Abstract
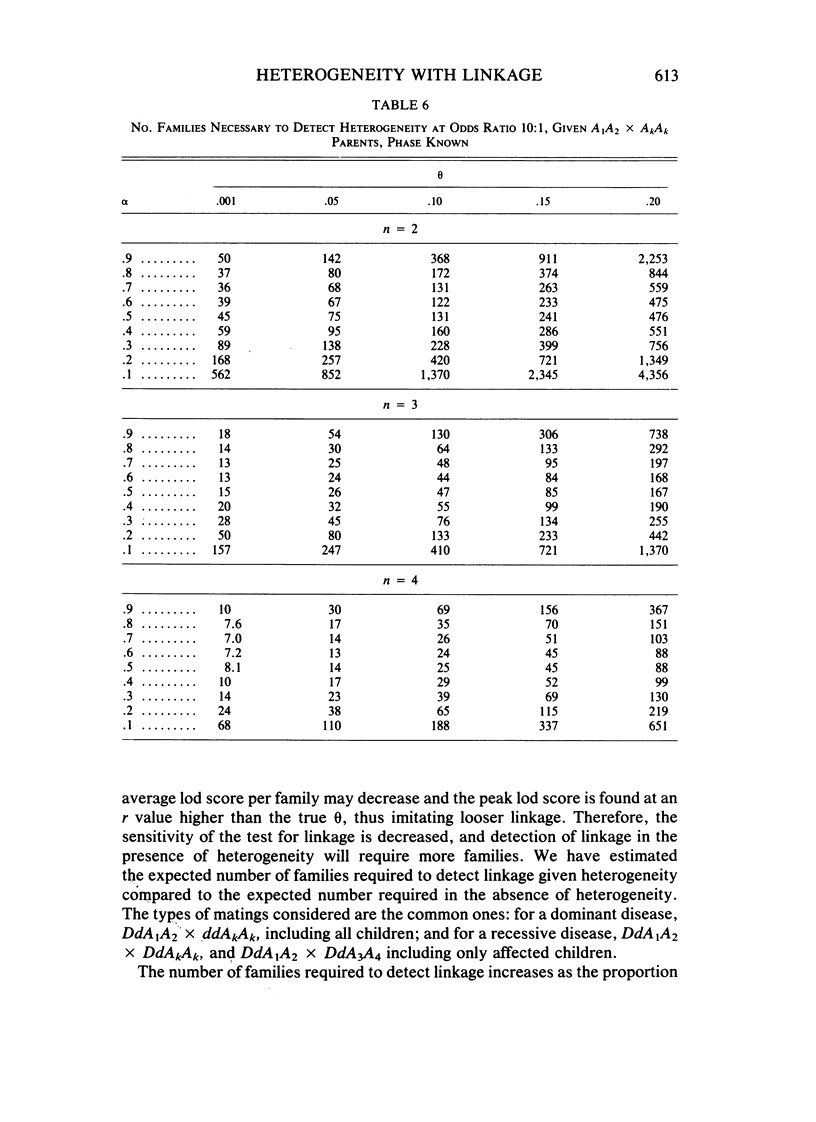
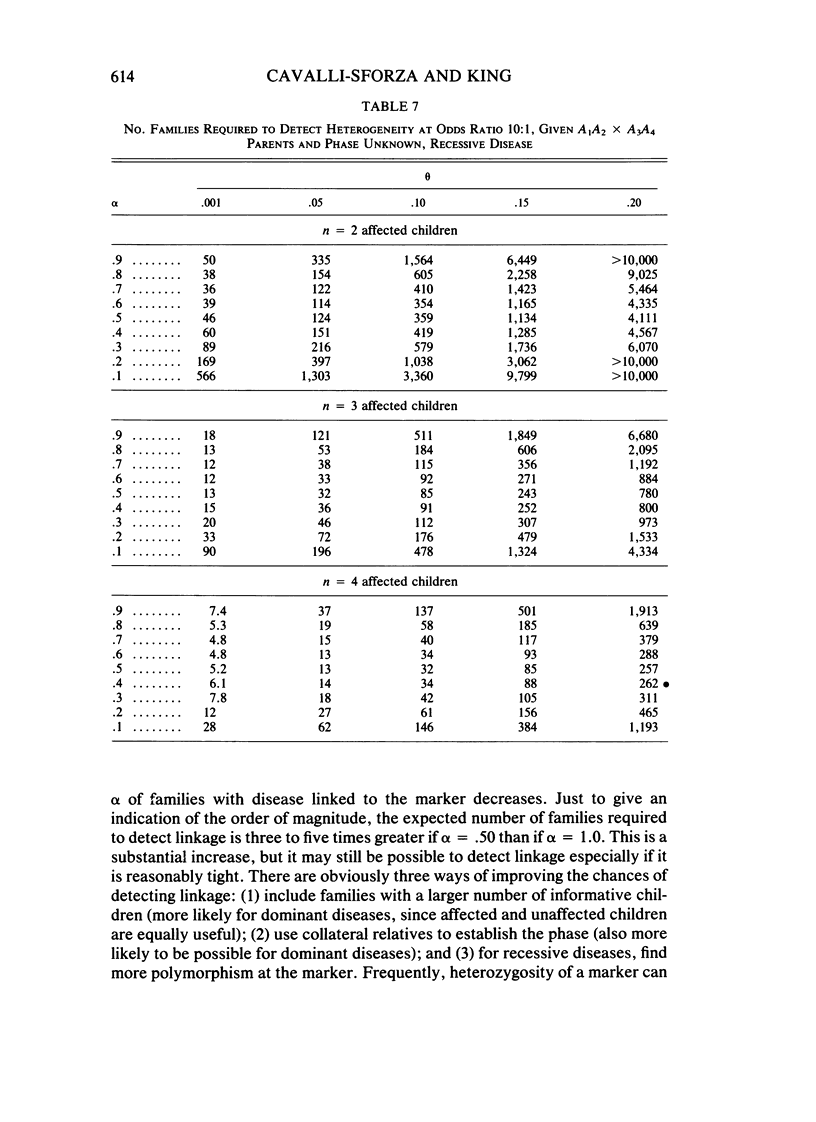
Interest in searching for genetic linkage between diseases and marker loci has been greatly increased by the recent introduction of DNA polymorphisms. However, even for the most well-behaved Mendelian disorders, those with clear-cut mode of inheritance, complete penetrance, and no phenocopies, genetic heterogeneity may exist; that is, in the population there may be more than one locus that