Abstract
The epidermal growth factor receptor (EGFR) signaling pathway is one of the most important pathways that regulate growth, survival, proliferation, and differentiation in mammalian cells. Reflecting this importance, it is one of the best-investigated signaling systems, both experimentally and computationally, and several computational models have been developed for dynamic analysis. A map of molecular interactions of the EGFR signaling system is a valuable resource for research in this area. In this paper, we present a comprehensive pathway map of EGFR signaling and other related pathways. The map reveals that the overall architecture of the pathway is a bow-tie (or hourglass) structure with several feedback loops. The map is created using CellDesigner software that enables us to graphically represent interactions using a well-defined and consistent graphical notation, and to store it in Systems Biology Markup Language (SBML).
Keywords: bow-tie structure, comprehensive pathway map, epidermal growth factor receptor, graphical notation, Systems Biology Markup Language (SBML)
Introduction
The epidermal growth factor receptor (EGFR) signaling pathway is one of the most important pathways that regulate growth, survival, proliferation, and differentiation in mammalian cells. It has been investigated in quite some depth, both experimentally and computationally (Wiley et al, 2003), and several computational models have been created to analyze its dynamics (Kholodenko et al, 1999; Schoeberl et al, 2002; Shvartsman et al, 2002). Further research is now needed to improve the model by incorporating various intracellular dynamics and expanding the scope where only a limited part of the signaling system has been modeled (Kholodenko, 2003). Recently, a consortium has been formed to specifically focus on the receptor tyrosine kinase signaling system, and the need for a shared model has been discussed. Despite its static nature, a comprehensive map of molecular interactions would serve as a useful reference, and greatly help research on EGFR signaling.
General characteristics of the EGFR signaling map
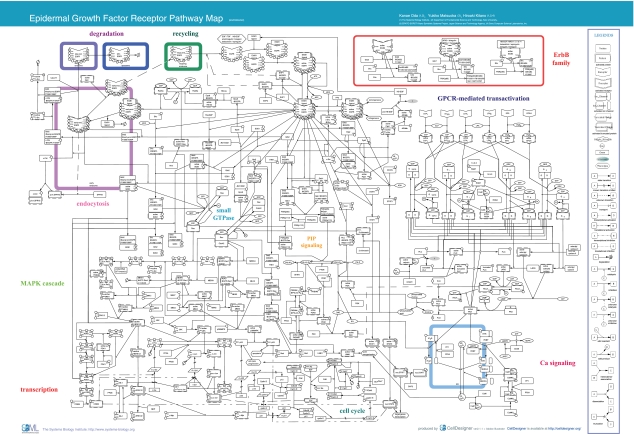
We manually constructed a comprehensive pathway map for EGFR-mediated signaling (Figure 1) based on published scientific papers. The map includes EGFR endocytosis followed by its degradation or recycling, small guanosine triphosphatase (GTPase)-mediated signal transduction such as mitogen-activated protein kinase (MAPK) cascade, phosphatidylinositol polyphosphate (PIP) signaling, cell cycle, and G protein-coupled receptor (GPCR)-mediated EGFR transactivation via intracellular Ca2+ signaling. The map was created using CellDesigner (http://celldesigner.org/), a software package that enables users to describe molecular interactions using a well-defined and consistent graphical notation (Funahashi et al, 2003; Kitano, 2003). The data of molecular interactions are stored in Systems Biology Markup Language (SBML; http://sbml.org/) (Hucka et al, 2003). Since SBML is a standard machine-readable model representation format, all the information can be used for a range of computational analysis, including computer simulation.
Figure 1.
EGFR Pathway Map. This map was created using CellDesigner ver. 2.0 (http://www.systems-biology.org/002/). A total of 219 reactions and 322 species were included. The map can be best viewed in the PDF format. Abi, abl-interactor; ADAM, a disintegrin and metalloproteinase; ADPR, ADP-ribose; Akt, v-akt murine thymoma viral oncogene homolog; AP-1, activator protein-1; Bad, BCL2-antagonist of cell death; cADPR, cyclic ADP-ribose; CAK, cyclin-dependent kinase-activating kinase; CaM, calmodulin; CaMK, calcium/calmodulin-dependent protein kinase; c-Cbl, Casitas B-lineage lymphoma proto-oncogene; CD, cluster of differentiation; Cdc, cell division cycle; Cdk, cyclin-dependent kinase; c-Fos, v-fos FBJ murine osteosarcoma viral oncogene; Chk, c-src tyrosine kinase (Csk) homologous kinase; c-Jun, v-jun sarcoma virus 17 oncogene homolog; c-Myc, v-myc myelocytomatosis viral oncogene homolog; CREB, cAMP response element-binding protein; c-Src, v-src sarcoma viral oncogene homolog; cyt., cytosol; DAG, diacylglycerol; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; Elk, Ets-like protein; end., endosome; EP, prostaglandin E receptor; Eps, EGF receptor pathway substrate; ER, endoplasmic reticulum; ErbB, erythroblastic leukemia viral (v-erb-b) oncogene homolog; ERK, extracellular signal-regulated kinase; Gab, GRB2-associated binding protein; GPCR, G protein-coupled receptor; Grb, growth factor receptor-bound protein; HB-EGF, heparin-binding EGF-like growth factor; IP3R, inositol 1,4,5-triphosphate receptor; IP3, inositol 1,4,5-triphosphate; JNK, c-Jun N-terminal kinase; KDI, kinase domain I; LARG, leukemia-associated rho guanine nucleotide exchange factor; LIMK, LIM (Lin-11 Isl-1 Mec-3) kinase; LPA, lysophosphatidic acid; LPA1/2, lysophosphatidic acid G protein-coupled receptor 1/2; lyso., lysosome; m., messenger; MAPK, mitogen-activated protein kinase; MEKK, MAPK/ERK kinase kinase; MKK, MAP kinase kinase; MKP, MAP kinase phosphatase; MLK, mixed lineage kinase; NAD, nicotinamide adenine dinucleotide; nuc., nucleus; PAK1, p21/Cdc42/Rac1-activated kinase; PDK, 3-phosphoinositide-dependent protein kinase; PGE2, prostaglandin E2; Pi, phosphoric ion; PI3,4,5-P3, phosphatidylinositol-3,4,5-triphosphate; PI3,4-P2, phosphatidylinositol-3,4-bisphosphate; PI3K, phosphatidylinositol-3-kinase; PI4,5-P2, phosphatidylinositol-4,5-bisphosphate; PI4-P, phosphatidylinositol-4-phosphate; PI5K, phosphatidylinositol-5-kinase; PIP, phosphatidylinositol polyphosphate; PKB, protein kinase B; PKC, protein kinase C; pl.m., plasma membrane; PLC, phospholipase C; PLD, phospholipase D; PP, protein phosphatase; PTB, phosphotyrosine-binding domain; PTEN, phosphatase and tensin homolog; Pyk, proline-rich tyrosine kinase; Rab5a, RAS-associated protein RAB5a; Raf, v-raf-1 murine leukemia viral oncogene homolog; Ras, rat sarcoma viral oncogene homolog; RasGAP, Ras GTPase-activating protein; Rb, retinoblastoma; RGS, regulator of G-protein signaling; Rin, Ras interaction; RN-tre, related to the N-terminus of tre; RSK, ribosomal protein S6 kinase; RYR, ryanodine receptor; S, serine; S1P, sphingosine-1-phosphate; S1P1/2/3, sphingolipid G protein-coupled receptor 1/2/3; SERCA, sarcoplasmic/endoplasmic reticulum calcium ATPase; Shc, Src homology 2 domain containing transforming protein; SHP, Shp-2 tyrosine phosphatase; SOS, son of sevenless homolog; SPRY, Sprouty; STAT, signal transducer and activator of transcription; T, threonine; TGFα, transforming growth factor alpha; Ubc, ubiquitin-conjugating enzyme; Y, tyrosine. This image is also available as high resolution PDF (see Supplementary PDF 1) or Scalable Vector Graphic (SVG) or SBML (see Supplementary SBML 1).
The map is based on the molecular interactions documented in 242 papers accessible from PubMed (see the list of references for EGFR Pathway Map). It comprises 211 reactions and 322 species. A 'species' is a term defined by SBML as 'an entity that takes part in reactions' and it is used to distinguish the different states that are caused by enzymatic modification, association, dissociation, and translocation.
The species shown on the EGFR map can be categorized as follows: 202 proteins, three ions, 21 simple molecules, 73 oligomers, seven genes, and seven RNAs. In the number of species, eight degraded products and one unknown molecule are also included. Among 202 protein species, we identified 122 molecules, among which are 10 ligands, 10 receptors, 61 enzymes (including 32 kinases), three ion channels, 10 transcription factors, six G protein subunits, and 22 adaptor proteins.
The reactions can be categorized as follows: 131 state transitions, 34 transportations, 32 associations, 11 dissociations, two truncations, and one unknown transition. Among these reactions, there are 247 interactions; these represent 206 catalyses, nine unknown catalyses, 16 inhibitions, 12 transcriptional activations, and four transcriptional inhibitions. There are clusters of reactions that are involved in specific functions, such as endocytosis, degradation, recycling of EGFR, small GTPase signaling, MAPK cascade, PIP signaling, cell cycle, Ca2+ signaling, and GPCR-mediated EGFR transactivation. Reactions within each cluster are visually collocated to improve readability of the map.
The architecture of ErbB and GPCR signaling networks
Bow-tie structure
While the EGFR map cannot yet be the basis for a dynamical simulation until a series of kinetic parameters have been identified, it can help us understand the architectural feature of the signaling network. Looking at the map displayed in Figure 1, a notable feature becomes apparent; a variety of ligands bind to corresponding subtypes of erythroblastic leukemia viral (v-erb-b) oncogene homolog (ErbB) receptors that activate molecules in an extensive network of receptor complexes, and then converge into a handful of molecules, such as nonreceptor tyrosine kinase (non-RTK), small GTPase, and PIPs, which activate a variety of cascades leading to diverse responses including transcriptional regulation. This architecture, also called a bow-tie (or hourglass) structure, is a characteristic feature for robust evolvable systems (Kitano, 2004). Typically, it has diverse molecules for input and output that are connected to the conserved core with highly redundant and extensively crosstalking pathways and feedback control loops in various places in the pathway.
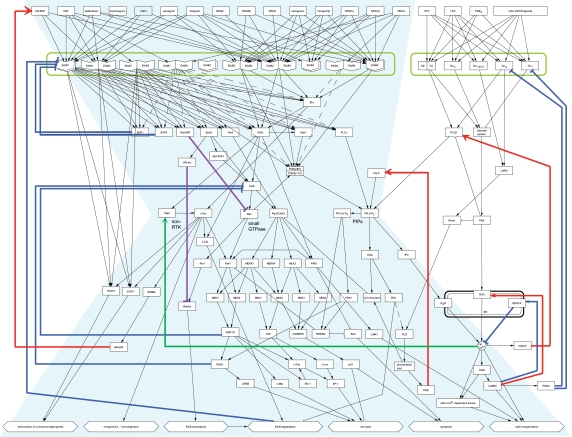
Figure 2 illustrates the overall bow-tie structure of molecular interactions included in the EGFR map ver. 2.0. The arrow in this figure represents the flow of a signal transduction. The ErbB receptor-mediated signaling network resembles a bow-tie structure with feedback control loops and inhibitory feed-forward paths. Positive and negative feedback controls are represented by red filled arrows and blue bar-headed lines, respectively. Inhibitory feed-forward paths are shown by purple bar-headed lines.
Figure 2.
