Abstract
We describe EnteriX, a suite of three web-based visualization tools for graphically portraying alignment information from comparisons among several fixed and user-supplied sequences from related enterobacterial species, anchored on a reference genome (http://bio.cse.psu.edu/). The first visualization, Enteric, displays stacked pairwise alignments between a reference genome and each of the related bacteria, represented schematically as PIPs (Percent Identity Plots). Encoded in the views are large-scale genomic rearrangement events and functional landmarks. The second visualization, Menteric, computes and displays 1 Kb views of nucleotide-level multiple alignments of the sequences, together with annotations of genes, regulatory sites and conserved regions. The third, a Java-based tool named Maj, displays alignment information in two formats, corresponding roughly to the Enteric and Menteric views, and adds zoom-in capabilities. The uses of such tools are diverse, from examining the multiple sequence alignment to infer conserved sites with potential regulatory roles, to scrutinizing the commonalities and differences between the genomes for pathogenicity or phylogenetic studies. The EnteriX suite currently includes >15 enterobacterial genomes, generates views centered on four different anchor genomes and provides support for including user sequences in the alignments.
INTRODUCTION
The enterobacterial system of microbial genomes provides one of the largest available collections of sequences from related species (Table 1). The availability of such a large volume of data opens up the opportunity for comparative studies to identify and interpret the common and divergent features among species and their phenotypic impact and brings the challenge of effectively organizing and presenting the data in a fashion that is both concise and informative. We describe EnteriX, a web visualization system that answers this need, consisting of three tools for graphically portraying alignment information from comparisons between a reference sequence [either Escherichia coli K-12 (ECO), E.coli O157:H7 (ECH), Salmonella typhimurium LT2 (STM) or Salmonella typhi CT18 (STY)] and several re-lated genomes (Table 1), together with integrated data and anno-tations of genomic re-organization events and functional sites.
Table 1. List of enterobacterial genomes included in the comparative views.
| Species name | Abbr. | Number of contigs | Ref. | Sequencing center and data source |
|---|---|---|---|---|
| E.coli K-12 | ECO | 1 chrom | (1) | U. of Wisconsin, Madison GenBank A#: U00096 |
| E.coli 0157:H7 EDL933 | ECH | 1 chrom | (2) | U. of Wisconsin, Madison GenBank A#: NC_0022655 |
| E.coli CFT073 | ECU | 1 chrom | (3) | U. of Wisconsin, Madison GenBank A#: AE014075 |
| Shigella flexneri | SHG | 1 chrom | (4) | Microbial Genome Center of Chinese Ministry of Public Health GenBank A#: AE005674 |
| S.typhimurium LT2 | STM | 1 chrom 1 plasmid | (5) | Washington U. St. Louis GenBank A#: AE006468 |
| S.typhimurium SL1344 | SSL | 426 | Sanger Center, UK http://www.sanger.ac.uk/Projects/Salmonella | |
| S.typhi CT18 | STY | 1 chrom 2 plasmids | (6) | Sanger Center, UK GenBank A#: AL513382 |
| S.typhi Ty2 | STT | 1 chrom 1 plasmid | (7) | U. of Wisconsin, Madison GenBank A#: NC_004631 |
| S.paratyphi A | SPA | 66 | Washington U. St Louis ftp://genome.wustl.edu/pub/seqmgr/bacterial/ | |
| S.paratyphi C | SPC | 2289 | U. of Calgary, Canada | |