can determine the disease, and these loci may not be linked. In such cases, two questions arise: (1) What sample size is necessary to detect linkage for a genetically heterogeneous disease? (2) What sample size is necessary to detect heterogeneity given linkage between a disease and a marker locus? We have answered these questions for the most important types of matings under specified conditions: linkage phase known or unknown, number of alleles involved in the cross at the marker locus, and different numbers of affected and unaffected children. In general, the presence of heterogeneity increases the recombination value at which lod scores peak, by an amount that increases with the degree of heterogeneity. There is a corresponding increase in the number of families necessary to establish linkage. For the specific case of backcrosses between disease and marker loci with two alleles, linkage can be detected at recombination fractions up to 20% with reasonable numbers of families, even if only half the families carry the disease locus linked to the marker. The task is easier if more than two informative children are available or if phase is known. For recessive diseases, highly polymorphic markers with four different alleles in the parents greatly reduce the number of families required.
Full text
PDF


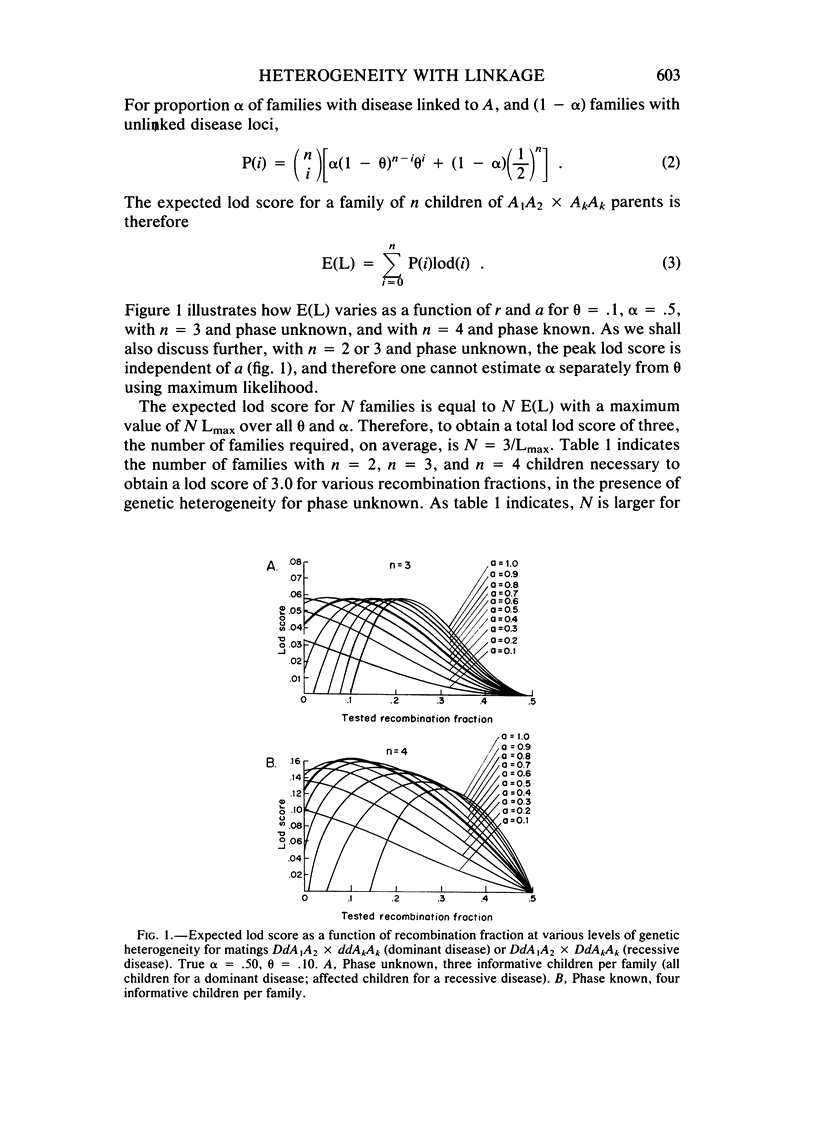
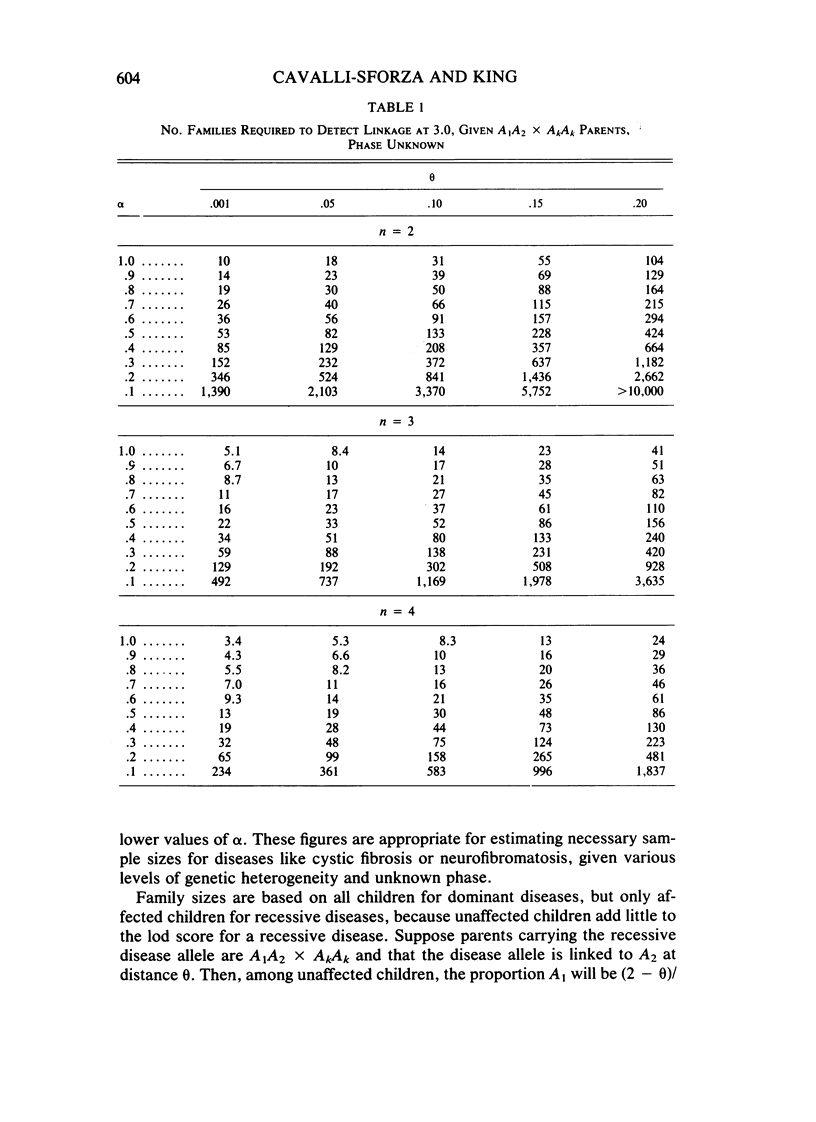

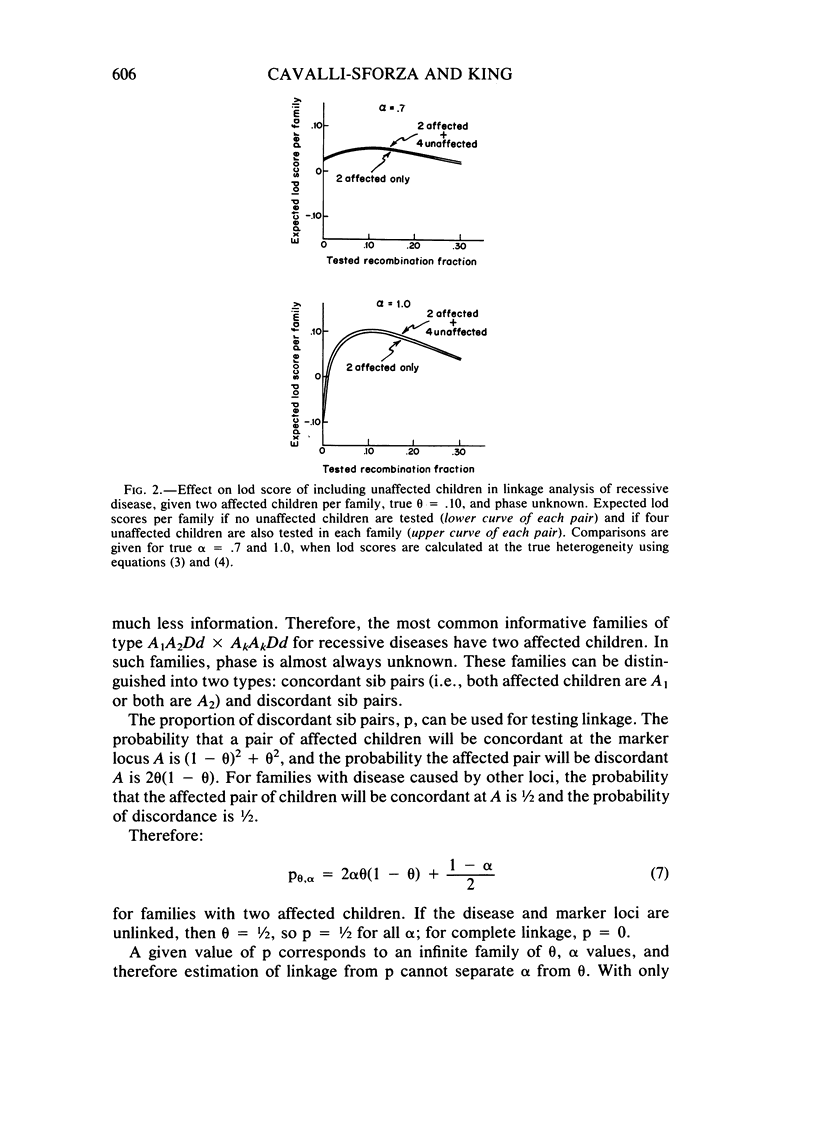

