Abstract
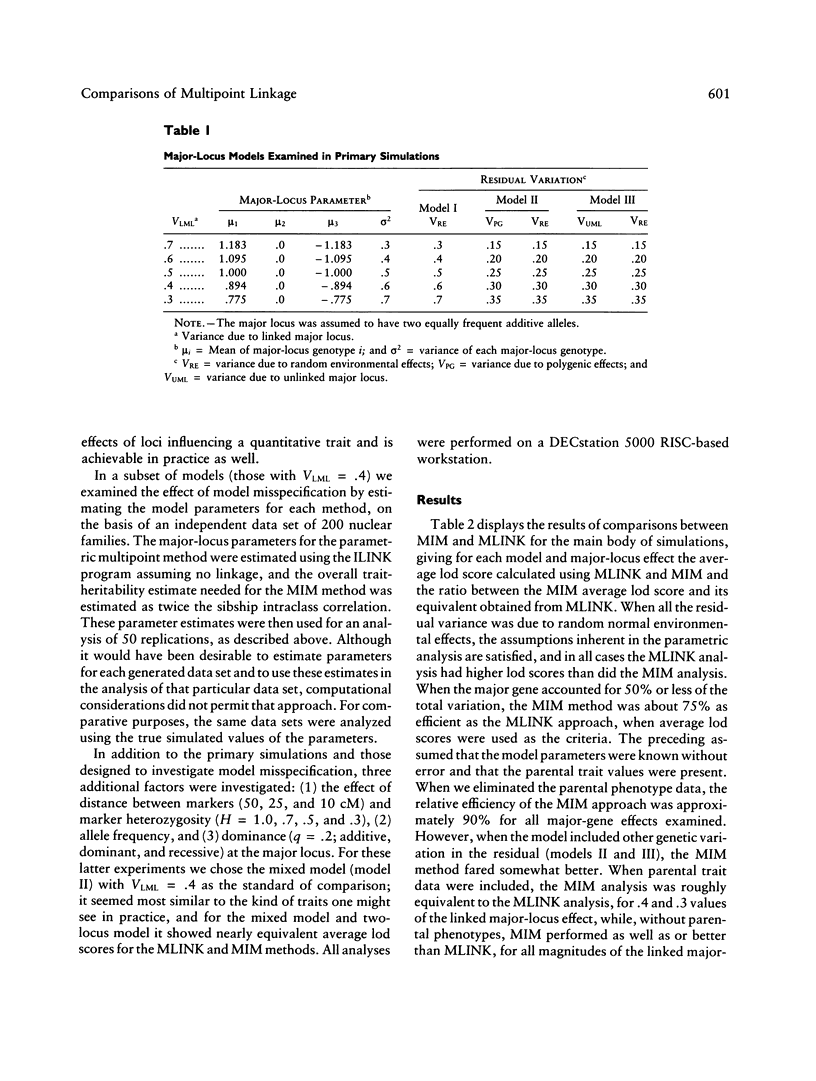
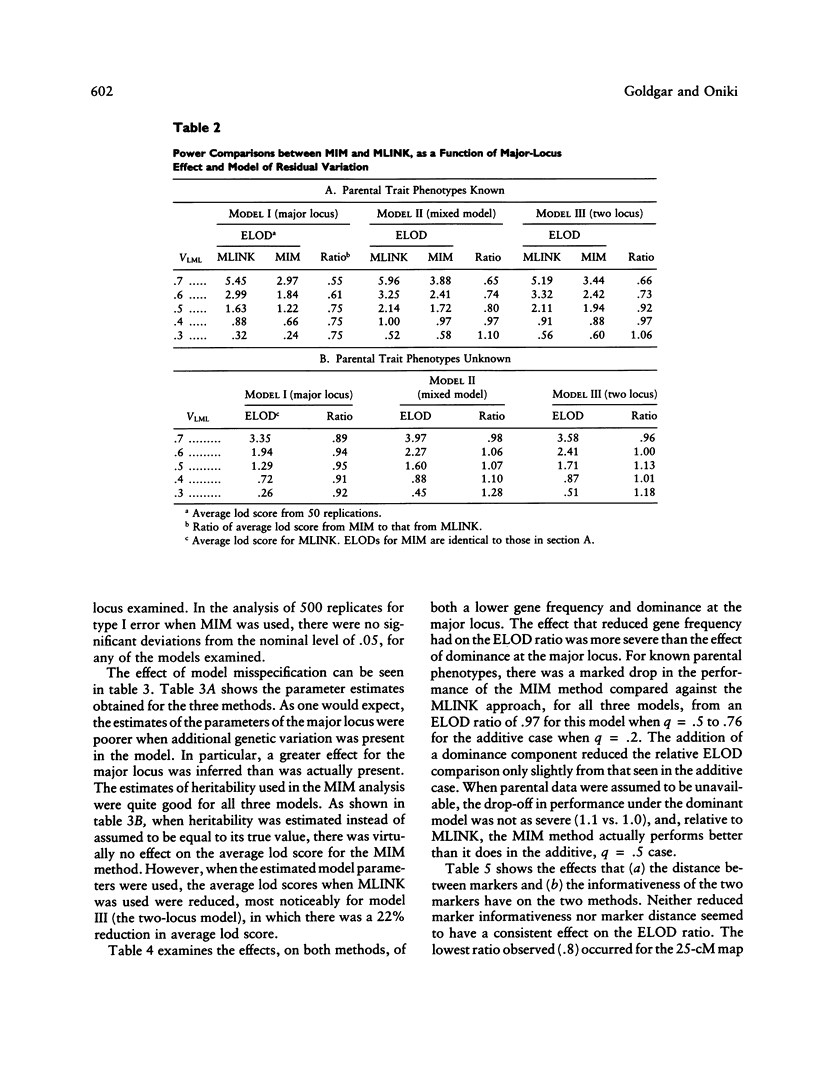
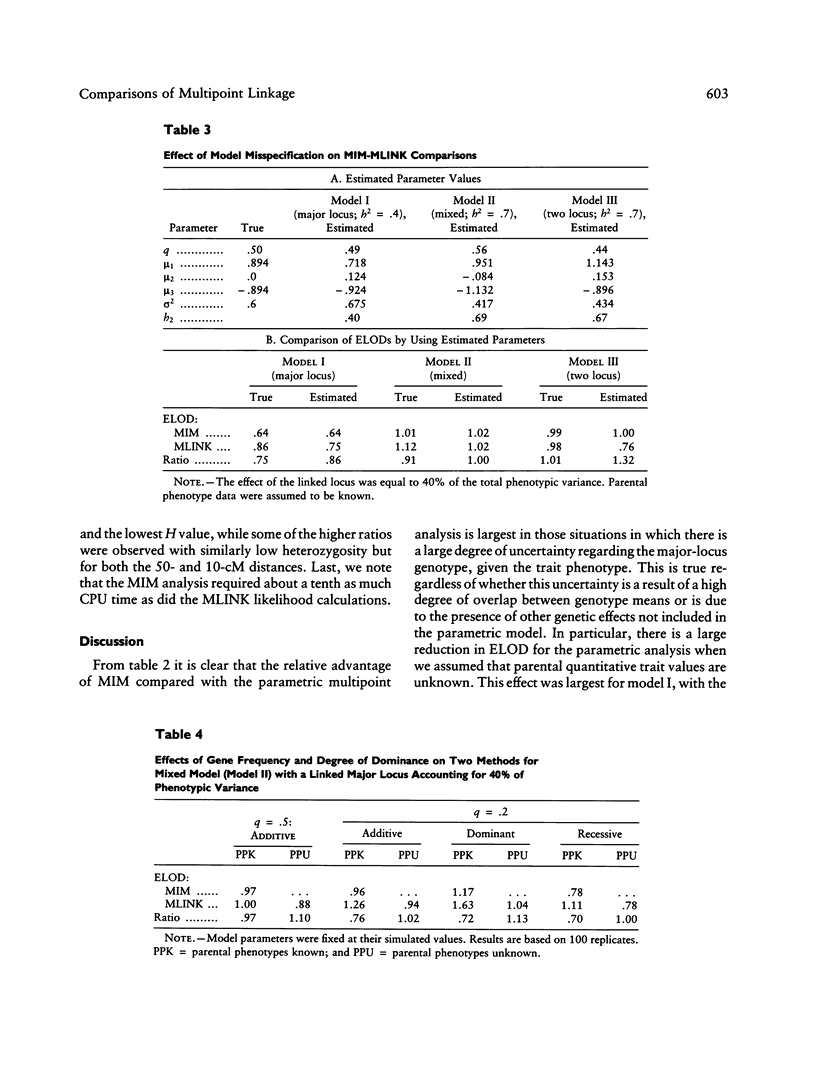
We previously developed a method of partitioning genetic variance of a quantitative trait to loci in specific chromosomal regions. In this paper, we compare this method--multipoint IBD (identical by descent) method (MIM)--with parametric multipoint linkage analysis (MLINK). A simulation study was performed comparing the methods for the major-locus, mixed, and two-locus models. The criterion for comparisons between MIM and MLINK was the average lod score from multiple replicates of simulated data sets. The effect of gene frequency, dominance, model misspecification, marker spacing, and informativeness are also considered in a smaller set of simulations. Within the context of the models examined, the MIM approach was found to be comparable in power with parametric multipoint linkage analysis when (a) parental data are unknown, (b) the effect of the major locus is small and there is additional genetic variation, or (c) the parameters of the major-locus model are misspecified. The performance of the MIM method relative to MLINK was markedly lower when the allele frequency at the trait locus was .2 versus .5, particularly for the case when parental data were assumed to be known. Dominance at the trait major locus, as well as marker spacing and heterozygosity, did not appear to have a large effect on the ELOD comparisons.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amos C. I., Elston R. C. Robust methods for the detection of genetic linkage for quantitative data from pedigrees. Genet Epidemiol. 1989;6(2):349–360. doi: 10.1002/gepi.1370060205. [DOI] [PubMed] [Google Scholar]