The bow-tie architecture of the EGFR signaling pathway. A simplified diagram was created based on the EGFR signaling map in Figure 1. Arrows in this figure represent an informal notation of 'flow of reaction'. Various ligands bind to diverse receptor heterodimers, which then converge into a handful of molecules building a conserved core. Activities of these molecules play important roles in controlling diverse responses. Notable interactions are color-coded: red, positive feedback loop; blue, negative feedback loop; purple, inhibitory feed-forward path; green, crosstalk from GPCR cascade to EGFR cascade via calcium release. This image is also available as high resolution PDF (see Supplementary PDF 2) or Scalable Vector Graphic (SVG).
As input signals, 15 members of the endogenous EGF ligand family have been identified, that is, amphiregulin, betacellulin, biregulin, EGF, epiregulin, HB-EGF, heregulin α/β, neuregulin (NRG) 1α/1β/2α/2β/3/4, and transforming growth factor alpha (TGFα) (Jones et al, 1999; Olayioye et al, 2000; Yarden and Sliwkowski, 2001). While the ligands overlap with respect to binding to ErbB receptors, they have their own specificities and affinities for the respective receptors. The redundant and overlapping nature of ligand receptor binding enhances robustness in sensing the molecules in the environment, as dysfunction in one of the receptors may be compensated for by other receptors that have an affinity for the overlapping ligand molecule.
The binding of ligands induces homo- and heterodimerization of four ErbB family receptors: EGFR (ErbB1), ErbB2, ErbB3, and ErbB4 (Yarden and Schlessinger, 1987; Yarden and Sliwkowski, 2001). Although 10 combinations of ErbB receptor dimers are mathematically possible, only a subset of these is biologically meaningful. Specifically, ErbB2 has no high-affinity ligand and is only activated by heterodimerization with another ErbB receptor (Holbro et al, 2003), and the ErbB3 homodimer is inactive (Chen et al, 1996; Olayioye et al, 2000; Yarden and Sliwkowski, 2001). ErbB heterodimers form a highly redundant group of receptor complexes and thereby add to the complexity of EGFR signaling. Dimerization stimulates ErbB cytoplasmic kinase activity leading to auto- and trans-phosphorylation on tyrosine residues (Qian et al, 1994; Heldin, 1995), which serve as docking sites for five adaptor proteins and five enzymes, as shown in Figure 2. Signals from ErbBs converge to molecules forming a bow-tie core and are supposed to represent a versatile and conserved group of molecules and interactions. Molecules such as non-RTK (proline-rich tyrosine kinase (Pyk) 2, v-src sarcoma viral oncogene homolog (c-Src)), small GTPase (rat sarcoma viral oncogene homolog (Ras), Rac/cell division cycle 42 (Cdc42)), and PIPs (phosphatidylinositol-4-phosphate (PI4-P), phosphatidylinositol-4,5-bisphosphate (PI4,5-P2), phosphatidylinositol-3,4,5-triphosphate (PI3,4,5-P3)) are candidate of components that constitute the conserved core. Each molecule in the bow-tie core plays a central role in downstream signaling cascades to produce various physiological events such as cell cycle progression and migration via actin reorganization.
Furthermore, there is crosstalk between the ErbB and G protein coupled-receptor (GPCR) signaling cascade. Phospholipase C (PLC) γ stimulated by ErbB dimer produces inositol 1,4,5-triphosphate (IP3) from PI4,5-P2, which binds to IP3 receptor and causes Ca2+ efflux, while GPCR signaling regulates cytosol Ca2+ concentration via two enzymes, PLCβ and adenylyl cyclase. Release of Ca2+ affects Pyk2 activity that is placed in the possible bow-tie core segment.
Network control
Several system controls define the overall behavior of the signaling network. There are two positive feedback loops in the ErbB bow-tie structure. Firstly, Pyk2/c-Src activates ADAMs, which shed pro-HB-EGF (Dikic et al, 1996; Li et al, 1996; Poghosyan et al, 2002), so that the amount of HB-EGF will be increased and enhance the signaling. This Pyk2/c-Src-mediated feedback loop is further enhanced by the Ca2+-mediated crosstalk from the GPCR signaling cascade (shown by a green line in Figure 2) (Prenzel et al, 1999; Carpenter, 2000; Shi et al, 2000; Schafer et al, 2004). Secondly, active PLCβ/γ produces diacylglycerol (DAG) from PI4,5-P2, which results in the cascading activation of protein kinase C (PKC) (Mellor and Parker, 1998), phospholipase D (PLD) (Exton, 2002), and phosphatidylinositol-5-kinase (PI5K) (Moritz et al, 1992). PI5K phosphorylates PI4-P resulting in an increase of PI4,5-P2.
There are six negative feedback loops. In two of these, protein tyrosine phosphatases (SHP-1 and SHP-2) inhibit EGFR at the input wing of the bow tie. In three others, a son of sevenless (SOS) homolog (Rozakis-Adcock et al, 1995; Douville and Downward, 1997) is inhibited (by extracellular signal-regulated kinase (ERK) 1, ERK2, or ribosomal protein S6 kinase (RSK 2)), starting from the output wing to SOS, which localizes near the core of the bow tie. In the sixth, ErbB is degraded (via the activity of Casitas B-lineage lymphoma proto-oncogene (c-Cbl), which is recruited by growth factor receptor-bound protein (Grb) 2) (Levkowitz et al, 1999; Yokouchi et al, 1999; Ravid et al, 2004); here, feedback starts from the very end of the output wing, moving toward the initial input wing of the bow tie. In addition, a number of local inhibitory control exist that use phosphatases to control kinase activities.
There are cases where both activation and inhibition are directed to the same protein. For example, EGFR provides both positive signaling to Ras activation, and negative regulation through recruitment of Ras GTPase-activating protein (RasGAP) (Agazie and Hayman, 2003). RAS-associated protein RAB5a (Rab5a) is influenced by both activation and inhibition signals from Ras interaction 1 (Rin1) (Tall et al, 2001) and related to the N-terminus of tre (RN-tre) (Lanzetti et al, 2000), respectively. EGFR essentially regulates both paths as it binds EGF receptor pathway substrate (Eps) 8 that activates RN-tre, and binds Grb2, which in turn stimulates Ras via SOS leading to Rin1 activation (Han et al, 1997). It is interesting to note that in both cases, the length of the path for inhibition is shorter than that of activation. It will be important to understand how such positive and negative controls are regulated.
In total, there are two positive feedback loops, six negative feedback controls, and inhibitory feed-forward paths in the ErbB bow-tie structure. In addition, there are a few positive and negative feedback loops in the GPCR cascade that affect ErbB pathway dynamics. As a whole, the ErbB signaling network forms an overall bow-tie structure with highly redundant and overlapping input pathways and feedback controls. We consider that such a bow-tie structure with feedback control is a typical architecture for signal transduction pathways that can be observed even in TLR and GPCR pathways. Understanding the dynamics of such an architecture is critically important for an in-depth knowledge of signaling systems in general. This includes understanding how such pathways have evolved, and how diverse input stimuli are encoded, converge, and differentially activate various reactions, including the transcription of downstream genes.
Graphical notations of the EGFR Pathway Map
Process diagram
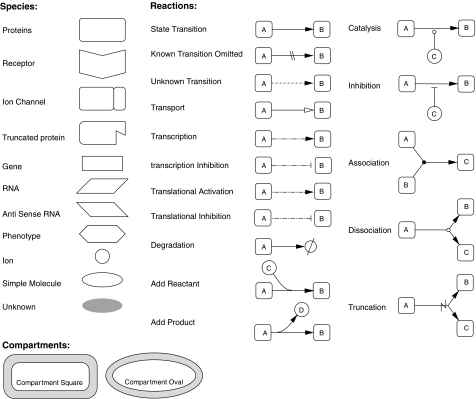
The main symbols used to represent molecules and interactions in this map are displayed in Figure 3. Kitano proposed a graphical notation system for biological networks designed to express sufficient information in a clearly visible and unambiguous way (Kitano, 2003). Several graphical notations for molecular interactions have been proposed previously (Kohn, 1999; Pirson et al, 2000; Cook et al, 2001; Kohn, 2001; Maimon and Browning, 2001), although none has been widely used. The Kohn Map is perhaps the most widely known of these. However, lack of software to support the notation has hampered its use. We have developed CellDesigner, a freely downloadable software tool. It has already been adopted by various research groups and databases such as the PANTHER pathway database (Mi et al, 2005). The current EGFR map is essentially a state transition diagram, in which one state of the system is represented in one node, and an arc from one node to another node represents a transition of the state of the system. This class of diagrams is often used in engineering and software development, and the schema avoids using symbols that directly point to molecules to indicate activation or inhibition. The arrow of state transition (a straight line with a filled arrowhead) represents the state changes that occur as a result of molecular interactions, instead of 'activation' in a traditional notation familiar to molecular biologists. The diagram directly indicates a transition from an inactive to an active state for activation, and a transition from an active state to an inactive state for inhibition. When these transitions are promoted or inhibited by other mediating molecules, such as active kinases, these reactions are represented by a catalysis arrow (circle-headed line) and inhibition arrow (bar-headed line), respectively. It is essential that such syntax and semantics are made clear and defined consistently, particularly for a large-scale map, so that the information presented is conveyed unambiguously.
Figure 3.
Main symbols adopted by CellDesigner ver. 2.1.1. These symbols are provided in CellDesigner ver. 2.1.1. Size and color of each module are configurable. CellDesigner also provides X-Y coordinates for each module and can distinguish between cellular compartments.
Notation on modification and localization of protein
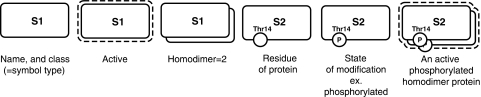
Figure 4 illustrates how the modification status of a protein is presented. Essentially, each state of a protein (i.e. phosphorylation, acetylation, etc.) can be represented such that it reflects its modification and oligomerization.
Figure 4.
Expression of the inner structures and states. The active state of the molecule is indicated by a dashed line surrounding the molecule. State changes of a component such as phosphorylation, acetylation, ubiquitination, and allosteric changes can be represented with specific information such as target residue and position.
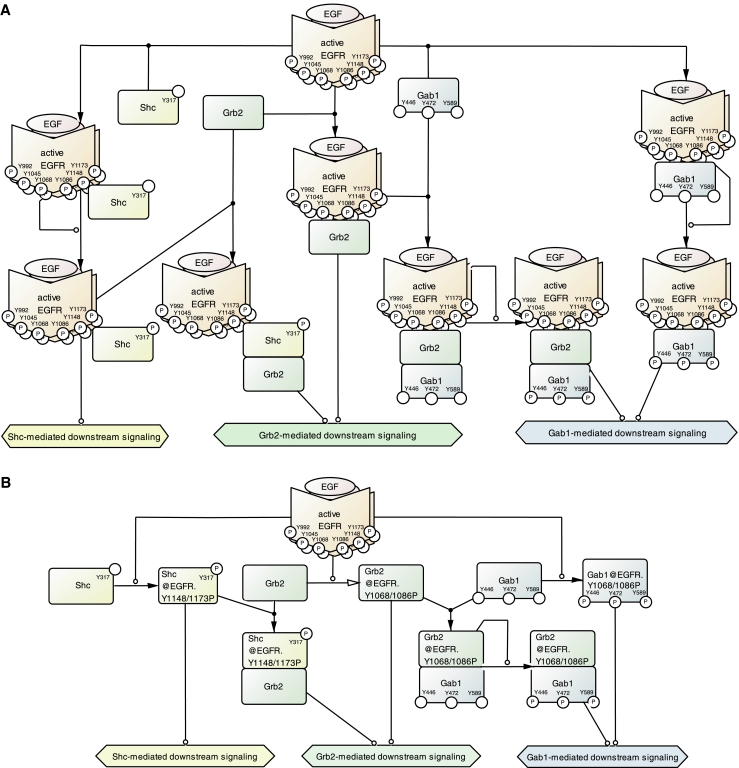
In this map, we employed a naming convention in which the localization of protein is indicated by a prefix to the protein name, such as 'cyt.XX' and 'pl.m.XX' for protein XX in the cytosol and protein XX at the plasma membrane, respectively. In addition, in order to provide a better overview and to understand pathways at a glance, we assigned unique names with an 'address' to a protein to express differences of combination states of protein species. For instance, Figure 5 provides the reader with a small part of the pathways illustrated in the map, namely interactions between EGFR and the three adaptor proteins, Src homology 2 domain containing transforming protein (Shc), Grb2, and GRB2-associated binding protein 1 (Gab1). Figure 5A shows the detailed scheme of combination states between EGFR and adaptors, while Figure 5B expresses combination states by assigning an 'address' to the name of a protein. The method of referring to proteins with an 'address' becomes clear using Grb2 as an example. Grb2 is recruited to the activated EGFR via the phosphotyrosine residues Tyr1068 or Tyr1086, and this event is denoted as 'Grb2@EGFR.Y1068/1086P'. The reaction of association between active EGFR and Grb2 is represented using an open-headed 'transport' arrow and a circle-headed 'catalysis' arrow as a local rule adopted in this map ver. 2.0. This convention allows for a more efficient presentation of signaling events and requires much less space, as illustrated in Figure 5B. It should be stressed that this convention is in accordance with the information provided by a full representation.
