martin SE, Druck T, et al. Proc Natl Acad Sci USA. 2000;97:4742–4747. doi: 10.1073/pnas.080063497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanesi N, Fidanza V, Fong LY, Mancini R, Druck T, Valtieri M, Rudiger T, McCue PA, Croce CM, Huebner K. Proc Natl Acad Sci USA. 2001;98:10250–10255. doi: 10.1073/pnas.191345898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumon KR, Ishii H, Fong LY, Zanesi N, Fidanza V, Mancini R, Vecchione A, Baffa R, Trapasso F, During MJ, et al. Proc Natl Acad Sci USA. 2001;98:3346–3351. doi: 10.1073/pnas.061020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishii H, Dumon KR, Vecchione A, Fong LY, Baffa R, Huebner K, Croce CM. J Am Med Assoc. 2001;286:2441–2449. doi: 10.1001/jama.286.19.2441. [DOI] [PubMed] [Google Scholar]
- 8.Pekarsky Y, Garrison PN, Palamarchuk A, Zanesi N, Aqeilan RI, Huebner K, Barnes LD, Croce CM. Proc Natl Acad Sci USA. 2004;101:3775–3779. doi: 10.1073/pnas.0400481101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maa M-C, Leu T-H, McCarley DJ, Schatzman RC, Parsons SJ. Proc Natl Acad Sci USA. 1995;92:6981–6985. doi: 10.1073/pnas.92.15.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belsches-Jablonski AP, Biscardi JS, Peavy DR, Tice DA, Romney DA, Parsons SJ. Oncogene. 2001;20:1465–1475. doi: 10.1038/sj.onc.1204205. [DOI] [PubMed] [Google Scholar]
- 11.Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SC, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 12.Biscardi JS, Ishizawar RC, Silva CM, Parsons SJ. Breast Cancer Res. 2000;2:203–210. doi: 10.1186/bcr55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrison PN, Robinson AK, Pekarsky Y, Croce CM, Barnes LD. Biochemistry. 2005;44:6286–6292. doi: 10.1021/bi047670s. [DOI] [PubMed] [Google Scholar]
- 14.Pace HC, Garrison PN, Robinson AK, Barnes LD, Draganescu A, Rosler A, Blackburn G. M, Siprashvili Z, Croce CM, Huebner K, et al. Proc Natl Acad Sci USA. 1998;95:5484–5489. doi: 10.1073/pnas.95.10.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casalini P, Botta L, Ménard S. J Biol Chem. 2001;276:12449–12453. doi: 10.1074/jbc.M009732200. [DOI] [PubMed] [Google Scholar]
- 16.Irby RB, Yeatman TJ. Oncogene. 2000;19:5636–5642. doi: 10.1038/sj.onc.1203912. [DOI] [PubMed] [Google Scholar]
- 17.Fang S, Weissman AM. Cell Mol Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ménard S, Pupa SM, Campiglio M, Tagliabue E. Oncogene. 2003;22:6570–6578. doi: 10.1038/sj.onc.1206779. [DOI] [PubMed] [Google Scholar]
- 19.Campiglio M, Pekarsky Y, Ménard S, Tagliabue E, Pilotti S, Croce CM. Cancer Res. 1999;59:3866–3869. [PubMed] [Google Scholar]
- 20.Pickart CM, Eddins MJ. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Mani A, Gelmann EP. J Clin Oncol. 2005;23:4776–4789. doi: 10.1200/JCO.2005.05.081. [DOI] [PubMed] [Google Scholar]
- 22.Cavazzoni A, Petronini PG, Galetti M, Roz L, Andriani F, Carbognani P, Rusca M, Fumarola C, Alfieri R, Sozzi G. Oncogene. 2004;23:8439–8446. doi: 10.1038/sj.onc.1207847. [DOI] [PubMed] [Google Scholar]
- 23.Trapasso F, Krakowiak A, Cesari R, Arkles J, Yendamuri S, Ishii H, Vecchione A, Kuroki T, Bieganowski P, Pace HC, et al. Proc Natl Acad Sci USA. 2003;100:1592–1597. doi: 10.1073/pnas.0437915100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semba S, Trapasso F, Fabbri M, McCorkell KA, Volinia S, Druck T, Iliopoulos D, Pekarsky Y, Ishii H, Garrison PN, et al. Oncogene. 2006;25:2860–2872. doi: 10.1038/sj.onc.1209323. [DOI] [PubMed] [Google Scholar]
- 25.Magnifico A, Ettenberg S, Yang C, Mariano J, Tiwari S, Fang S, Lipkowitz S, Weissman AM. J Biol Chem. 2003;278:43169–43177. doi: 10.1074/jbc.M308009200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.