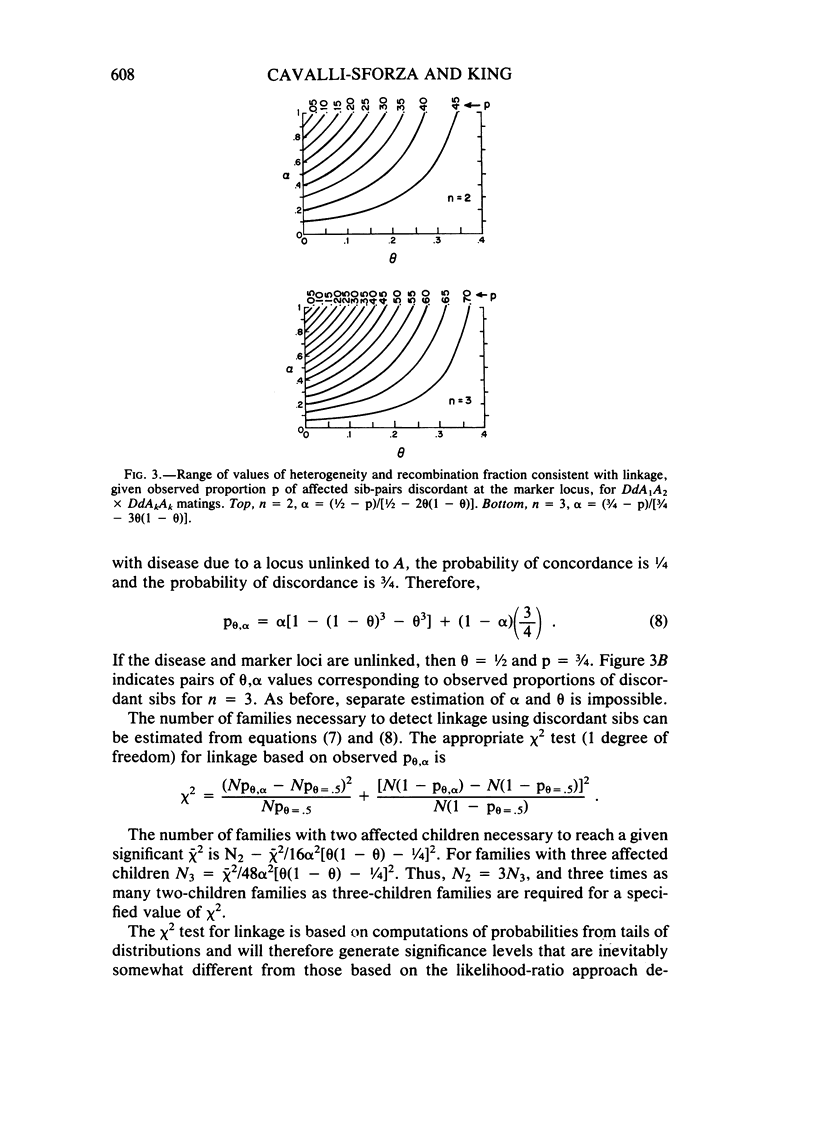

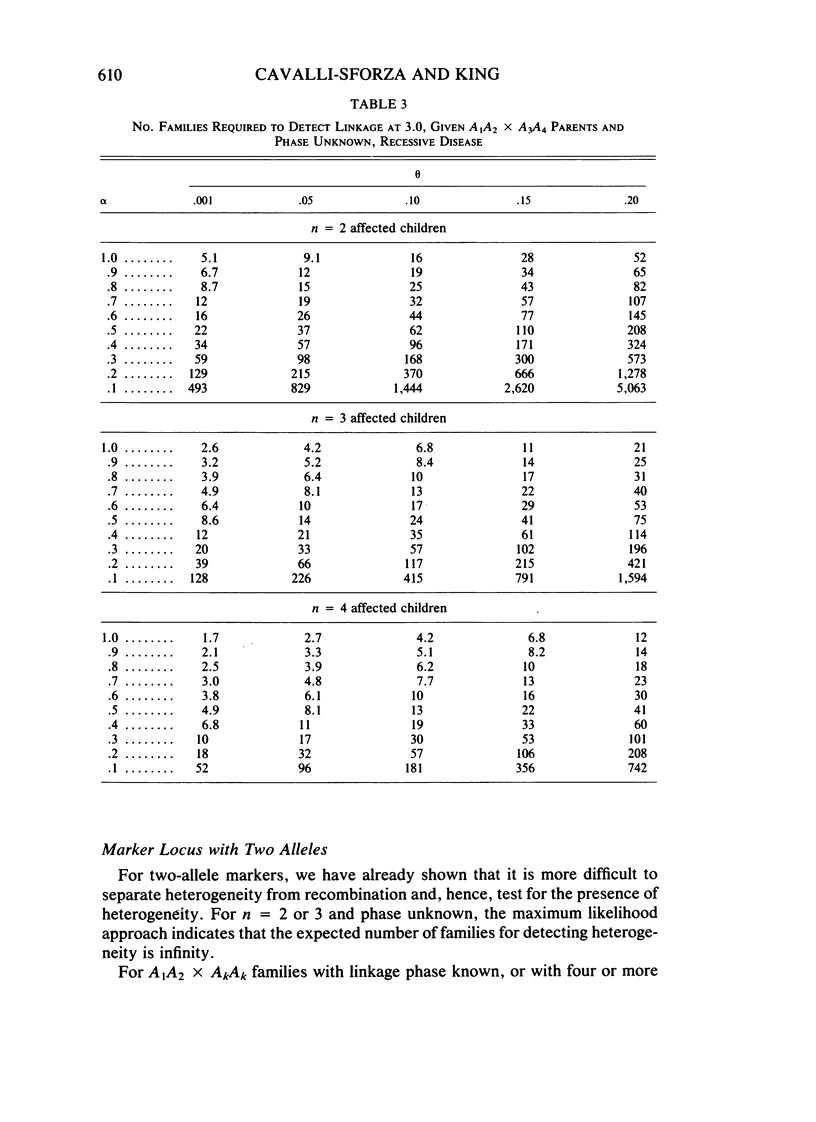
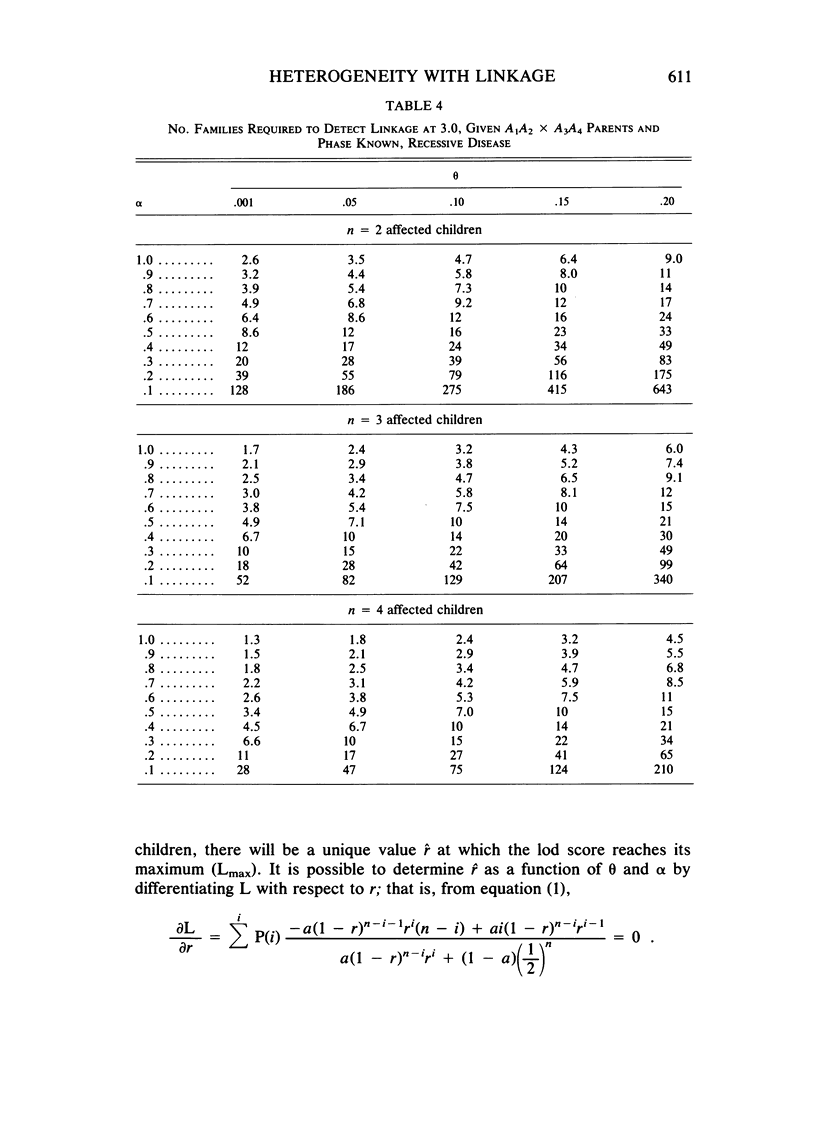
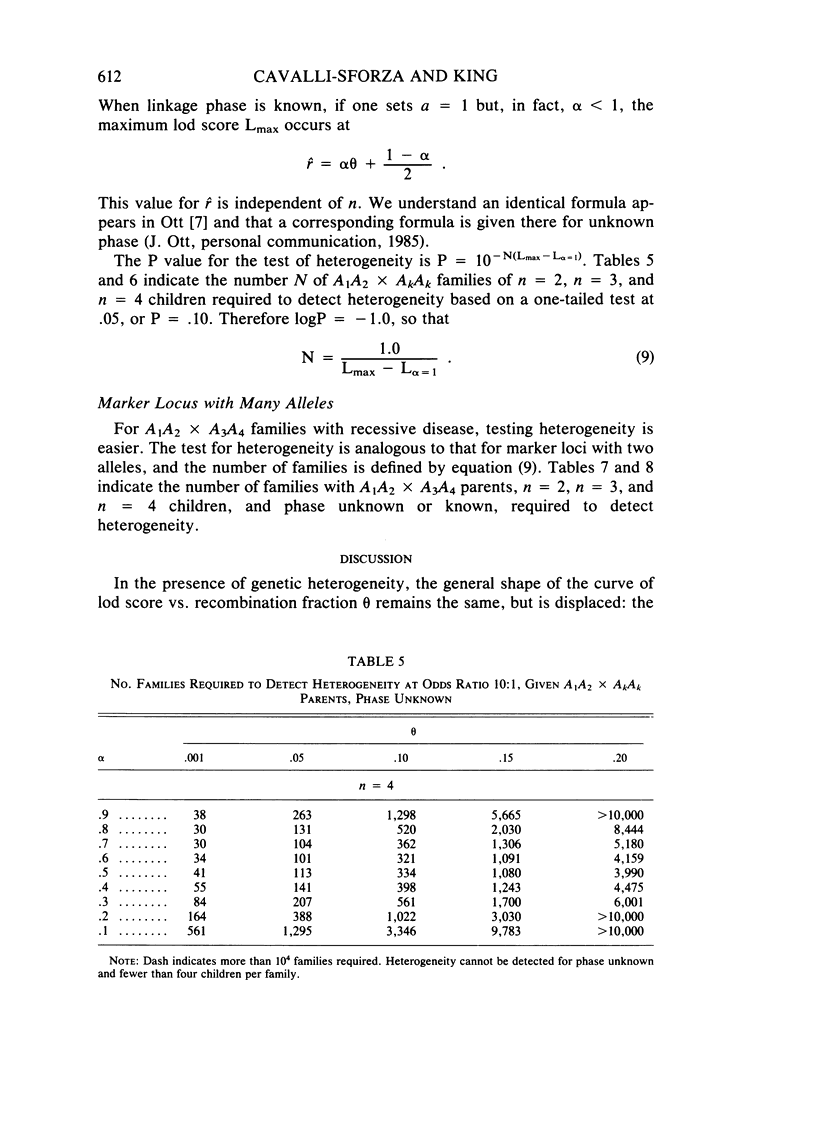


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hodge S. E., Anderson C. E., Neiswanger K., Sparkes R. S., Rimoin D. L. The search for heterogeneity in insulin-dependent diabetes mellitus (IDDM): linkage studies, two-locus models, and genetic heterogeneity. Am J Hum Genet. 1983 Nov;35(6):1139–1155. [PMC free article] [PubMed] [Google Scholar]
- MORTON N. E. Sequential tests for the detection of linkage. Am J Hum Genet. 1955 Sep;7(3):277–318. [PMC free article] [PubMed] [Google Scholar]
- Ott J. Counting methods (EM algorithm) in human pedigree analysis: linkage and segregation analysis. Ann Hum Genet. 1977 May;40(4):443–454. [PubMed] [Google Scholar]
- Ott J. Linkage analysis and family classification under heterogeneity. Ann Hum Genet. 1983 Oct;47(Pt 4):311–320. doi: 10.1111/j.1469-1809.1983.tb01001.x. [DOI] [PubMed] [Google Scholar]
- Risch N., Baron M. X-linkage and genetic heterogeneity in bipolar-related major affective illness: reanalysis of linkage data. Ann Hum Genet. 1982 May;46(Pt 2):153–166. doi: 10.1111/j.1469-1809.1982.tb00706.x. [DOI] [PubMed] [Google Scholar]
- SMITH C. A. TESTING FOR HETEROGENEITY OF RECOMBINATION FRACTION VALUES IN HUMAN GENETICS. Ann Hum Genet. 1963 Nov;27:175–182. doi: 10.1111/j.1469-1809.1963.tb00210.x. [DOI] [PubMed] [Google Scholar]


