Abstract
Trichomonas vaginalis is a unicellular eukaryote that lacks mitochondria and contains a specialized organelle, the hydrogenosome, involved in carbohydrate metabolism and iron-sulfur cluster assembly. We report the identification of two glycine cleavage H proteins and a dihydrolipoamide dehydrogenase (L protein) of the glycine decarboxylase complex in T. vaginalis with predicted N-terminal hydrogenosomal presequences. Immunofluorescence analyses reveal that both H and L proteins are localized in hydrogenosomes, providing the first evidence for amino acid metabolism in this organelle. All three proteins were expressed in Escherichia coli and purified to homogeneity. The experimental Km of L protein for the two H proteins were 2.6 μM and 3.7 μM, consistent with both H proteins serving as substrates of L protein. Analyses using purified hydrogenosomes showed that endogenous H proteins exist as monomers and endogenous L protein as a homodimer in their native states. Phylogenetic analyses of L proteins revealed that the T. vaginalis homologue shares a common ancestry with dihydrolipoamide dehydrogenases from the firmicute bacteria, indicating its acquisition via a horizontal gene transfer event independent of the origins of mitochondria and hydrogenosomes.
Hydrogenosomes are double membrane-bound organelles involved in iron-sulfur cluster assembly that produce molecular hydrogen and ATP via carbohydrate metabolism (30, 40, 41). These organelles were originally discovered in trichomonads and subsequently were detected in a variety of unrelated organisms, such as certain ciliates and fungi. Although the evolutionary origin(s) of hydrogenosomes is under debate, they appear to be polyphyletic, with ciliate hydrogenosomes being derived directly from mitochondria (3) and the origin of those in trichomonads and fungi less clear (11, 17). Functional and phylogenetic data do, however, indicate that all hydrogenosomes are related to mitochondria (5, 14, 34, 36, 41).
Trichomonas vaginalis is a deep-branching, microaerophilic, unicellular protist that lacks mitochondria and contains hydrogenosomes (30, 39). The lack of a genome in the hydrogenosomes of trichomonads precludes reconstruction of their ancestry directly as has been convincingly done for mitochondria (18). However, the phylogeny of genes that encode hydrogenosomal proteins, the hydrogenosomal targeting signals, and protein import machinery can provide indirect evidence for the evolutionary history of hydrogenosomes (4, 12, 43). Regardless of whether a protomitochondrion was modified to become a hydrogenosome or was lost during the adaptation of Trichomonas to anoxic environments, the absence of the tricarboxylic acid (TCA) cycle and many metabolic enzymes underscore the independent evolution of the T. vaginalis hydrogenosome as a unique organelle.
Carbohydrates are the preferred source of nutrients for T. vaginalis; however, under conditions where carbohydrates are limiting, amino acids have been shown to sustain trichomonad growth and survival. Evidence of uptake of amino acids, especially arginine, threonine, and leucine, are found when T. vaginalis is grown in the absence of maltose. Under normal culture conditions, T. vaginalis consumes large amounts of arginine and smaller amounts of methionine for use in energy production (reviewed in reference 32).
Serine-glycine interconversion is an essential and ubiquitous step in primary metabolism. In Escherichia coli, 15% of all the carbon atoms assimilated from glucose are estimated to pass through the glycine-serine pathway (44). The glycine decarboxylase complex (GDC) together with serine hydroxymethyl transferase (SHMT) is responsible for this interconversion. GDC is a multienzyme complex which occurs in all organisms. In eukaryotes, GDC is present exclusively in the mitochondria and catalyzes the oxidative decarboxylation and deamination of glycine. It is comprised of four proteins, namely, the glycine cleavage H protein, the glycine decarboxylase P protein, the dihydrolipoamide dehydrogenase L protein, and the aminomethyl transferase T protein. The H protein plays a pivotal role in the mechanism of the reaction, since its prosthetic group, 5[3-(1,2)-dithiolanyl]pentanoic acid (lipoic acid), interacts successively with the three other components of the complex. The term glycine-serine interconversion might suggest that the central importance of this pathway is simply the synthesis of serine from glycine and vice versa. However, in both directions of the concerted reactions of GDC and SHMT, tetrahydrofolate becomes N5,N10-methylenated, making these reactions the most important source of active one-carbon units for a number of biosynthetic processes, such as methionine, pyrimidine, and purine biosynthesis. Moreover, glycine and serine are precursors of glutathione, tryptophan, phosphatidyl choline, and related phospholipids and ethanolamine (reviewed in reference 2). GDC has been extensively studied and characterized in plants, Saccharomyces cerevisiae, and mammals (9).