- Amos C. I., Elston R. C., Wilson A. F., Bailey-Wilson J. E. A more powerful robust sib-pair test of linkage for quantitative traits. Genet Epidemiol. 1989;6(3):435–449. doi: 10.1002/gepi.1370060306. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Sample-size guidelines for linkage analysis of a dominant locus for a quantitative trait by the method of lod scores. Am J Hum Genet. 1990 Aug;47(2):218–227. [PMC free article] [PubMed] [Google Scholar]
- Clerget-Darpoux F., Bonaïti-Pellié C., Hochez J. Effects of misspecifying genetic parameters in lod score analysis. Biometrics. 1986 Jun;42(2):393–399. [PubMed] [Google Scholar]
- Demenais F., Lathrop G. M., Lalouel J. M. Detection of linkage between a quantitative trait and a marker locus by the lod score method: sample size and sampling considerations. Ann Hum Genet. 1988 Jul;52(Pt 3):237–246. doi: 10.1111/j.1469-1809.1988.tb01101.x. [DOI] [PubMed] [Google Scholar]
- Goldgar D. E. Multipoint analysis of human quantitative genetic variation. Am J Hum Genet. 1990 Dec;47(6):957–967. [PMC free article] [PubMed] [Google Scholar]
- Haseman J. K., Elston R. C. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972 Mar;2(1):3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Lander E. S., Botstein D. Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989 Jan;121(1):185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K., Spence M. A., Frank M. B. Application of the lod method to the detection of linkage between a quantitative trait and a qualitative marker: a simulation experiment. Am J Hum Genet. 1976 Mar;28(2):167–173. [PMC free article] [PubMed] [Google Scholar]