Figure 5.
Ellipsis in drawing association states of proteins using an 'address'. (A) Precise association states between EGFR and adaptors. Three adaptor proteins, Shc, Grb2, and Gab1, bind to the activated EGFR via its autophosphorylated tyrosine residues. Shc binds to activated EGFR and is phosphorylated on its tyrosine 317. Grb2 binds to activated EGFR either directly or via Shc bound to activated EGFR. Gab1 also binds to activated EGFR either directly or via Grb2 bound to activated EGFR, and is phosphorylated on its tyrosine 446, 472, and 589. (B) The same signaling pathway as in panel A using an 'address' such as 'Grb2@EGFR.Y1068/1086P', thereby achieving a presentation of the pathway details. EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; Gab, GRB2-associated binding protein; Grb, growth factor receptor-bound protein; Shc, Src homology 2 domain containing transforming protein.
Omissions in notation
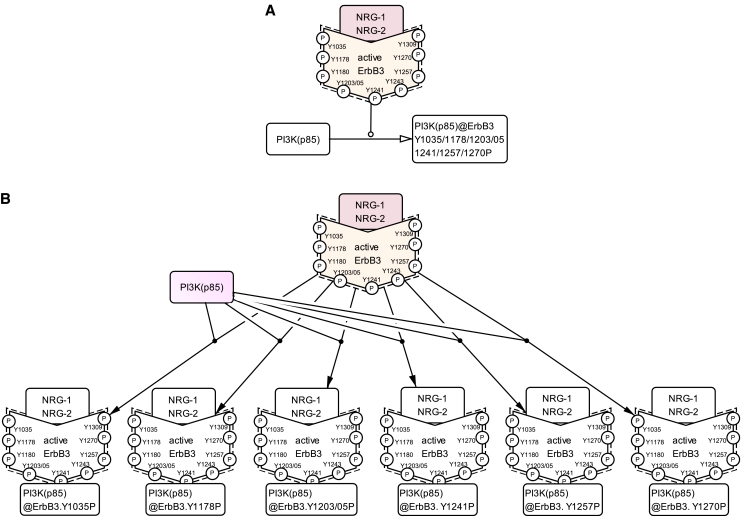
For ease of readability and in order to save space, we adopted to omit notation from this version of the EGFR Pathway Map (ver. 2.0). While simulation studies require precise representation of pathways, such representation has to deal with the complicated issue of multiple states of complexes. Figure 6 shows a simple example. The 85 kDa regulatory subunit of phosphatidylinositol 3-kinase (PI3K (p85)) binds to active ErbB3 receptor via its phosphorylated tyrosine residues: Tyr1035, Tyr1178, Tyr1203/05, Tyr1241, Tyr1257, and Tyr1270 (Olayioye et al, 2000). To distinguish the complexes according to differences of phosphotyrosine residues, Figure 6A should be redrawn as Figure 6B.
Figure 6.
Combinations of multiple states of complexes. (A) Omitted notation in the EGFR Pathway Map. The 85 kDa regulatory subunit of phosphatidylinositol 3-kinase (PI3K (p85)) binds to active ErbB3 receptor via its phosphorylated tyrosine residues: Tyr1035, Tyr1178, Tyr1203/05, Tyr1241, Tyr1257, and Tyr1270. (B) Detailed portrayal of panel A for distinguishing the complexes according to differences of phosphotyrosine residues. ErbB3, erythroblastic leukemia viral (v-erb-b) oncogene homolog 3; PI3K (p85), 85 kDa regulatory subunit of phosphatidylinositol 3-kinase.
Another type of omission concerns the case in which many pathways are represented by fewer pathways. For example, it has been reported that Grb2 and Shc bind to activated EGFR via their phosphotyrosine residues and function as adaptors of downstream signaling. They are recruited to endosomes during stimulation by EGF where they form complexes with endocytosed EGFR and activate Ras signaling (see the list of references for EGFR Pathway Map). Although some other proteins such as PI3K (p85/p110) are reported to be translocated to endosomes with growth factor receptors (Christoforidis et al, 1999), it is not clear whether all other EGF-induced interactions occur similarly in endosomes as well as at the cell surface. To conserve space in the current version of the EGFR Pathway Map, we made Grb2 represent interactions with endosomal EGFR.
In addition to sphingosine-1-phosphate (S1P), lysophosphatidic acid (LPA), and prostaglandin E2 (PGE2), other ligands such as endothelin-1 (Vacca et al, 2000) and angiotensin II (Hama et al, 2004) have been reported to be involved in GPCR-mediated EGFR signal transactivation.
Ambiguity
A number of ambiguous cases of protein-protein interactions came up during the construction of the map. For example, EGF simulation induces activation of protein kinase B (PKB/Akt) via PIPs, which have multiple functions including antiapoptotic properties. However, the mechanistic details as to its activation are controversial. It has been reported that PKB/Akt is phosphorylated at two sites for its full activation: Thr308 in the activation T-loop of kinase domain and Ser473 in the C-terminal hydrophobic motif. While phosphoinositide-dependent kinase 1 (PDK1) has been unambiguously identified as Thr308 kinase, Ser473 kinase named PDK2 remains elusive. Although it has recently been reported that the conventional isoforms of PKC could phosphorylate at Ser473 by distinct stimulation (Kawakami et al, 2004), PKC inhibitors including PKCbeta inhibitor LY 379196 caused PKB/Akt phosphorylation at Ser473 (Wen et al, 2003). Moreover, Toker and Newton (2000) reported that the PDK2 site, namely Ser473, was regulated by autophosphorylation. Because it is not clear whether Ser473 undergoes autophosphorylation, phosphorylation by PDK2, or both, the pathway is represented by unknown catalysis arrows (circle-headed dashed line).
CellDesigner
The EGFR Pathway Map was created using CellDesigner ver. 2.1.1. Compliance of CellDesigner with SBML enables researchers to store models and to use them for analyses by other SBML-compliant applications.
CellDesigner is also a Systems Biology Workbench (SBW)-enabled application. With SBW installed, CellDesigner can integrate with all SBW-enabled modules, including simulation and other analysis packages.
The most recent version of CellDesigner (ver. 2.2) enables users to store data of each molecule and reaction in the species and reaction <notes>, respectively, to link directly to the database such as PubMed simply by clicking. CellDesigner can thus be a portal software platform as well as information organizer for systems biology research.
Updating of the EGFR Pathway Map
This version of the map (ver. 2.0) is intended to be comprehensive but is not necessarily exhaustive. We will periodically update and expand the map on our website using experimental data derived from further studies and through interactions with researchers specialized in certain modules of the EGFR signaling network. To facilitate such interaction and updating of the map, we are currently designing community-support web-based tools that will allow a community-based collaborative development process. Addition and correction of the original map can be made through comments and feedback from experts in specific molecules and interactions, while kinetic constants and other experimentally obtained data can be incorporated into the map.
In systems biology research, both molecular details and a system-wide network structure must be taken into account. Thus, data resources and tools that enable flexible and updated access to various levels of information are essential. The EGFR signaling map presented in this article is one attempt to seed such effort.
Supplementary Material
Supplementary PDF 1
Supplementary PDF 2
Supplementary SBML 1
Acknowledgments
This research is, in part, supported by the Exploratory Research for Advanced Technology (ERATO) and the Solution-Oriented Research for Science and Technology (SORST) programs (Japan Science and Technology Organization), the NEDO Grant (New Energy and Industrial Technology Development Organization) of the Japanese Ministry of Economy, Trade and Industry (METI), the Special Coordination Funds for Promoting Science and Technology and the Center of Excellence Program for Keio University (Ministry of Education, Culture, Sports, Science, and Technology), The Genome Network Project (the Japanese Ministry of Education, Culture, Sports, Science and Technology), and the Air Force Office of Scientific Research (AFOSR).