In this paper we report the identification and analyses of glycine cleavage H proteins and dihydrolipoamide dehydrogenase L protein from T. vaginalis. The proteins were found to possess a hydrogenosomal presequence and to be targeted to the hydrogenosome. Their in vitro and in vivo interactions were also studied, and their phylogeny was determined.
MATERIALS AND METHODS
Parasite cell culture.
T. vaginalis strains T1 and G3 were grown in Diamond's medium, supplemented with 10% (vol/vol) horse serum and iron as described previously (35).
Plasmid construction.
We designed primers H1NF, H1XR, H2NF, H2XR, LNF, LXR (see the table in the supplemental material) to amplify the complete open reading frame of the genes encoding the H1, H2, and L proteins, respectively, from the T. vaginalis genomic DNA for in-frame cloning into the pET-29B expression vector (Novagen) using NdeI and XhoI restriction sites. The resulting recombinant C-terminally His-tagged fusion protein was expressed in Escherichia coli BL21(DE3) cells (Invitrogen). For expressing the proteins in T. vaginalis, we PCR amplified the respective genes with primer pairs H1NF/H1TvKR, H2NF/H2TvKR, and LNF/LTvKR (see the table in the supplemental material). The PCR fragments were then cloned into master-neo-(HA)2 plasmid (12) to generate constructs to transfect T. vaginalis.
RNA extraction and RT-PCR.
Total cellular RNA was extracted using Trizol reagent (Invitrogen) from T. vaginalis cells. For reverse transcriptase-PCR (RT-PCR), cDNA was synthesized from 1 μg of total RNA of T. vaginalis (G3 strain) with reverse transcriptase using gene-specific (H1F/H1R and H2F/H2R) (see the table in the supplemental material) and oligo(dT) primers. The reaction mixture was then subjected to PCR amplification with specific forward and reverse primers for 30 cycles consisting of heat denaturation, annealing, and extension steps. PCR products were size fractionated on 1% agarose gels and visualized under UV light.
Selectable transfection of T. vaginalis.
Electroporation of T. vaginalis strain T1 was carried out as described previously (7) with 50 μg of circular plasmid DNA. Transfectants were selected with 100 μg/ml of G418 (Sigma) prior to crude fractionation and organelle purification.
Crude fractionation of T. vaginalis transfectants.
Cells from cultures of the transfectants were broken in a cell disruptor (Energy Service Co.) and fractionated by centrifugation at 12,000 × g into a crude cytosolic fraction and a crude organellar fraction as described previously (12).
Isolation of hydrogenosomes.
Hydrogenosomes were prepared as previously described by Bradley et al. (4). The organelles were then resuspended in a buffer containing 1% Triton X-100, 20 mM Tris-HCl, pH 7.5, 2% sucrose, 250 mM NaCl, 5 mg/ml leupeptin, and 25 mg/ml Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK) and incubated on ice for 30 min, with occasional vortexing every 5 min. The clear hydrogenosomal lysate was finally obtained after centrifugation at 16,000 × g for 30 min at 4°C.
Immunofluorescence microscopy.