| S.dublin | SDU | 2442 | U. of Illinois Urbana-Champaign http://salmonella.utmem.edu/ | |
| S.enteriditis | SEN | 2303 | U. of Illinois Urbana-Champaign http://salmonella.utmem.edu/ | |
| S.bongori | SBO | 66 | Sanger Center, UK http://www.sanger.ac.uk/Projects/Salmonella | |
| S.diarizonae | SDI | 576 | Sanger Center, UK http://www.sanger.ac.uk/Projects/Salmonella | |
| Klebsiella pneumoniae | KPN | 111 | Washington U. St. Louis ftp://genome.wustl.edu/pub/seqmgr/bacterial/ | |
| Yersinia pestis | YPE | 1 chrom 3 plasmids | (8) | Sanger Center, UK GenBank A#: NC_003143 |
| Vibrio cholerae | VCH | 2 chrom | (9) | TIGR GenBank A#: NC_002505 and NC_002506 |
| Pseudomonas aeruginosa | PAE | 1 chrom | (10) | Pseudomonas Genome Project GenBank A#: AE004091 |
RESULTS
Enteric
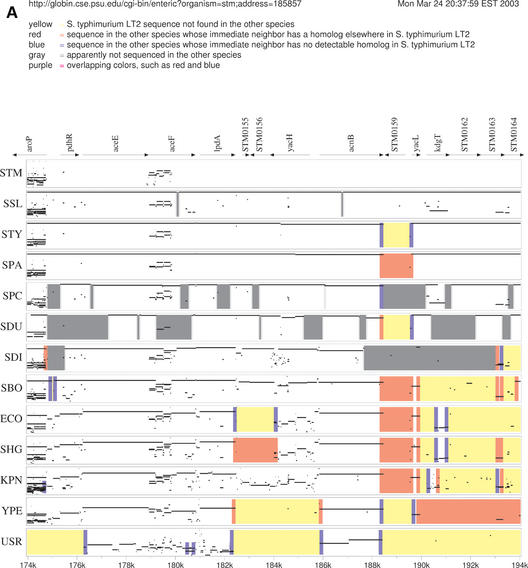
The first visualization component, Enteric, presents pairwise alignments between a reference genome and each of the related bacteria, in a 20 Kb region centered at a user-specified address or gene in the reference sequence. Alignments are represented schematically as PIPs (Percent Identity Plots; Fig. 1). A PIP is a 2D plot in which positions along the horizontal axis correspond to locations in the reference genome, and coordinates on the vertical axis correspond to alignment percent sequence identity levels, restricted to the 50–100% range. The ungapped segments within each alignment are represented as horizontal lines spanning the corresponding range in the reference genome and at a vertical position equal to the ungapped alignment's percent sequence identity value. The ends of alignments in Enteric PIP views are marked with color-coded bars that indicate deletion, insertion and rearrangement events between the genomes. Additional information, such as the length of deletion or the location of the nearest neighbor in the other genome, is revealed by placing the mouse pointer on the feature. Genes annotated in the reference genome are shown with arrows above the PIPs. Using an embedded-hyperlink mechanism, their names contain links to the associated COG category pages maintained at NCBI (11) in the ECO and ECH centered views or to a page containing information on E.coli orthologs of S.typhimurium genes at the web site of the Washington University Salmonella Sequencing Center, for the STM-centered views. The output is presented in PDF format. The alignments of fixed genomes are pre-computed using a locally developed program called blastz (12,13), an independent implementation of the Gapped BLAST algorithm that was specifically designed to compare two long sequences, and stored for fast retrieval. The server also has support for incorporating one user-supplied sequence, for which the alignment and annotations are computed on-the-fly.
Figure 1.
(A and B) Enteric display at the S.typhimurium acnB gene location. The P.aeruginosa genome is used as the user-supplied sequence (USR). Alignments were pre-computed using the program blastz with parameters match=10, mismatch=−10, gap_open=60, gap_ext=2, starting from exact words of size w=8 (12 for genomes from the same species). Gap-free alignment segments are shown as horizontal bars in the PIPs, at a vertical coordinate corresponding to the alignment segment's percent sequence identity, and genomic re-arrangement events are marked with color bars at the ends of alignments. Multiple horizontal matches at the same location in the reference genome indicate paralogues in the other sequence (e.g. the 174–174.8 K region in the STM PIP).