References
- Agazie YM, Hayman MJ (2003) Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol 23: 7875–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G (2000) EGF receptor transactivation mediated by the proteolytic production of EGF-like agonists. Sci STKE 2000: PE1 [DOI] [PubMed] [Google Scholar]
- Chen X, Levkowitz G, Tzahar E, Karunagaran D, Lavi S, Ben-Baruch N, Leitner O, Ratzkin BJ, Bacus SS, Yarden Y (1996) An immunological approach reveals biological differences between the two NDF/heregulin receptors, ErbB-3 and ErbB-4. J Biol Chem 271: 7620–7629 [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M (1999) Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol 1: 249–252 [DOI] [PubMed] [Google Scholar]
- Cook DL, Farley JF, Tapscott SJ (2001) A basis for a visual language for describing, archiving and analyzing functional models of complex biological systems. Genome Biol 2, RESEARCH0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Tokiwa G, Lev S, Courtneidge SA, Schlessinger J (1996) A role for Pyk2 and Src in linking G-protein-coupled receptors with MAP kinase activation. Nature 383: 547–550 [DOI] [PubMed] [Google Scholar]
- Douville E, Downward J (1997) EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene 15: 373–383 [DOI] [PubMed] [Google Scholar]
- Exton JH (2002) Regulation of phospholipase D. FEBS Lett 531: 58–61 [DOI] [PubMed] [Google Scholar]
- Funahashi A, Morohashi M, Tanimura N, Kitano H (2003) CellDesigner: a process diagram editor for gene-regulatory and biochemical networks. BioSilico 1: 159–162 [Google Scholar]
- Hama K, Ohnishi H, Yasuda H, Ueda N, Mashima H, Satoh Y, Hanatsuka K, Kita H, Ohashi A, Tamada K, Sugano K (2004) Angiotensin II stimulates DNA synthesis of rat pancreatic stellate cells by activating ERK through EGF receptor transactivation. Biochem Biophys Res Commun 315: 905–911 [DOI] [PubMed] [Google Scholar]
- Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J (1997) Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci USA 94: 4954–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldin CH (1995) Dimerization of cell surface receptors in signal transduction. Cell 80: 213–223 [DOI] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE (2003) The ErbB receptors and their role in cancer progression. Exp Cell Res 284: 99–110 [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, Dronov S, Gilles ED, Ginkel M, Gor V, Goryanin II, Hedley WJ, Hodgman TC, Hofmeyr JH, Hunter PJ, Juty NS, Kasberger JL, Kremling A, Kummer U, Le Novere N, Loew LM, Lucio D, Mendes P, Minch E, Mjolsness ED, Nakayama Y, Nelson MR, Nielsen PF, Sakurada T, Schaff JC, Shapiro BE, Shimizu TS, Spence HD, Stelling J, Takahashi K, Tomita M, Wagner J, Wang J, SBML Forum (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 19: 524–531 [DOI] [PubMed] [Google Scholar]
- Jones JT, Akita RW, Sliwkowski MX (1999) Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett 447: 227–231 [DOI] [PubMed] [Google Scholar]
- Kawakami Y, Nishimoto H, Kitaura J, Maeda-Yamamoto M, Kato RM, Littman DR, Leitges M, Rawlings DJ, Kawakami T (2004) Protein kinase C betaII regulates Akt phosphorylation on Ser-473 in a cell type- and stimulus-specific fashion. J Biol Chem 279: 47720–47725 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN (2003) Four-dimensional organization of protein kinase signaling cascades: the roles of diffusion, endocytosis and molecular motors. J Exp Biol 206: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Kholodenko BN, Demin OV, Moehren G, Hoek JB (1999) Quantification of short term signaling by the epidermal growth factor receptor. J Biol Chem 274: 30169–30181 [DOI] [PubMed] [Google Scholar]
- Kitano H (2003) A graphical notation for biochemical networks. BioSilico 1: 169–176 [Google Scholar]
- Kitano H (2004) Biological robustness. Nat Rev Genet 5: 826–837 [DOI] [PubMed] [Google Scholar]
- Kohn K (2001) Molecular Interaction Maps as information organizers and simulation guides. Chaos 11: 84–97 [DOI] [PubMed] [Google Scholar]
- Kohn KW (1999) Molecular interaction map of the mammalian cell cycle control and DNA repair systems. Mol Biol Cell 10: 2703–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP (2000) The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408: 374–377 [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Li J, Avraham H, Rogers RA, Raja S, Avraham S (1996) Characterization of RAFTK, a novel focal adhesion kinase, and its integrin-dependent phosphorylation and activation in megakaryocytes. Blood 88: 417–428 [PubMed] [Google Scholar]
- Maimon R, Browning S (2001) Diagrammatic notation and computational structure of gene networks. Proceedings of the Second International Conference on Systems Biology, Pasadena, CA [Google Scholar]
- Mellor H, Parker PJ (1998) The extended protein kinase C superfamily. Biochem J 332: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, Kitano H, Thomas PD (2005) The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res 33 (Database Issue): D284–D288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz A, De Graan PN, Gispen WH, Wirtz KW (1992) Phosphatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase. J Biol Chem 267: 7207–7210 [PubMed] [Google Scholar]
- Olayioye MA, Neve RM, Lane HA, Hynes NE (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 19: 3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirson I, Fortemaison N, Jacobs C, Dremier S, Dumont JE, Maenhaut C (2000) The visual display of regulatory information and networks. Trends Cell Biol 10: 404–408 [DOI] [PubMed] [Google Scholar]
- Poghosyan Z, Robbins SM, Houslay MD, Webster A, Murphy G, Edwards DR (2002) Phosphorylation-dependent interactions between ADAM15 cytoplasmic domain and Src family protein-tyrosine kinases. J Biol Chem 277: 4999–5007 [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A (1999) EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 402: 884–888 [DOI] [PubMed] [Google Scholar]
- Qian X, LeVea CM, Freeman JK, Dougall WC, Greene MI (1994) Heterodimerization of epidermal growth factor receptor and wild-type or kinase-deficient Neu: a mechanism of interreceptor kinase activation and transphosphorylation. Proc Natl Acad Sci USA 91: 1500–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid T, Heidinger JM, Gee P, Khan EM, Goldkorn T (2004) c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J Biol Chem 279: 37153–37162 [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, van der Geer P, Mbamalu G, Pawson T (1995) MAP kinase phosphorylation of mSos1 promotes dissociation of mSos1-Shc and mSos1-EGF receptor complexes. Oncogene 11: 1417–1426 [PubMed] [Google Scholar]
- Schafer B, Marg B, Gschwind A, Ullrich A (2004) Distinct ADAM metalloproteinases regulate G protein-coupled receptor-induced cell proliferation and survival. J Biol Chem 279: 47929–47938 [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G (2002) Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nat Biotechnol 20: 370–375 [DOI] [PubMed] [Google Scholar]
- Shi CS, Sinnarajah S, Cho H, Kozasa T, Kehrl JH (2000) G13alpha-mediated PYK2 activation. PYK2 is a mediator of G13alpha-induced serum response element-dependent transcription. J Biol Chem 275: 24470–24476 [DOI] [PubMed] [Google Scholar]
- Shvartsman SY, Muratov CB, Lauffenburger DA (2002) Modeling and computational analysis of EGF receptor-mediated cell communication in Drosophila oogenesis. Development 129: 2577–2589 [DOI] [PubMed] [Google Scholar]
- Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF (2001) Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell 1: 73–82 [DOI] [PubMed] [Google Scholar]
- Toker A, Newton AC (2000) Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J Biol Chem 275: 8271–8274 [DOI] [PubMed] [Google Scholar]
- Vacca F, Bagnato A, Catt KJ, Tecce R (2000) Transactivation of the epidermal growth factor receptor in endothelin-1-induced mitogenic signaling in human ovarian carcinoma cells. Cancer Res 60: 5310–5317 [PubMed] [Google Scholar]
- Wen HC, Huang WC, Ali A, Woodgett JR, Lin WW (2003) Negative regulation of phosphatidylinositol 3-kinase and Akt signalling pathway by PKC. Cell Signal 15: 37–45 [DOI] [PubMed] [Google Scholar]
- Wiley HS, Shvartsman SY, Lauffenburger DA (2003) Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol 13: 43–50 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Schlessinger J (1987) Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry 26: 1443–1451 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R (1999) Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem 274: 31707–31712 [DOI] [PubMed] [Google Scholar]
[References for EGFR Pathway Map] Epidermal growth factor (EGFR)
- Agazie YM, Hayman MJ (2003) Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol 23: 7875–7886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham H, Park SY, Schinkmann K, Avraham S (2000) RAFTK/Pyk2-mediated cellular signalling. Cell Signal 12: 123–133 [DOI] [PubMed] [Google Scholar]
- Batzer AG, Rotin D, Urena JM, Skolnik EY, Schlessinger J (1994) Hierarchy of binding sites for Grb2 and Shc on the epidermal growth factor receptor. Mol Cell Biol 14: 5192–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biscardi JS, Maa MC, Tice DA, Cox ME, Leu TH, Parsons SJ (1999) c-Src-mediated phosphorylation of the epidermal growth factor receptor on Tyr845 and Tyr1101 is associated with modulation of receptor function. J Biol Chem 274: 8335–8343 [DOI] [PubMed] [Google Scholar]
- Bishayee A, Beguinot L, Bishayee S (1999) Phosphorylation of tyrosine 992, 1068, and 1086 is required for conformational change of the human epidermal growth factor receptor C-terminal tail. Mol Biol Cell 10: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger C, Nagel-Wolfrum K, Kunz C, Wittig I, Butz K, Hoppe-Seyler F, Groner B (2003) Sequence-specific peptide aptamers, interacting with the intracellular domain of the epidermal growth factor receptor, interfere with Stat3 activation and inhibit the growth of tumor cells. J Biol Chem 278: 37610–37621 [DOI] [PubMed] [Google Scholar]
- Castagnino P, Biesova Z, Wong WT, Fazioli F, Gill GN, Di Fiore PP (1995) Direct binding of eps8 to the juxtamembrane domain of EGFR is phosphotyrosine- and SH2-independent. Oncogene 10: 723–729 [PubMed] [Google Scholar]
- Chattopadhyay A, Vecchi M, Ji Q, Mernaugh R, Carpenter G (1999) The role of individual SH2 domains in mediating association of phospholipase C-gamma1 with the activated EGF receptor. J Biol Chem 274: 26091–26097 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr (1997) STATs and gene regulation. Science 277: 1630–1635 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Scita G (2002) Eps8 in the midst of GTPases. Int J Biochem Cell Biol 34: 1178–1183 [DOI] [PubMed] [Google Scholar]
- Dikic I, Tokiwa G, Lev S, Courtneidge SA, Schlessinger J (1996) A role for Pyk2 and Src in linking G-protein-coupled receptors with MAP kinase activation. Nature 383: 547–550 [DOI] [PubMed] [Google Scholar]
- Downward J, Parker P, Waterfield MD (1984) Autophosphorylation sites on the epidermal growth factor receptor. Nature 311: 483–485 [DOI] [PubMed] [Google Scholar]
- Heldin CH (1995) Dimerization of cell surface receptors in signal transduction. Cell 80: 213–223 [DOI] [PubMed] [Google Scholar]
- Jones JT, Akita RW, Sliwkowski MX (1999) Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett 447: 227–231 [DOI] [PubMed] [Google Scholar]