Live T. vaginalis cells were washed with warm Diamond's media and allowed to attach to coverslips for 30 min in a humidifying chamber. The cells were then washed with warm 1× phosphate-buffered saline and fixed with 3.5% formalin for 20 min at room temperature. The C-terminally hemagglutinin (HA)-tagged H and L proteins and Hsp70 were visualized using mouse anti-HA monoclonal antibody (Sigma) and rabbit anti-Hsp70 polyclonal antibody as primary antibodies and secondary Alexa Fluor-488 donkey anti-mouse (green) and Alexa Fluor-594 donkey anti-rabbit (red) antibodies (Invitrogen). Cells were visualized at ×100 magnification using an Axioscop2 (Zeiss). Images were processed with Axiovision v. 3.2 software (Zeiss).
Expression of recombinant proteins and antibody production.
The pET-H1, pET-H2, and pET-L plasmids were transformed into E. coli BL21(DE3) cells and selected with 30 μg/ml kanamycin in LB media. For the expression of H1 and H2 proteins, 500-ml cultures were grown at 37°C in the presence or absence of 0.1 mM lipoic acid to an A600 of 0.8 before induction with 0.5 mM isopropyl-1-thio-β-d-galactopyranoside for 2 h at 37°C. Induction of recombinant L protein was carried out at 22°C for 7 h in presence of 0.5 mM isopropyl-1-thio-β-d-galactopyranoside to obtain the expressed protein in soluble form. Harvested cells were then resuspended in a buffer containing 1 mg/ml of lysozyme, 0.05 M NaH2PO4, 0.3 M NaCl, 10 mM imidazole, protease inhibitor cocktail (Roche Diagnostics), 5 mg/ml leupeptin, and 25 mg/ml TLCK for 20 min at 4°C and sonicated. The insoluble material was pelleted at 10,000 × g for 30 min at 4°C, and the resulting lysate was applied to a nickel-nitrilotriacetic acid-agarose column. The column was washed four times with a buffer containing 0.05 M NaH2PO4, 0.3 M NaCl, and 20 mM imidazole, pH 8.0. Specifically bound proteins were eluted with the above buffer containing 0.25 M imidazole. Recombinant proteins were sent to Animal Pharm Services, Inc. (California) to raise polyclonal antibodies in rabbits.
Analysis of the native mass of the proteins by gel filtration chromatography.
The molecular size of endogenous, native H and L proteins were calculated using 1 mg of hydrogenosomal extract by Superdex 200 column calibrated with molecular mass standards (Bio-Rad). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out with the different eluted fractions according to the method of Laemmli followed by Western blotting using respective protein antibodies. The masses of the native proteins were ascertained by plotting the band intensities of the respective proteins, as visualized by Western blotting, against fraction numbers. The apparent molecular masses of the proteins were estimated by comparison with the logarithmic plot of molecular mass standards (soybean trypsin inhibitor, 20.1 kDa; bovine serum albumin, 67 kDa; alcohol dehydrogenase, 150 kDa; catalase, 232 kDa; ferritin, 440 kDa; thyroglobuin, 669 kDa;).
In vitro activity of H and L proteins.
The interaction of recombinant H and L proteins were measured by monitoring the reaction of H protein with 5,5′-dithiobis(2-nitrobenzoic acid) (Nbs2) in the presence of L protein (28, 29) with minor modifications. Briefly, in the presence of NADH, dihydrolipoamide dehydrogenase (L protein) catalyzes the conversion of the lipoamide of H protein into two SH groups. A large excess of Nbs2 rapidly catalyzes the conversion of the SH groups into disulfide bonds with the formation of two molecules of 2-nitro-5-thiobenzoate (Nbs). The L protein activity was assayed following the reduction of the disulfide bond of the lipoamide of H protein, resulting in the formation of 2-nitro-5-thiobenzoate. The reaction is monitored spectrophotometrically at 412 nm (maximum absorbance of Nbs; ɛmax, 13,600 M−1/cm−1), and L protein activity was expressed as nmol of Nbs formed/s. The assay was performed at 25°C in a buffer containing 40 mM Tris, pH 7.4, 0.1 mM EDTA, 100 μM L protein, 0.5 μM to 30 μM H proteins (or 0.01 mM to 1.0 mM lipoamide) (Sigma) and 1 mM NADH. The reaction was initiated by adding 2 mM Nbs2.