Menteric
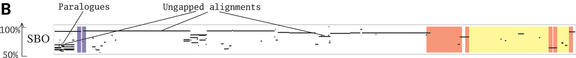
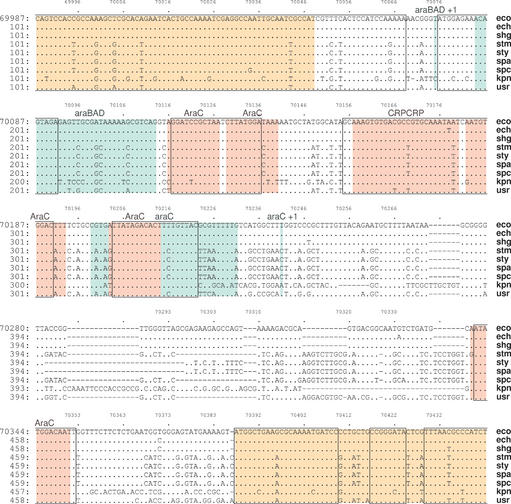
The second tool, Menteric, computes and displays 1 Kb views of annotated multiple alignments of the same sequences, starting at a user specified address or gene in the reference genome, shown at nucleotide-level resolution (Fig. 2). Known functional sites and other characteristic regions are marked on the alignment with a combination of graphical symbols. The user can select from five different criteria for determining conserved regions in the multiple alignment, ranging from consensus majority rule, phylogenetic distance, information content and distance from a fixed or unknown center sequence (14). The conserved regions thus identified are shown enclosed in boxes. In addition, known or predicted regulatory sites are marked with color-coded underlays (light chocolate, ORFs; green, promoters; red, regulatory protein binding sites) and may contain links to the annotation data source. Currently, only the GenBank annotation is available (1,2,5,6). Links embedded in the sequence labels on the right-hand side of the multiple alignment can be used to download sequence data in the restricted range displayed in that view. The output can be presented in either PDF or PostScript format. The multiple alignment is produced dynamically at run time, from sequences retrieved based on the pre-computed pairwise similarities. Like Enteric, Menteric also provides support for one user-specified sequence to be included in the alignment.
Figure 2.
Menteric-type view in the common regulatory region of the E.coli K-12 araC and araBAD operons. Color-coded underlays are used to mark known or predicted functional elements in the E.coli genome (light chocolate, ORFs; green, promoters; red, binding sites of regulatory proteins). The user sequence (usr) is the concatenation of S.bongori contigs.
Maj
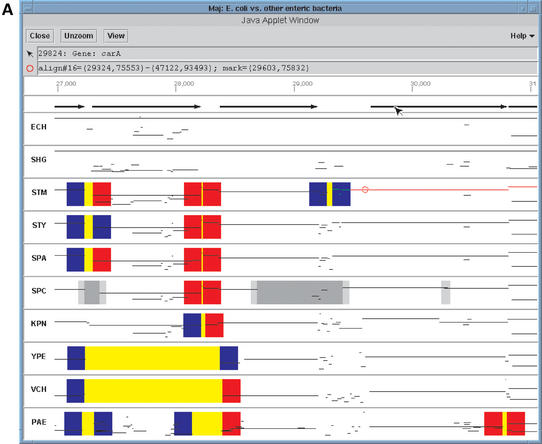
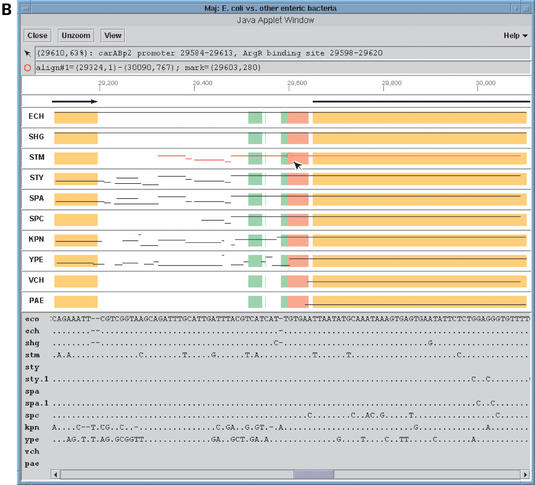
The third EnteriX component is a Java-based tool named Maj. It displays alignment information in two formats, ‘wide’ and ‘close-up’, corresponding roughly to the Enteric and Menteric views, respectively, and adds interactive zoom-in capabilities. The Maj wide view (Fig. 3A) uses Enteric's paradigm for presenting alignments and associated information. However, unlike Enteric, information about the various features that was previously provided via labels and embedded pseudo-links is now displayed in the two message boxes at the top of the window. The top box displays details about the mouse location (PIP coordinates, contig name, properties associated with the color bands), while the bottom one is used to show information about the local alignment that the user has selected by clicking on its horizontal line in the PIP. Maj's close-up view (Fig. 3B) emulates Menteric's nucleotide-level multiple alignment views, but the information is organized somewhat differently. The multiple alignment is now shown in a scrollable bar at the bottom of the frame, while the main panel displays interactive PIPs of pairwise alignments projected directly from the multiple alignment. Using a Java applet, Maj allows the user to ‘zoom in’ on a sub-region of the view, selected by dragging the mouse in any PIP panel. It also provides the ability to toggle between the wide and close-up views for the current region, using the View button located at the top of the window.