- Keilhack H, Tenev T, Nyakatura E, Godovac-Zimmermann J, Nielsen L, Seedorf K, Bohmer FD (1998) Phosphotyrosine 1173 mediates binding of the protein-tyrosine phosphatase SHP-1 to the epidermal growth factor receptor and attenuation of receptor signaling. J Biol Chem 273: 24839–24846 [DOI] [PubMed] [Google Scholar]
- Kim JW, Sim SS, Kim UH, Nishibe S, Wahl MI, Carpenter G, Rhee SG (1990) Tyrosine residues in bovine phospholipase C-gamma phosphorylated by the epidermal growth factor receptor in vitro . J Biol Chem 265: 3940–3943 [PubMed] [Google Scholar]
- Lanzetti L, Rybin V, Malabarba MG, Christoforidis S, Scita G, Zerial M, Di Fiore PP (2000) The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature 408: 374–377 [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y (1999) Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell 4: 1029–1040 [DOI] [PubMed] [Google Scholar]
- Li J, Avraham H, Rogers RA, Raja S, Avraham S (1996) Characterization of RAFTK, a novel focal adhesion kinase, and its integrin-dependent phosphorylation and activation in megakaryocytes. Blood 88: 417–428 [PubMed] [Google Scholar]
- Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J (1993) Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 363: 85–88 [DOI] [PubMed] [Google Scholar]
- Liu Y, Rohrschneider LR (2002) The gift of Gab. FEBS Lett 515: 1–7 [DOI] [PubMed] [Google Scholar]
- Lock LS, Royal I, Naujokas MA, Park M (2000) Identification of an atypical Grb2 carboxyl-terminal SH3 domain binding site in Gab docking proteins reveals Grb2-dependent and -independent recruitment of Gab1 to receptor tyrosine kinases. J Biol Chem 275: 31536–31545 [DOI] [PubMed] [Google Scholar]
- Margolis BL, Lax I, Kris R, Dombalagian M, Honegger AM, Howk R, Givol D, Ullrich A, Schlessinger J (1989) All autophosphorylation sites of epidermal growth factor (EGF) receptor and HER2/neu are located in their carboxyl-terminal tails. Identification of a novel site in EGF receptor. J Biol Chem 264: 10667–10671 [PubMed] [Google Scholar]
- Mattoon DR, Lamothe B, Lax I, Schlessinger J (2004) The docking protein Gab1 is the primary mediator of EGF-stimulated activation of the PI-3K/Akt cell survival pathway. BMC Biol 2: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishibe S, Wahl MI, Hernandez-Sotomayor SM, Tonks NK, Rhee SG, Carpenter G (1990) Increase of the catalytic activity of phospholipase C-gamma 1 by tyrosine phosphorylation. Science 250: 1253–1256 [DOI] [PubMed] [Google Scholar]
- Okabayashi Y, Kido Y, Okutani T, Sugimoto Y, Sakaguchi K, Kasuga M (1994) Tyrosines 1148 and 1173 of activated human epidermal growth factor receptors are binding sites of Shc in intact cells. J Biol Chem 269: 18674–18678 [PubMed] [Google Scholar]
- Okutani T, Okabayashi Y, Kido Y, Sugimoto Y, Sakaguchi K, Matuoka K, Takenawa T, Kasuga M (1994) Grb2/Ash binds directly to tyrosines 1068 and 1086 and indirectly to tyrosine 1148 of activated human epidermal growth factor receptors in intact cells. J Biol Chem 269: 31310–31314 [PubMed] [Google Scholar]
- Olayioye MA, Beuvink I, Horsch K, Daly JM, Hynes NE (1999) ErbB receptor-induced activation of stat transcription factors is mediated by Src tyrosine kinases. J Biol Chem 274: 17209–17218 [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Neve RM, Lane HA, Hynes NE (2000) The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J 19: 3159–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues GA, Falasca M, Zhang Z, Ong SH, Schlessinger J (2000) A novel positive feedback loop mediated by the docking protein Gab1 and phosphatidylinositol 3-kinase in epidermal growth factor receptor signaling. Mol Cell Biol 20: 1448–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Margolis B, Mohammadi M, Daly RJ, Daum G, Li N, Fischer EH, Burgess WH, Ullrich A, Schlessinger J (1992) SH2 domains prevent tyrosine dephosphorylation of the EGF receptor: identification of Tyr992 as the high-affinity binding site for SH2 domains of phospholipase C gamma. EMBO J 11: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D (1993) The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature 363: 83–85 [DOI] [PubMed] [Google Scholar]
- Sakaguchi K, Okabayashi Y, Kido Y, Kimura S, Matsumura Y, Inushima K, Kasuga M (1998) Shc phosphotyrosine-binding domain dominantly interacts with epidermal growth factor receptors and mediates Ras activation in intact cells. Mol Endocrinol 12: 536–543 [DOI] [PubMed] [Google Scholar]
- Sato K, Nagao T, Iwasaki T, Nishihira Y, Fukami Y (2003) Src-dependent phosphorylation of the EGF receptor Tyr-845 mediates Stat-p21waf1 pathway in A431 cells. Genes Cells 8: 995–1003 [DOI] [PubMed] [Google Scholar]
- Sato K, Nagao T, Kakumoto M, Kimoto M, Otsuki T, Iwasaki T, Tokmakov AA, Owada K, Fukami Y (2002) Adaptor protein Shc is an isoform-specific direct activator of the tyrosine kinase c-Src. J Biol Chem 277: 29568–29576 [DOI] [PubMed] [Google Scholar]
- Sato K, Sato A, Aoto M, Fukami Y (1995) c-Src phosphorylates epidermal growth factor receptor on tyrosine 845. Biochem Biophys Res Commun 215: 1078–1087 [DOI] [PubMed] [Google Scholar]
- Scita G, Nordstrom J, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegard M, Betsholtz C, Di Fiore PP (1999) EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401: 290–293 [DOI] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH (2004) Pyk2 amplifies epidermal growth factor and c-Src-induced Stat3 activation. J Biol Chem 279: 17224–17231 [DOI] [PubMed] [Google Scholar]
- Shoelson SE (1997) SH2 and PTB domain interactions in tyrosine kinase signal transduction. Curr Opin Chem Biol 1: 227–234 [DOI] [PubMed] [Google Scholar]
- Silvennoinen O, Schindler C, Schlessinger J, Levy DE (1993) Ras-independent growth factor signaling by transcription factor tyrosine phosphorylation. Science 261: 1736–1739 [DOI] [PubMed] [Google Scholar]
- Stover DR, Becker M, Liebetanz J, Lydon NB (1995) Src phosphorylation of the epidermal growth factor receptor at novel sites mediates receptor interaction with Src and P85 alpha. J Biol Chem 270: 15591–15597 [DOI] [PubMed] [Google Scholar]
- Sudol M (1998) From Src Homology domains to other signaling modules: proposal of the 'protein recognition code'. Oncogene 17 (11 Reviews): 1469–1474 [DOI] [PubMed] [Google Scholar]
- Tice DA, Biscardi JS, Nickles AL, Parsons SJ (1999) Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc Natl Acad Sci USA 96: 1415–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton GM, Chen WS, Rosenfeld MG, Gill GN (1990) Analysis of deletions of the carboxyl terminus of the epidermal growth factor receptor reveals self-phosphorylation at tyrosine 992 and enhanced in vivo tyrosine phosphorylation of cell substrates. J Biol Chem 265: 1750–1754 [PubMed] [Google Scholar]
- Weiss A, Schlessinger J (1998) Switching signals on or off by receptor dimerization. Cell 94: 277–280 [DOI] [PubMed] [Google Scholar]
- Wu TR, Hong YK, Wang XD, Ling MY, Dragoi AM, Chung AS, Campbell AG, Han ZY, Feng GS, Chin YE (2002) SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem 277: 47572–47580 [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX (2001) Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2: 127–137 [DOI] [PubMed] [Google Scholar]
- Yu CL, Meyer DJ, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R (1995) Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269: 81–83 [DOI] [PubMed] [Google Scholar]
- Zhang SQ, Tsiaras WG, Araki T, Wen G, Minichiello L, Klein R, Neel BG (2002) Receptor-specific regulation of phosphatidylinositol 3′-kinase activation by the protein tyrosine phosphatase Shp2. Mol Cell Biol 22: 4062–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wen Z, Darnell JE Jr (1994) Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264: 95–98 [DOI] [PubMed] [Google Scholar]
EGFR endocytosis followed by degradation or recycling
- Bao J, Alroy I, Waterman H, Schejter ED, Brodie C, Gruenberg J, Yarden Y (2000) Threonine phosphorylation diverts internalized epidermal growth factor receptors from a degradative pathway to the recycling endosome. J Biol Chem 275: 26178–26186 [DOI] [PubMed] [Google Scholar]
- Barbieri MA, Roberts RL, Gumusboga A, Highfield H, Alvarez-Dominguez C, Wells A, Stahl PD (2000) Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J Cell Biol 151: 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Wong D, Dhaka A, Afar D, White M, Xie W, Herschman H, Witte O, Colicelli J (1997) Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci USA 94: 4954–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Sorkin A (2002) Coordinated traffic of Grb2 and Ras during epidermal growth factor receptor endocytosis visualized in living cells. Mol Biol Cell 13: 1522–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassenbrock CK, Hunter S, Garl P, Johnson GL, Anderson SM (2002) Inhibition of Src family kinases blocks epidermal growth factor (EGF)-induced activation of Akt, phosphorylation of c-Cbl, and ubiquitination of the EGF receptor. J Biol Chem 277: 24967–24975 [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev 12: 3663–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Wong ES, Ong SH, Yusoff P, Low BC, Guy GR (2000) Sprouty proteins are targeted to membrane ruffles upon growth factor receptor tyrosine kinase activation. Identification of a novel translocation domain. J Biol Chem 275: 32837–32845 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Benjamin DR (1996) A novel family of Ras-binding domains. Trends Biochem Sci 21: 422–425 [DOI] [PubMed] [Google Scholar]
- Ravid T, Heidinger JM, Gee P, Khan EM, Goldkorn T (2004) c-Cbl-mediated ubiquitinylation is required for epidermal growth factor receptor exit from the early endosomes. J Biol Chem 279: 37153–37162 [DOI] [PubMed] [Google Scholar]
- Sorkin A, McClure M, Huang F, Carter R (2000) Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Curr Biol 10: 1395–1398 [DOI] [PubMed] [Google Scholar]
- Tall GG, Barbieri MA, Stahl PD, Horazdovsky BF (2001) Ras-activated endocytosis is mediated by the Rab5 guanine nucleotide exchange activity of RIN1. Dev Cell 1: 73–82 [DOI] [PubMed] [Google Scholar]
- Wong ES, Lim J, Low BC, Chen Q, Guy GR (2001) Evidence for direct interaction between Sprouty and Cbl. J Biol Chem 276: 5866–5875 [DOI] [PubMed] [Google Scholar]
- Yokouchi M, Kondo T, Houghton A, Bartkiewicz M, Horne WC, Zhang H, Yoshimura A, Baron R (1999) Ligand-induced ubiquitination of the epidermal growth factor receptor involves the interaction of the c-Cbl RING finger and UbcH7. J Biol Chem 274: 31707–31712 [DOI] [PubMed] [Google Scholar]
- Yokouchi M, Kondo T, Sanjay A, Houghton A, Yoshimura A, Komiya S, Zhang H, Baron R (2001) Src-catalyzed phosphorylation of c-Cbl leads to the interdependent ubiquitination of both proteins. J Biol Chem 276: 35185–35193 [DOI] [PubMed] [Google Scholar]
Small GTPase
- Ahmed S, Lee J, Kozma R, Best A, Monfries C, Lim L (1993) A novel functional target for tumor-promoting phorbol esters and lysophosphatidic acid. The p21rac-GTPase activating protein n-chimaerin. J Biol Chem 268: 10709–10712 [PubMed] [Google Scholar]
- Burbelo PD, Drechsel D, Hall A (1995) A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem 270: 29071–29074 [DOI] [PubMed] [Google Scholar]
- Caloca MJ, Wang H, Delemos A, Wang S, Kazanietz MG (2001) Phorbol esters and related analogs regulate the subcellular localization of beta 2-chimaerin, a non-protein kinase C phorbol ester receptor. J Biol Chem 276: 18303–18312 [DOI] [PubMed] [Google Scholar]
- Caloca MJ, Wang H, Kazanietz MG (2003) Characterization of the Rac-GAP (Rac-GTPase-activating protein) activity of beta2-chimaerin, a 'non-protein kinase C' phorbol ester receptor. Biochem J 375: 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox AD, Der CJ (2003) The dark side of Ras: regulation of apoptosis. Oncogene 22: 8999–9006 [DOI] [PubMed] [Google Scholar]
- Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR (1997) Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385: 169–172 [DOI] [PubMed] [Google Scholar]
- Diekmann D, Brill S, Garrett MD, Totty N, Hsuan J, Monfries C, Hall C, Lim L, Hall A (1991) Bcr encodes a GTPase-activating protein for p21rac. Nature 351: 400–402 [DOI] [PubMed] [Google Scholar]
- Downward J (1998) Ras signalling and apoptosis. Curr Opin Genet Dev 8: 49–54 [DOI] [PubMed] [Google Scholar]
- Edwards DC, Sanders LC, Bokoch GM, Gill GN (1999) Activation of LIM-kinase by Pak1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat Cell Biol 1: 253–259 [DOI] [PubMed] [Google Scholar]
- Fanger GR, Johnson NL, Johnson GL (1997) MEK kinases are regulated by EGF and selectively interact with Rac/Cdc42. EMBO J 16: 4961–4972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffer ZM, Chernoff J (2002) p21-activated kinases: three more join the Pak. Int J Biochem Cell Biol 34: 713–717 [DOI] [PubMed] [Google Scholar]
- Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L (1994) A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367: 40–46 [DOI] [PubMed] [Google Scholar]
- Menna PL, Skilton G, Leskow FC, Alonso DF, Gomez DE, Kazanietz MG (2003) Inhibition of aggressiveness of metastatic mouse mammary carcinoma cells by the beta2-chimaerin GAP domain. Cancer Res 63: 2284–2291 [PubMed] [Google Scholar]
- Moodie SA, Willumsen BM, Weber MJ, Wolfman A (1993) Complexes of Ras.GTP with Raf-1 and mitogen-activated protein kinase kinase. Science 260: 1658–1661 [DOI] [PubMed] [Google Scholar]
- Puto LA, Pestonjamasp K, King CC, Bokoch GM (2003) p21-activated kinase 1 (PAK1) interacts with the Grb2 adapter protein to couple to growth factor signaling. J Biol Chem 278: 9388–9393 [DOI] [PubMed] [Google Scholar]
- Schurmann A, Mooney AF, Sanders LC, Sells MA, Wang HG, Reed JC, Bokoch GM (2000) p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol 20: 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Pruitt K, McFall A, Shaub A, Der CJ (2000) Understanding Ras: 'it ain't over til it's over'. Trends Cell Biol 10: 147–154 [DOI] [PubMed] [Google Scholar]
- Tamas P, Solti Z, Bauer P, Illes A, Sipeki S, Bauer A, Farago A, Downward J, Buday L (2003) Mechanism of epidermal growth factor regulation of Vav2, a guanine nucleotide exchange factor for Rac. J Biol Chem 278: 5163–5171 [DOI] [PubMed] [Google Scholar]
- Zenke FT, King CC, Bohl BP, Bokoch GM (1999) Identification of a central phosphorylation site in p21-activated kinase regulating autoinhibition and kinase activity. J Biol Chem 274: 32565–32573 [DOI] [PubMed] [Google Scholar]
Phosphatidylinositol phosphate (PIP) signaling
- Alessi DR, Deak M, Casamayor A, Caudwell FB, Morrice N, Norman DG, Gaffney P, Reese CB, MacDougall CN, Harbison D, Ashworth A, Bownes M (1997) 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr Biol 7: 776–789 [DOI] [PubMed] [Google Scholar]
- Belham C, Wu S, Avruch J (1999) Intracellular signalling: PDK1–a kinase at the hub of things. Curr Biol 9: R93–R96 [DOI] [PubMed] [Google Scholar]
- Carpenter G, Ji Q (1999) Phospholipase C-gamma as a signal-transducing element. Exp Cell Res 253: 15–24 [DOI] [PubMed] [Google Scholar]
- Casamayor A, Morrice NA, Alessi DR (1999) Phosphorylation of Ser-241 is essential for the activity of 3-phosphoinositide-dependent protein kinase-1: identification of five sites of phosphorylation in vivo . Biochem J 342: 287–292 [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME (1997) Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91: 231–241 [DOI] [PubMed] [Google Scholar]
- Downward J (2004) PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol 15: 177–182 [DOI] [PubMed] [Google Scholar]
- Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller RD, Krishna UM, Falck JR, White MA, Broek D (1998) Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279: 558–560 [DOI] [PubMed] [Google Scholar]
- Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, Scita G (2003) Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol 160: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H, Parker PJ (1998) The extended protein kinase C superfamily. Biochem J 332: 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit GY, Sun H, Kerfant BG, Crackower MA, Penninger JM, Backx PH (2004) The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J Mol Cell Cardiol 37: 449–471 [DOI] [PubMed] [Google Scholar]
- Reif K, Nobes CD, Thomas G, Hall A, Cantrell DA (1996) Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol 6: 1445–1455 [DOI] [PubMed] [Google Scholar]
- Sato S, Fujita N, Tsuruo T (2000) Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci USA 97: 10832–10837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid MP, Woodgett JR (2003) Unravelling the activation mechanisms of protein kinase B/Akt. FEBS Lett 546: 108–112 [DOI] [PubMed] [Google Scholar]
- Stephens LR, Jackson TR, Hawkins PT (1993) Agonist-stimulated synthesis of phosphatidylinositol(3,4,5)-trisphosphate: a new intracellular signalling system? Biochim Biophys Acta 1179: 27–75 [DOI] [PubMed] [Google Scholar]
- Tolias KF, Cantley LC (1999) Pathways for phosphoinositide synthesis. Chem Phys Lipids 98: 69–77 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD (2001) Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem 70: 535–602 [DOI] [PubMed] [Google Scholar]
- Zha J, Harada H, Yang E, Jockel J, Korsmeyer SJ (1996) Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 87: 619–628 [DOI] [PubMed] [Google Scholar]
Mitogen-activated protein kinase (MAPK) cascade
- Carey KD, Watson RT, Pessin JE, Stork PJ (2003) The requirement of specific membrane domains for Raf-1 phosphorylation and activation. J Biol Chem 278: 3185–3196 [DOI] [PubMed] [Google Scholar]
- Chi H, Sarkisian MR, Rakic P, Flavell RA (2005) Loss of mitogen-activated protein kinase kinase kinase 4 (MEKK4) results in enhanced apoptosis and defective neural tube development. Proc Natl Acad Sci USA 102: 3846–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrestensen CA, Sturgill TW (2002) Characterization of the p90 ribosomal S6 kinase 2 carboxyl-terminal domain as a protein kinase. J Biol Chem 277: 27733–27741 [DOI] [PubMed] [Google Scholar]
- Cleghon V, Morrison DK (1994) Raf-1 interacts with Fyn and Src in a non-phosphotyrosine-dependent manner. J Biol Chem 269: 17749–17755 [PubMed] [Google Scholar]
- Crews CM, Alessandrini A, Erikson RL (1992) The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science 258: 478–480 [DOI] [PubMed] [Google Scholar]
- Dent P, Haser W, Haystead TA, Vincent LA, Roberts TM, Sturgill TW (1992) Activation of mitogen-activated protein kinase kinase by v-Raf in NIH 3T3 cells and in vitro . Science 257: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Douville E, Downward J (1997) EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene 15: 373–383 [DOI] [PubMed] [Google Scholar]
- Franklin CC, Kraft AS (1995) Constitutively active MAP kinase kinase (MEK1) stimulates SAP kinase and c-Jun transcriptional activity in U937 human leukemic cells. Oncogene 11: 2365–2374 [PubMed] [Google Scholar]
- Frodin M, Gammeltoft S (1999) Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol Cell Endocrinol 151: 65–77 [DOI] [PubMed] [Google Scholar]
- Frodin M, Jensen CJ, Merienne K, Gammeltoft S (2000) A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J 19: 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Buckman SY, Pentland AP, Templeton DJ, Morrison AR (1998) Induction of cyclooxygenase-2 by the activated MEKK1 → SEK1/MKK4 → p38 mitogen-activated protein kinase pathway. J Biol Chem 273: 12901–12908 [DOI] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M (1993) Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev 7: 2135–2148 [DOI] [PubMed] [Google Scholar]
- Horgan AM, Stork PJ (2003) Examining the mechanism of Erk nuclear translocation using green fluorescent protein. Exp Cell Res 285: 208–220 [DOI] [PubMed] [Google Scholar]
- Jensen CJ, Buch MB, Krag TO, Hemmings BA, Gammeltoft S, Frodin M (1999) 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J Biol Chem 274: 27168–27176 [DOI] [PubMed] [Google Scholar]
- King AJ, Wireman RS, Hamilton M, Marshall MS (2001) Phosphorylation site specificity of the Pak-mediated regulation of Raf-1 and cooperativity with Src. FEBS Lett 497: 6–14 [DOI] [PubMed] [Google Scholar]
- Kishimoto H, Nakagawa K, Watanabe T, Kitagawa D, Momose H, Seo J, Nishitai G, Shimizu N, Ohata S, Tanemura S, Asaka S, Goto T, Fukushi H, Yoshida H, Suzuki A, Sasaki T, Wada T, Penninger JM, Nishina H, Katada T (2003) Different properties of SEK1 and MKK7 in dual phosphorylation of stress-induced activated protein kinase SAPK/JNK in embryonic stem cells. J Biol Chem 278: 16595–16601 [DOI] [PubMed] [Google Scholar]
- Kolch W (2000) Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J 351: 289–305 [PMC free article] [PubMed] [Google Scholar]
- Kyriakis JM, App H, Zhang XF, Banerjee P, Brautigan DL, Rapp UR, Avruch J (1992) Raf-1 activates MAP kinase-kinase. Nature 358: 417–421 [DOI] [PubMed] [Google Scholar]
- Lange-Carter CA, Pleiman CM, Gardner AM, Blumer KJ, Johnson GL (1993) A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science 260: 315–319 [DOI] [PubMed] [Google Scholar]
- Langlois WJ, Sasaoka T, Saltiel AR, Olefsky JM (1995) Negative feedback regulation and desensitization of insulin- and epidermal growth factor-stimulated p21ras activation. J Biol Chem 270: 25320–25323 [DOI] [PubMed] [Google Scholar]
- Lawler S, Cuenda A, Goedert M, Cohen P (1997) SKK4, a novel activator of stress-activated protein kinase-1 (SAPK1/JNK). FEBS Lett 414: 153–158 [DOI] [PubMed] [Google Scholar]
- Lu X, Nemoto S, Lin A (1997) Identification of c-Jun NH2-terminal protein kinase (JNK)-activating kinase 2 as an activator of JNK but not p38. J Biol Chem 272: 24751–24754 [DOI] [PubMed] [Google Scholar]
- Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T (2002) The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell 9: 945–956 [DOI] [PubMed] [Google Scholar]
- Mason CS, Springer CJ, Cooper RG, Superti-Furga G, Marshall CJ, Marais R (1999) Serine and tyrosine phosphorylations cooperate in Raf-1, but not B-Raf activation. EMBO J 18: 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Fukuda M, Nishida E (2001) Evidence for existence of a nuclear pore complex-mediated, cytosol-independent pathway of nuclear translocation of ERK MAP kinase in permeabilized cells. J Biol Chem 276: 41755–41760 [DOI] [PubMed] [Google Scholar]
- Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E (1997) A novel SAPK/JNK kinase, MKK7, stimulated by TNFalpha and cellular stresses. EMBO J 16: 7045–7053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Cohen P, Wu J, Sturgill T (1992) MAP kinase activator from insulin-stimulated skeletal muscle is a protein threonine/tyrosine kinase. EMBO J 11: 2123–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porfiri E, McCormick F (1996) Regulation of epidermal growth factor receptor signaling by phosphorylation of the ras exchange factor hSOS1. J Biol Chem 271: 5871–5877 [DOI] [PubMed] [Google Scholar]