Sucrose density gradient analysis.
One milligram of hydrogenosomal extracts (1.0 ml) was loaded onto linear sucrose gradients (2 to 20%) in 20 mM Tris-HCl at pH 7.5 containing 1% Triton X-100, 250 mM NaCl, TLCK, and leupeptin prepared in 25- by 89-mm centrifugation tubes (Beckman) made with a Gradient Master (BioComp Instruments) and were immediately subjected to centrifugation at 38,000 rpm for 16 h at 4°C with an SW41 rotor (Beckman). Ten serial fractions were collected from the top of the gradients using a Piston Gradient Fractionator (BioComp Instruments), TCA precipitated, and subjected to 15% SDS-PAGE followed by immunoblot analysis using anti-H and anti-L antibodies. Marker proteins (Amersham Bioscience) soybean trypsin inhibitor (20.1 kDa), bovine serum albumin (67 kDa), lactate dehydrogenase (140 kDa), catalase (232 kDa), and ferritin (440 kDa) were subjected to sucrose gradients under conditions similar to those described above for the marker proteins and detected by Coomassie staining after SDS-PAGE.
Phylogenetic analysis.
Homologues of the T. vaginalis glycine cleavage system H and L proteins were identified by BLAST searching the NCBI nonredundant protein database and other genome databases. Alignments were produced using MUSCLE (13) with manual curation. Gaps and highly divergent or ambiguous regions of the alignments were excluded from phylogenetic analysis, resulting in final alignments of 109 and 395 amino acid positions for H and L proteins, respectively. The alignments are available upon request. Phylogenetic relationships for each protein were assessed using a Bayesian statistical procedure, as implemented by the computer program MrBayes (21). MrBayes performs a Metropolis-coupled Markov chain Monte Carlo (MC3) estimation of posterior probabilities (20, 25, 38). We performed MC3 estimation of posterior probabilities using noninformative prior probabilities, the JTT+I+Γ (22) substitution model with inclusion of unequal amino acid frequencies, and four incrementally heated Markov chains with different random starting trees. The Metropolis-coupled Markov chains were run to 10,000,000 generations with sampling every 100 generations. Posterior probabilities of topologies, clades, and parameters were estimated from the sampled topologies after removal of MC3 burn-in.
RESULTS
Identification of putative genes encoding components of the GDC.
In an attempt to obtain genes encoding the glycine cleavage enzymes involved in the glycine-serine metabolism, we searched an ∼7× coverage of the Trichomonas vaginalis genome database (http://tigrblast.tigr.org/er-blast/index.cgi?project=tvg) with homologous sequences of GDC proteins. Two independent contigs were found (TVAG_100550 and TVAG_177600) encoding glycine cleavage H protein with similar but not identical sequences. Another contig (TVAG_272760) encoding the dihydrolipoamide dehydrogenase (L protein) was also found. We also searched for orthologues of the other two members of the glycine decarboxylase complex, the glycine dehydrogenase (P protein) and aminomethyltranferase (T protein), using BLAST and a hidden Markov model using a Pfam database (16), and none were identified.
Sequence alignments of glycine cleavage H1 and H2 proteins and the L protein were performed using CLUSTALX.
Comparison of the deduced amino acid sequences of the glycine cleavage H1 and H2 proteins from the T. vaginalis revealed 28% and 36% identity and 48% and 61% similarity to the glycine cleavage H protein from Pisum sativum, respectively (Fig. 1). A 42% identity and 61% similarity with the dihydrolipoamide dehydrogenase from P. sativum were observed when the deduced amino acids of the L protein from T. vaginalis was compared. All 3 genes were also shown to be expressed using RT-PCR (data not shown).