Figure 3.
(A and B) Maj wide and close-up views centered at the carAB operon in E.coli.
New features
Since its public release in 2000 (15), the EnteriX suite has expanded to include >15 enterobacterial species, including the completely sequenced E.coli K-12, O157:H7 and CFT073, S.typhimurium LT2 and S.typhi strains CT18 and Ty2, Vibrio cholerae, Pseudomonas aeruginosa, Yersinia pestis and a number of partially sequenced Salmonella and Klebsiella species. It has also been adapted to present alternative reference genomes (E.coli K-12 and O157:H7, S.typhimurium LT2 and S.typhi CT18). With the increase in the number of genomes scheduled to be partially or completely sequenced over the next years, particularly from among the Salmonella species, storing, organizing and presenting the information efficiently will become increasingly difficult. To answer the need for flexibility and compactness, EnteriX now provides the user with the ability to select the genomes to be included in the views from among those available in the data store. Perhaps the most notable new feature is that the Enteric and Menteric tools have acquired the capability to include a user-provided sequence in their comparative views. The third tool, Maj, is currently being updated to incorporate some of these new features as well.
AVAILABILITY
The EnteriX servers are available from http://bio.cse.psu.edu and from the Salmonella Sequencing Center site at Washington University, St Louis (http://genome.wustl.edu/projects/bacterial/styphimurium/).
DISCUSSION
Recent years have brought a tremendous increase in the amount of sequence data from various genome sequencing projects, an increase that is projected to accelerate over the next years. As a result, the task of organizing and summarizing the data to extract the most informative features has become a challenging yet critical endeavor. Visualization is an effective way of structuring and presenting such information effectively, in a concise and eloquent fashion. The software we describe, EnteriX, has been developed to present alignment information and inferred or associated properties in an integrated framework, as an instrument for discovery and analysis.
The uses of such tools are diverse. Examination of the multiple alignment in a region may reveal conserved sites with potential regulatory roles, such as binding sites of regulatory proteins or non-coding RNAs. The large-scale views unveil commonalities and differences between the genomes that may shed light on their evolutionary relationships, or may be characteristic of pathogenicity.
To aid in the processes of structural and functional annotation, as well as in selecting the most promising candidates for experimental validation, integration of data from various complementary resources is essential. The PDF files produced by Enteric and Menteric, as well as Maj's Java views, contain hyperlinks to related repositories of information on the internet, such as GenBank entry pages, COG and the Washington University list of orthologous ECO and STM genes. Using the same mechanism, hyperlinks incorporated in Menteric's sequence labels allow one to download contig sequence data for further analyses.