- Poteet-Smith CE, Smith JA, Lannigan DA, Freed TA, Sturgill TW (1999) Generation of constitutively active p90 ribosomal S6 kinase in vivo. Implications for the mitogen-activated protein kinase-activated protein kinase family. J Biol Chem 274: 22135–22138 [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Whitmarsh AJ, Barrett T, Derijard B, Davis RJ (1996) MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol 16: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusch HP, Zimmermann S, Schaefer M, Paul M, Moelling K (2001) Regulation of Raf by Akt controls growth and differentiation in vascular smooth muscle cells. J Biol Chem 276: 33630–33637 [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, van der Geer P, Mbamalu G, Pawson T (1995) MAP kinase phosphorylation of mSos1 promotes dissociation of mSos1-Shc and mSos1-EGF receptor complexes. Oncogene 11: 1417–1426 [PubMed] [Google Scholar]
- Sun H, Charles CH, Lau LF, Tonks NK (1993) MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo . Cell 75: 487–493 [DOI] [PubMed] [Google Scholar]
- Tibbles LA, Ing YL, Kiefer F, Chan J, Iscove N, Woodgett JR, Lassam NJ (1996) MLK-3 activates the SAPK/JNK and p38/RK pathways via SEK1 and MKK3/6. EMBO J 15: 7026–7035 [PMC free article] [PubMed] [Google Scholar]
- Tran NH, Frost JA (2003) Phosphorylation of Raf-1 by p21-activated kinase 1 and Src regulates Raf-1 autoinhibition. J Biol Chem 278: 11221–11226 [DOI] [PubMed] [Google Scholar]
- Waters SB, Holt KH, Ross SE, Syu LJ, Guan KL, Saltiel AR, Koretzky GA, Pessin JE (1995) Desensitization of Ras activation by a feedback disassociation of the SOS-Grb2 complex. J Biol Chem 270: 20883–20886 [DOI] [PubMed] [Google Scholar]
- Witowsky JA, Johnson GL (2003) Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways. J Biol Chem 278: 1403–1406 [DOI] [PubMed] [Google Scholar]
- Wu J, Harrison JK, Dent P, Lynch KR, Weber MJ, Sturgill TW (1993) Identification and characterization of a new mammalian mitogen-activated protein kinase kinase, MKK2. Mol Cell Biol 13: 4539–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Robbins D, Frost J, Dang A, Lange-Carter C, Cobb MH (1995) MEKK1 phosphorylates MEK1 and MEK2 but does not cause activation of mitogen-activated protein kinase. Proc Natl Acad Sci USA 92: 6808–6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Diener K, Wang XS, Zukowski M, Matsumoto G, Zhou G, Mo R, Sasaki T, Nishina H, Hui CC, Tan TH, Woodgett JP, Penninger JM (1997) Activation of stress-activated protein kinases/c-Jun N-terminal protein kinases (SAPKs/JNKs) by a novel mitogen-activated protein kinase kinase. J Biol Chem 272: 32378–32383 [DOI] [PubMed] [Google Scholar]
- Yujiri T, Sather S, Fanger GR, Johnson GL (1998) Role of MEKK1 in cell survival and activation of JNK and ERK pathways defined by targeted gene disruption. Science 282: 1911–1914 [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Moelling K (1999) Phosphorylation and regulation of Raf by Akt (protein kinase B). Science 286: 1741–1744 [DOI] [PubMed] [Google Scholar]
Transcription
- Alvarez E, Northwood IC, Gonzalez FA, Latour DA, Seth A, Abate C, Curran T, Davis RJ (1991) Pro-Leu-Ser/Thr-Pro is a consensus primary sequence for substrate protein phosphorylation. Characterization of the phosphorylation of c-myc and c-jun proteins by an epidermal growth factor receptor threonine 669 protein kinase. J Biol Chem 266: 15277–15285 [PubMed] [Google Scholar]
- Aplin AE, Stewart SA, Assoian RK, Juliano RL (2001) Integrin-mediated adhesion regulates ERK nuclear translocation and phosphorylation of Elk-1. J Cell Biol 153: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavigelli M, Dolfi F, Claret FX, Karin M (1995) Induction of c-fos expression through JNK-mediated TCF/Elk-1 phosphorylation. EMBO J 14: 5957–5964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronella-Wood J, Terrand J, Sun H, Chen QM (2004) c-Fos phosphorylation induced by H2O2 prevents proteasomal degradation of c-Fos in cardiomyocytes. J Biol Chem 279: 33567–33574 [DOI] [PubMed] [Google Scholar]
- De Cesare D, Jacquot S, Hanauer A, Sassone-Corsi P (1998) Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Natl Acad Sci USA 95: 12202–12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginty DD, Bonni A, Greenberg ME (1994) Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell 77: 713–725 [DOI] [PubMed] [Google Scholar]
- Gupta S, Seth A, Davis RJ (1993) Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc Natl Acad Sci USA 90: 3216–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E (1997) AP-1 function and regulation. Curr Opin Cell Biol 9: 240–246 [DOI] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R (1993) The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73: 381–393 [DOI] [PubMed] [Google Scholar]
- Minden A, Lin A, Smeal T, Derijard B, Cobb M, Davis R, Karin M (1994) c-Jun N-terminal phosphorylation correlates with activation of the JNK subgroup but not the ERK subgroup of mitogen-activated protein kinases. Mol Cell Biol 14: 6683–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A, Alvarez E, Gupta S, Davis RJ (1991) A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem 266: 23521–23524 [PubMed] [Google Scholar]
- Smeal T, Binetruy B, Mercola DA, Birrer M, Karin M (1991) Oncogenic and transcriptional cooperation with Ha-Ras requires phosphorylation of c-Jun on serines 63 and 73. Nature 354: 494–496 [DOI] [PubMed] [Google Scholar]
- Tian J, Karin M (1999) Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin). J Biol Chem 274: 15173–15180 [DOI] [PubMed] [Google Scholar]
- Ueno Y, Kume N, Miyamoto S, Morimoto M, Kataoka H, Ochi H, Nishi E, Moriwaki H, Minami M, Hashimoto N, Kita T (1999) Lysophosphatidylcholine phosphorylates CREB and activates the jun2TRE site of c-jun promoter in vascular endothelial cells. FEBS Lett 457: 241–245 [DOI] [PubMed] [Google Scholar]
- Vanhoutte P, Barnier JV, Guibert B, Pages C, Besson MJ, Hipskind RA, Caboche J (1999) Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinase-dependent pathway in brain slices. Mol Cell Biol 19: 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz A, Rosales R (1990) Identification of an estrogen response element upstream of the human c-fos gene that binds the estrogen receptor and the AP-1 transcription factor. Nucleic Acids Res 18: 5097–5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Kornhauser JM, Xia Z, Thiele EA, Greenberg ME (1998) Nerve growth factor activates extracellular signal-regulated kinase and p38 mitogen-activated protein kinase pathways to stimulate CREB serine 133 phosphorylation. Mol Cell Biol 18: 1946–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
Cell cycle
- Alkarain A, Jordan R, Slingerland J (2004) p27 deregulation in breast cancer: prognostic significance and implications for therapy. J Mammary Gland Biol Neoplasia 9: 67–80 [DOI] [PubMed] [Google Scholar]
- Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M (2000) Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J 19: 2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rajpert-De Meyts E, Skakkebaek NE, Lukas J, Bartek J (2003) Deregulation of the G1/S-phase control in human testicular germ cell tumours. Apmis 111: 252–265, discussion 265–256 [DOI] [PubMed] [Google Scholar]
- Bates S, Bonetta L, MacAllan D, Parry D, Holder A, Dickson C, Peters G (1994) CDK6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin D1. Oncogene 9: 71–79 [PubMed] [Google Scholar]
- Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Durr P (1996) Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol 16: 3401–3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr (1999) Stat3 as an oncogene. Cell 98: 295–303 [DOI] [PubMed] [Google Scholar]
- Bulavin DV, Saito S, Hollander MC, Sakaguchi K, Anderson CW, Appella E, Fornace AJ Jr (1999) Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J 18: 6845–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR (1991) The E2F transcription factor is a cellular target for the RB protein. Cell 65: 1053–1061 [DOI] [PubMed] [Google Scholar]
- Claassen GF, Hann SR (2000) A role for transcriptional repression of p21CIP1 by c-Myc in overcoming transforming growth factor beta-induced cell-cycle arrest. Proc Natl Acad Sci USA 97: 9498–9503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clurman BE, Sheaff RJ, Thress K, Groudine M, Roberts JM (1996) Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev 10: 1979–1990 [DOI] [PubMed] [Google Scholar]
- Coqueret O (2002) Linking cyclins to transcriptional control. Gene 299: 35–55 [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75: 817–825 [DOI] [PubMed] [Google Scholar]
- Gartel AL, Shchors K (2003) Mechanisms of c-myc-mediated transcriptional repression of growth arrest genes. Exp Cell Res 283: 17–21 [DOI] [PubMed] [Google Scholar]
- Grana X, Reddy EP (1995) Cell cycle control in mammalian cells: role of cyclins, cyclin dependent kinases (CDKs), growth suppressor genes and cyclin-dependent kinase inhibitors (CKIs). Oncogene 11: 211–219 [PubMed] [Google Scholar]
- Gu Y, Rosenblatt J, Morgan DO (1992) Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J 11: 3995–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Beach D (1994) p15INK4B is a potential effector of TGF-beta-induced cell cycle arrest. Nature 371: 257–261 [DOI] [PubMed] [Google Scholar]
- Harbour JW, Luo RX, Dei Santi A, Postigo AA, Dean DC (1999) Cdk phosphorylation triggers sequential intramolecular interactions that progressively block Rb functions as cells move through G1. Cell 98: 859–869 [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ, Keyomarsi K, Dynlacht B, Tsai LH, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, Fox MP, Wei N (1995) Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell 6: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato JY, Matsuoka M, Strom DK, Sherr CJ (1994) Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol Cell Biol 14: 2713–2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi H, Nakagawa K, Matsumoto M, Suga M, Ando M, Taya Y, Yamaizumi M (2001) Osmotic shock induces G1 arrest through p53 phosphorylation at Ser33 by activated p38MAPK without phosphorylation at Ser15 and Ser20. J Biol Chem 276: 39115–39122 [DOI] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ (2001) Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294: 173–177 [DOI] [PubMed] [Google Scholar]
- Koff A, Giordano A, Desai D, Yamashita K, Harper JW, Elledge S, Nishimoto T, Morgan DO, Franza BR, Roberts JM (1992) Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science 257: 1689–1694 [DOI] [PubMed] [Google Scholar]
- Lee RJ, Albanese C, Stenger RJ, Watanabe G, Inghirami G, Haines GK III, Webster M, Muller WJ, Brugge JS, Davis RJ, Pestell RG (1999) pp60(v-src) induction of cyclin D1 requires collaborative interactions between the extracellular signal-regulated kinase, p38, and Jun kinase pathways. A role for cAMP response element-binding protein and activating transcription factor-2 in pp60(v-src) signaling in breast cancer cells. J Biol Chem 274: 7341–7350 [DOI] [PubMed] [Google Scholar]
- Ortega S, Malumbres M, Barbacid M (2002) Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta 1602: 73–87 [DOI] [PubMed] [Google Scholar]
- Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M (1995) Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science 269: 682–685 [DOI] [PubMed] [Google Scholar]
- Rousseau D, Cannella D, Boulaire J, Fitzgerald P, Fotedar A, Fotedar R (1999) Growth inhibition by CDK-cyclin and PCNA binding domains of p21 occurs by distinct mechanisms and is regulated by ubiquitin-proteasome pathway. Oncogene 18: 4313–4325 [DOI] [PubMed] [Google Scholar]
- Sanchez-Prieto R, Rojas JM, Taya Y, Gutkind JS (2000) A role for the p38 mitogen-acitvated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Res 60: 2464–2472 [PubMed] [Google Scholar]