FIG.1.
Sequence comparison of glycine cleavage H proteins and dihydrolipoamide dehydrogenase (L protein) from different organisms using CLUSTALX. (A) Alignment of amino acid sequences deduced from two genes encoding glycine cleavage H proteins (TvH1 and TvH2) from Trichomonas vaginalis compared with H proteins from S. cerevisiae (Sc; P39726), B. subtilis (Bc; O32174), and P. sativum (Ps; P16048). The lysine (K) that binds lipoic acid is boxed. (B) Alignment of amino acid sequences deduced from the gene encoding dihydrolipoamide dehydrogenase (L protein) from T. vaginalis (Tv) compared with L proteins from S. cerevisiae (Sc; P09624), P. sativum (Ps; P31023), and B. subtilis (Bs; AAC05585). Asterisks and dots represent identities and similarities between the sequences, respectively. The hydrogenosomal presequence is marked in boldface type in the T. vaginalis protein sequences.
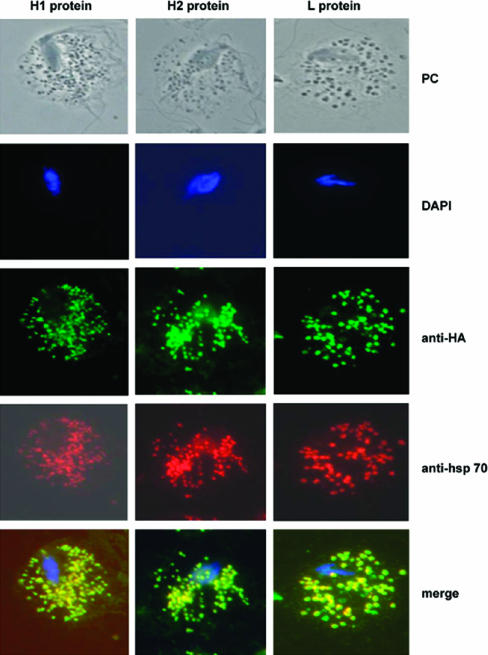
H1, H2, and L localize in hydrogenosomes.
To investigate the subcellular localization of the H and L proteins, C-terminally HA-tagged proteins were expressed in T. vaginalis, and immunofluorescence labeling of trichomonads expressing these proteins was performed. The labeling of the HA-tagged proteins was found to colocalize with Hsp70, a marker for the hydrogenosomes, demonstrating that the proteins are localized to the organelle (Fig. 2). Western analysis of the crude cytosolic and organellar fractions of the T. vaginalis cells expressing the tagged proteins with anti-HA antibody further confirmed these results (data not shown).
FIG. 2.
Cellular localization of HA-tagged H1, H2, and L proteins in T. vaginalis transfectants. Cells were transfected with plasmids encoding HA-tagged H1, H2, or L proteins and were stained for immunofluorescence microscopy using mouse HA-tagged antibody. T. vaginalis anti-Hsp70 was used as a control for hydrogenosomal localization (5). Merge images are provided. Nuclei (blue) were stained with 4′,6′-diamidino-2-phenylindole (DAPI), H1, H2, and L were stained with mouse anti-HA (green), and Hsp70 was stained with rabbit anti-Hsp70 (red). PC indicates phase-contrast image.
Characterization of recombinant proteins.