To increase the applicability of these tools, we plan to incorporate additional reference genomes and to provide more extensive access to external sources of data, such as RegulonDB's database of experimentally validated E.coli regulatory sites, using our established mechanisms of embedded hyperlinks. With the ongoing effort to sequence a number of Salmonella serovars and related genomes, including S.paratyphi A and K.pneumoniae, at the Washington University Genome Sequencing Center in St Louis, it is anticipated that EnteriX will provide a complex and multi-faceted view of the genomics of the Enterobacteriaceae and will prove a valuable resource in the area of visualizing integrated annotation for the bacterial genomics community.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported in part by grant HG-02238 from the National Human Genome Research Institute to W.M. and grant AI34829 to M.M.
REFERENCES
- 1.Blattner F.R., Plunkett,G.III, Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F. et al. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 2.Perna N.T., Plunkett,G.III, Burland,V., Mau,B., Glasner,J.D., Rose,D.J., Mayhew,G.F., Evans,P.S., Gregor,J., Kirkpatrick,H.A. et al. (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature, 409, 529–533. [DOI] [PubMed] [Google Scholar]
- 3.Welch R.A., Burland,V., Plunkett,G.D.III, Redford,P., Roesch,P., Rasko,D.A., Buckles,E.L., Liou,S.-R., Boutin,A., Hackett,J. et al. (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli.Proc. Natl Acad. Sci. USA, 99, 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Q., Yuan,Z.H., Xu,J.G., Wang,Y., Shen,Y., Lu,W.C., Wang,J.H., Liu,H., Yang,J., Yang,F. et al. (2002) Genome sequence of Shigella flexneri 2a, insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res., 30, 4432–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClelland M., Sanderson,K.E., Spieth,J., Clifton,S.W., Latreille,P., Courtney,L., Porwollik,S., Ali,J., Dante,M., Du,F. et al. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature, 413, 852–856. [DOI] [PubMed] [Google Scholar]
- 6.Parkhill J., Dougan,G., James,K.D., Thomson,N.R., Pickard,D., Wain,J., Churcher,C., Mungall,K.L., Bentley,S.D., Holden,T.G. et al. (2001) Complete genome sequence of a multiple drug resistant Salmonella enterica serovar typhi CT18. Nature, 413, 848–852. [DOI] [PubMed] [Google Scholar]
- 7.Deng W., Liou,S.R., Plunkett,G.III, Mayhew,G.F., Rose,D.J., Burland,V., Kodoyianni,V., Schwartz,D.C. and Blattner,F.R. (2003) Comparative genomics of Salmonella enterica serovar typhi Strains Ty2 and CT18. J. Bacteriol., 185, 2330–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkhill J., Wren,B.W., Thomson,N.R., Titball,R.W., Holden,M.T.G., Prentice,M.B., Sebaihia,M., James,K.D., Churcher,C., Mungall,K.L. et al. (2001) Genome sequence of Yersinia pestis, the causative agent of plague. Nature, 413, 523–527. [DOI] [PubMed] [Google Scholar]
- 9.Heidelberg J.F., Eisen,J.A., Nelson,W.C., Clayton,R.A., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Umayam,L.A. et al. (2000) DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature, 406, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stover C.K., Pham,X.-Q.T., Erwin,A.L., Mizoguchi,S.D., Warrener,P., Hickey,M.J., Brinkman,F.S.L., Hufnagle,W.O., Kowalik,D.J., Lagrou,M. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- 11.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartz S., Zhang,Z., Frazer,K.A., Smit,A., Riemer,C., Bouck,J., Gibbs,R., Hardison,R. and Miller,W. (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res., 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz S., Kent,W.J., Smit,A., Zhang,Z., Baertsch,R., Hardison,R.C., Haussler,D. and Miller,W. (2003) Human-mouse alignments with Blastz. Genome Res., 13, 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stojanovic N., Florea,L., Riemer,C., Gumucio,D., Slightom,J., Goodman,M., Miller,W. and Hardison,R. (1999) Comparison of five methods for finding conserved sequences in multiple alignments of gene regulatory regions. Nucleic Acids Res., 27, 3899–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Florea L., Riemer,C., Schwartz,S., Zhang,Z., Stojanovic,N., Miller,W. and McClelland,M. (2000) Web-based visualization tools for bacterial genome alignments. Nucleic Acids Res., 28, 3486–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]