- Schreiber M, Kolbus A, Piu F, Szabowski A, Mohle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF (1999) Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev 13: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE (1997) Cyclin E-CDK2 is a regulator of p27Kip1. Genes Dev 11: 1464–1478 [DOI] [PubMed] [Google Scholar]
- Sherr CJ (1993) Mammalian G1 cyclins. Cell 73: 1059–1065 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1995) Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev 9: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM (1999) CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev 13: 1501–1512 [DOI] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Amati B (1997) Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J 16: 5334–5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner BJ, Blain SW, Seoane J, Massague J (1999) Myc downregulation by transforming growth factor beta required for activation of the p15(Ink4b) G(1) arrest pathway. Mol Cell Biol 19: 5913–5922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, Pestell RG (1998) Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol 18: 3212–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub SJ, Chow KN, Luo RX, Zhang SH, He S, Dean DC (1995) Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 375: 812–815 [DOI] [PubMed] [Google Scholar]
- Won KA, Reed SI (1996) Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J 15: 4182–4193 [PMC free article] [PubMed] [Google Scholar]
G protein-coupled receptor (GPCR)-mediated EGFR transactivation
- Beebe SJ (1994) The cAMP-dependent protein kinases and cAMP signal transduction. Semin Cancer Biol 5: 285–294 [PubMed] [Google Scholar]
- Benzing T, Yaffe MB, Arnould T, Sellin L, Schermer B, Schilling B, Schreiber R, Kunzelmann K, Leparc GG, Kim E, Walz G (2000) 14-3-3 interacts with regulator of G protein signaling proteins and modulates their activity. J Biol Chem 275: 28167–28172 [DOI] [PubMed] [Google Scholar]
- Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA (1997) Phospholipase D2, a distinct phospholipase D isoform with novel regulatory properties that provokes cytoskeletal reorganization. Curr Biol 7: 191–201 [DOI] [PubMed] [Google Scholar]
- Dzimiri N (2002) Receptor crosstalk. Implications for cardiovascular function, disease and therapy. Eur J Biochem 269: 4713–4730 [DOI] [PubMed] [Google Scholar]
- Exton JH (2002) Regulation of phospholipase D. FEBS Lett 531: 58–61 [DOI] [PubMed] [Google Scholar]
- Gether U, Kobilka BK (1998) G protein-coupled receptors. II. Mechanism of agonist activation. J Biol Chem 273: 17979–17982 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, Nozawa Y, Prestwich GD, Frohman MA, Morris AJ (1997) Characterization of two alternately spliced forms of phospholipase D1. Activation of the purified enzymes by phosphatidylinositol 4,5-bisphosphate, ADP-ribosylation factor, and Rho family monomeric GTP-binding proteins and protein kinase C-alpha. J Biol Chem 272: 3860–3868 [DOI] [PubMed] [Google Scholar]
- Ishii M, Kurachi Y (2003) Physiological actions of regulators of G-protein signaling (RGS) proteins. Life Sci 74: 163–171 [DOI] [PubMed] [Google Scholar]
- Jang MJ, Lee MJ, Park HY, Bae YS, Min do S, Ryu SH, Kwak JY (2004) Phosphorylation of phospholipase D1 and the modulation of its interaction with RhoA by cAMP-dependent protein kinase. Exp Mol Med 36: 172–178 [DOI] [PubMed] [Google Scholar]
- Johnson EN, Druey KM (2002) Heterotrimeric G protein signaling: role in asthma and allergic inflammation. J Allergy Clin Immunol 109: 592–602 [DOI] [PubMed] [Google Scholar]
- Johnston CA, Watts VJ (2003) Sensitization of adenylate cyclase: a general mechanism of neuroadaptation to persistent activation of Galpha(i/o)-coupled receptors? Life Sci 73: 2913–2925 [DOI] [PubMed] [Google Scholar]
- Kim JW, Sim SS, Kim UH, Nishibe S, Wahl MI, Carpenter G, Rhee SG (1990) Tyrosine residues in bovine phospholipase C-gamma phosphorylated by the epidermal growth factor receptor in vitro . J Biol Chem 265: 3940–3943 [PubMed] [Google Scholar]
- Lee SB, Shin SH, Hepler JR, Gilman AG, Rhee SG (1993) Activation of phospholipase C-beta 2 mutants by G protein alpha q and beta gamma subunits. J Biol Chem 268: 25952–25957 [PubMed] [Google Scholar]
- Litosch I (1997) G-protein betagamma subunits antagonize protein kinase C-dependent phosphorylation and inhibition of phospholipase C-beta1. Biochem J 326: 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Armant DR (2004) Lysophosphatidic acid regulates murine blastocyst development by transactivation of receptors for heparin-binding EGF-like growth factor. Exp Cell Res 296: 317–326 [DOI] [PubMed] [Google Scholar]
- Moritz A, De Graan PN, Gispen WH, Wirtz KW (1992) Phosphatidic acid is a specific activator of phosphatidylinositol-4-phosphate kinase. J Biol Chem 267: 7207–7210 [PubMed] [Google Scholar]
- Neer EJ (1995) Heterotrimeric G proteins: organizers of transmembrane signals. Cell 80: 249–257 [DOI] [PubMed] [Google Scholar]
- Pai R, Soreghan B, Szabo IL, Pavelka M, Baatar D, Tarnawski AS (2002) Prostaglandin E2 transactivates EGF receptor: a novel mechanism for promoting colon cancer growth and gastrointestinal hypertrophy. Nat Med 8: 289–293 [DOI] [PubMed] [Google Scholar]
- Poghosyan Z, Robbins SM, Houslay MD, Webster A, Murphy G, Edwards DR (2002) Phosphorylation-dependent interactions between ADAM15 cytoplasmic domain and Src family protein-tyrosine kinases. J Biol Chem 277: 4999–5007 [DOI] [PubMed] [Google Scholar]
- Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A (1999) EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature 402: 884–888 [DOI] [PubMed] [Google Scholar]
- Schafer B, Marg B, Gschwind A, Ullrich A (2004) Distinct ADAM metalloproteinases regulate G protein-coupled receptor-induced cell proliferation and survival. J Biol Chem 279: 47929–47938 [DOI] [PubMed] [Google Scholar]
- Shi CS, Sinnarajah S, Cho H, Kozasa T, Kehrl JH (2000) G13alpha-mediated PYK2 activation. PYK2 is a mediator of G13alpha-induced serum response element-dependent transcription. J Biol Chem 275: 24470–24476 [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Sternweis PC (1993) Regulation of purified subtypes of phosphatidylinositol-specific phospholipase C beta by G protein alpha and beta gamma subunits. J Biol Chem 268: 9667–9674 [PubMed] [Google Scholar]
- Suzuki N, Nakamura S, Mano H, Kozasa T (2003) Galpha 12 activates Rho GTPase through tyrosine-phosphorylated leukemia-associated RhoGEF. Proc Natl Acad Sci USA 100: 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt S, Grosse R, Schultz G, Offermanns S (2003) Receptor-dependent RhoA activation in G12/G13-deficient cells: genetic evidence for an involvement of Gq/G11. J Biol Chem 278: 28743–28749 [DOI] [PubMed] [Google Scholar]
- Yue C, Ku CY, Liu M, Simon MI, Sanborn BM (2000) Molecular mechanism of the inhibition of phospholipase C beta 3 by protein kinase C. J Biol Chem 275: 30220–30225 [DOI] [PubMed] [Google Scholar]
- Zhong H, Neubig RR (2001) Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther 297: 837–845 [PubMed] [Google Scholar]
Ca2+ signaling
- Banno Y, Asano T, Nozawa Y (1994) Proteolytic modification of membrane-associated phospholipase C-beta by mu-calpain enhances its activation by G-protein beta gamma subunits in human platelets. FEBS Lett 340: 185–188 [DOI] [PubMed] [Google Scholar]
- Bruce JI, Shuttleworth TJ, Giovannucci DR, Yule DI (2002) Phosphorylation of inositol 1,4,5-trisphosphate receptors in parotid acinar cells. A mechanism for the synergistic effects of cAMP on Ca2+ signaling. J Biol Chem 277: 1340–1348 [DOI] [PubMed] [Google Scholar]
- Danila CI, Hamilton SL (2004) Phosphorylation of ryanodine receptors. Biol Res 37: 521–525 [DOI] [PubMed] [Google Scholar]
- DeSouza N, Reiken S, Ondrias K, Yang YM, Matkovich S, Marks AR (2002) Protein kinase A and two phosphatases are components of the inositol 1,4,5-trisphosphate receptor macromolecular signaling complex. J Biol Chem 277: 39397–39400 [DOI] [PubMed] [Google Scholar]
- Ishida A, Shigeri Y, Taniguchi T, Kameshita I (2003) Protein phosphatases that regulate multifunctional Ca2+/calmodulin-dependent protein kinases: from biochemistry to pharmacology. Pharmacol Ther 100: 291–305 [DOI] [PubMed] [Google Scholar]
- Narayanan N, Xu A (1997) Phosphorylation and regulation of the Ca(2+)-pumping ATPase in cardiac sarcoplasmic reticulum by calcium/calmodulin-dependent protein kinase. Basic Res Cardiol 92: 25–35 [DOI] [PubMed] [Google Scholar]
- Noguchi N, Takasawa S, Nata K, Tohgo A, Kato I, Ikehata F, Yonekura H, Okamoto H (1997) Cyclic ADP-ribose binds to FK506-binding protein 12.6 to release Ca2+ from islet microsomes. J Biol Chem 272: 3133–3136 [DOI] [PubMed] [Google Scholar]
- Okamoto H, Takasawa S (2002) Recent advances in the Okamoto model: the CD38-cyclic ADP-ribose signal system and the regenerating gene protein (Reg)-Reg receptor system in beta-cells. Diabetes 51: S462–S473 [DOI] [PubMed] [Google Scholar]
- Toyofuku T, Curotto Kurzydlowski K, Narayanan N, MacLennan DH (1994) Identification of Ser38 as the site in cardiac sarcoplasmic reticulum Ca(2+)-ATPase that is phosphorylated by Ca2+/calmodulin-dependent protein kinase. J Biol Chem 269: 26492–26496 [PubMed] [Google Scholar]
- Werry TD, Wilkinson GF, Willars GB (2003) Mechanisms of cross-talk between G-protein-coupled receptors resulting in enhanced release of intracellular Ca2+ . Biochem J 374: 281–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcikiewicz RJ, Luo SG (1998) Phosphorylation of inositol 1,4,5-trisphosphate receptors by cAMP-dependent protein kinase. Type I, II, and III receptors are differentially susceptible to phosphorylation and are phosphorylated in intact cells. J Biol Chem 273: 5670–5677 [DOI] [PubMed] [Google Scholar]
ErbB family
- Cohen BD, Green JM, Foy L, Fell HP (1996) HER4-mediated biological and biochemical properties in NIH 3T3 cells. Evidence for HER1-HER4 heterodimers. J Biol Chem 271: 4813–4818 [DOI] [PubMed] [Google Scholar]
- Elenius K, Choi CJ, Paul S, Santiestevan E, Nishi E, Klagsbrun M (1999) Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene 18: 2607–2615 [DOI] [PubMed] [Google Scholar]
- Fiddes RJ, Campbell DH, Janes PW, Sivertsen SP, Sasaki H, Wallasch C, Daly RJ (1998) Analysis of Grb7 recruitment by heregulin-activated erbB receptors reveals a novel target selectivity for erbB3. J Biol Chem 273: 7717–7724 [DOI] [PubMed] [Google Scholar]
- Hazan R, Margolis B, Dombalagian M, Ullrich A, Zilberstein A, Schlessinger J (1990) Identification of autophosphorylation sites of HER2/neu. Cell Growth Differ 1: 3–7 [PubMed] [Google Scholar]
- Prigent SA, Gullick WJ (1994) Identification of c-erbB-3 binding sites for phosphatidylinositol 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J 13: 2831–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci A, Lanfrancone L, Chiari R, Belardo G, Pertica C, Natali PG, Pelicci PG, Segatto O (1995) Analysis of protein-protein interactions involved in the activation of the Shc/Grb-2 pathway by the ErbB-2 kinase. Oncogene 11: 1519–1529 [PubMed] [Google Scholar]
- Segatto O, Lonardo F, Pierce JH, Bottaro DP, Di Fiore PP (1990) The role of autophosphorylation in modulation of erbB-2 transforming function. New Biol 2: 187–195 [PubMed] [Google Scholar]
- Zrihan-Licht S, Deng B, Yarden Y, McShan G, Keydar I, Avraham H (1998) Csk homologous kinase, a novel signaling molecule, directly associates with the activated ErbB-2 receptor in breast cancer cells and inhibits their proliferation. J Biol Chem 273: 4065–4072 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary PDF 1
Supplementary PDF 2
Supplementary SBML 1