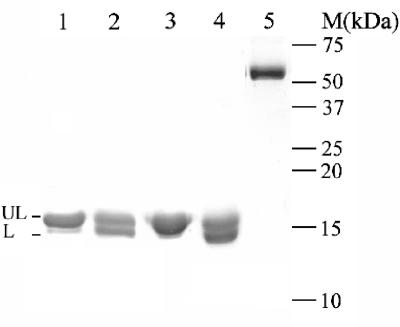
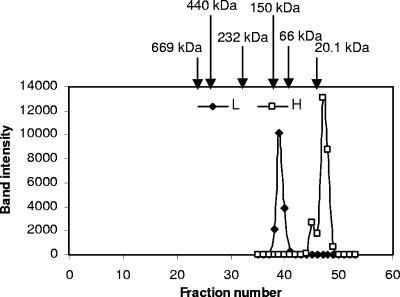
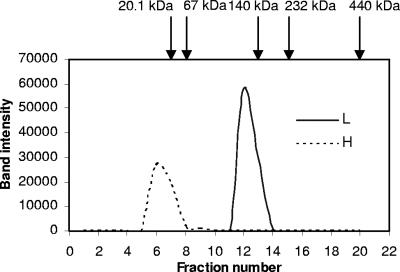
To understand the biochemical properties of the two H proteins and L protein, recombinant proteins were produced. The recombinant proteins (rTvH1, rTvH2, rTvL) were assessed to be >95% pure with Coomassie-stained SDS-PAGE (Fig. 3). The apparent molecular masses of rTvH1, rTvH2, and rTvL were 14 kDa, 15 kDa, and 52 kDa which agreed well with their predicted values (13.9 kDa, 14.8 kDa, and 51.8 kDa, respectively). Furthermore, both anti-H1 and anti-L antibodies produced were shown to detect their respective recombinant proteins; however, anti-H1 antibody also detected the recombinant H2 protein (data not shown). To ascertain the native form of the endogenous proteins, gel filtration chromatography was performed with hydrogenosomal extracts prepared from untransfected cells, and the eluted fractions were analyzed by Western blotting using anti-H and anti-L antibody, respectively. As shown in Fig. 4, these data indicate that endogenous H proteins exist as monomers and endogenous L protein exists as a homodimer.
FIG. 3.
SDS-PAGE analysis of the recombinant proteins. C-terminally His-tagged H1, H2, and L proteins were expressed in BL21(DE3) in the absence and presence of lipoic acid, as indicated. The recombinant proteins were then purified on a nickel-nitrilotriacetic acid-agarose column, analyzed by 15% SDS-PAGE, and stained with Coomassie blue. Lanes 1 and 3 and lanes 2 and 4 represent H1 and H2 proteins purified from bacterial cultures grown in absence and presence of lipoic acid, respectively. Lane 5 represents purified L protein. UL and L represent unlipoylated and lipoylated forms of the H proteins, respectively.
FIG. 4.
Gel filtration elution profiles of endogenous H and L proteins. An extract of purified hydrogenosomes (1.5 ml) was loaded on a Superdex 200 column and run at 0.2 ml/min. Two-milliliter fractions were collected, TCA precipitated, and subjected to 15% SDS-PAGE, followed by immunoblotting using anti-L and anti-H antibody, respectively. Band intensity was plotted against fraction number. Arrows indicate the elution profiles of marker proteins (soybean trypsin inhibitor, 20.1 kDa; bovine serum albumin, 67 kDa; alcohol dehydrogenase, 150 kDa; catalase, 232 kDa; ferritin, 440 kDa; thyroglobuin, 669 kDa;). The apparent molecular masses of native H and L proteins are 15 kDa and 100 kDa, respectively.
Interactions of recombinant glycine cleavage H and L proteins in vitro.