- Lange K. The affected sib-pair method using identity by state relations. Am J Hum Genet. 1986 Jul;39(1):148–150. [PMC free article] [PubMed] [Google Scholar]
- Lathrop G. M., Lalouel J. M., Julier C., Ott J. Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORTON N. E. Sequential tests for the detection of linkage. Am J Hum Genet. 1955 Sep;7(3):277–318. [PMC free article] [PubMed] [Google Scholar]
- Ott J. Estimation of the recombination fraction in human pedigrees: efficient computation of the likelihood for human linkage studies. Am J Hum Genet. 1974 Sep;26(5):588–597. [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Lander E. S., Hewitt J. D., Peterson S., Lincoln S. E., Tanksley S. D. Resolution of quantitative traits into Mendelian factors by using a complete linkage map of restriction fragment length polymorphisms. Nature. 1988 Oct 20;335(6192):721–726. doi: 10.1038/335721a0. [DOI] [PubMed] [Google Scholar]
- Suarez B. K., Rice J., Reich T. The generalized sib pair IBD distribution: its use in the detection of linkage. Ann Hum Genet. 1978 Jul;42(1):87–94. doi: 10.1111/j.1469-1809.1978.tb00933.x. [DOI] [PubMed] [Google Scholar]
- Weeks D. E., Lange K. The affected-pedigree-member method of linkage analysis. Am J Hum Genet. 1988 Feb;42(2):315–326. [PMC free article] [PubMed] [Google Scholar]
- Wilson A. F., Elston R. C., Tran L. D., Siervogel R. M. Use of the robust sib-pair method to screen for single-locus, multiple-locus, and pleiotropic effects: application to traits related to hypertension. Am J Hum Genet. 1991 May;48(5):862–872. [PMC free article] [PubMed] [Google Scholar]