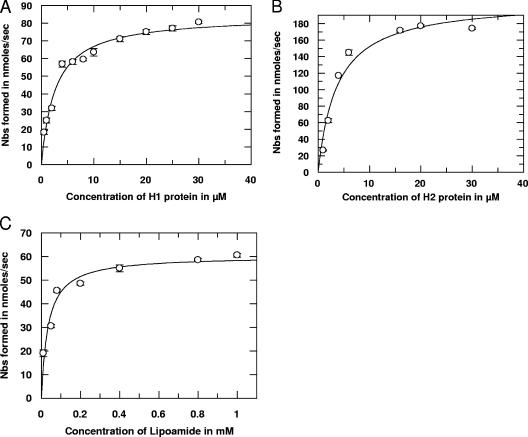
We have determined the kinetic parameters of the recombinant L protein (rTvL) for lipoylated recombinant H proteins (rTvH). As indicated in Fig. 5, the interactions between lipoylated H and L proteins follow Michaelis-Menten kinetics. The apparent Km values of rTvL for binding rTvH1 and rTvH2 proteins were found to be 2.6 μM and 3.7 μM (Table 1). However, we observed that the apparent Km value (34.0 μM) for the free lipoamide was ∼13 times higher than that for rTvH1 protein and ∼9 times higher than rTvH2. These results confirm an affinity of the rTvL protein for both lipoylated rTvH1 rTvH2 proteins. Furthermore, the increase in the specificity constant (kcat/Km) for the two H proteins rather than for the free lipoamide indicates that the lipoylated rTvH1 and rTvH2 proteins serve as effective substrates for the rTvL protein.
FIG. 5.
Substrate kinetics of the reaction catalyzed by L protein as a function of lipoylated H1, H2, and lipoamide as substrates. Purified recombinant proteins were used in reactions conducted at 25°C in a buffer containing 40 mM Tris, pH 7.4, 0.1 mM EDTA, 100 μM L protein, and 0.5 μM to 30 μM H1 protein (A), 0.5 μM to 30 μM H2 protein (B), or 0.01 mM to 1.0 mM lipoamide (C). The reaction was initiated by adding 2 mM Nbs2. L protein activity was measured spectrophotometrically at 415 nm and expressed as nmol of Nbs formed/s.
TABLE 1.
Kinetics parameters for L protein, using different concentrations of H protein and lipoamide as substratesa
| Compound | Apparent Km (μM) | Apparent Vmax (nmol/s) | kcat (s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|---|
| rTvH1 protein | 2.6 | 84.07 | 0.84 | 3.2 × 105 |
| rTvH2 protein | 3.7 | 208.03 | 2.08 | 5.6 × 105 |
| Lipoamide | 34.0 | 60.26 | 0.602 | 1.7 × 104 |
The enzymatic assay was performed as described in Materials and Methods. Kinetic parameters were calculated by using varying concentrations of the two H proteins and lipoamide.
Association of H and L proteins in the hydrogenosomes.
To assess the in vivo interaction of H and L proteins in the hydrogenosomes, we adapted two different techniques: the sucrose density gradient ultracentrifugation and blue native-PAGE. Isolated hydrogenosomes from untransfected cells were solubilized in nonionic, nondenaturing detergent, Triton X-100, and the cleared lysates were subjected to 2 to 20% sucrose gradient ultracentrifugation. Organellar fractions from different sucrose density gradient fractions were then analyzed on SDS-PAGE followed by immunoblotting using both anti-H and anti-L antisera (Fig. 6). H and L proteins were found to fractionate with 6 to 8% and 12% sucrose, respectively, indicating that these proteins do not form a stable complex in the hydrogenosomes and are found as monomers and homodimers, respectively. BN-PAGE analysis of the organellar fraction followed by immunoblotting with anti-H and anti-L antibodies confirmed these results (data not shown).
FIG. 6.
Sucrose density gradient analysis of hydrogenosomal lysates. One milligram of hydrogenosomal extract (1.0 ml) was loaded onto linear sucrose gradients (2 to 20%) in 20 mM Tris-HCl at pH 7.5 containing 1% Triton X-100, 250 mM NaCl, TLCK, and leupeptin. One-milliliter fractions were collected, TCA precipitated, subjected to 15% SDS-PAGE, and analyzed by immunoblotting using anti-L and anti-H antibodies. Band intensity was plotted against fraction number. Arrows indicate the positions of molecular mass standards (soybean trypsin inhibitor, 20.1 kDa; bovine serum albumin, 67 kDa; lactate dehydrogenase, 140 kDa; catalase, 232 kDa; ferritin, 440 kDa).
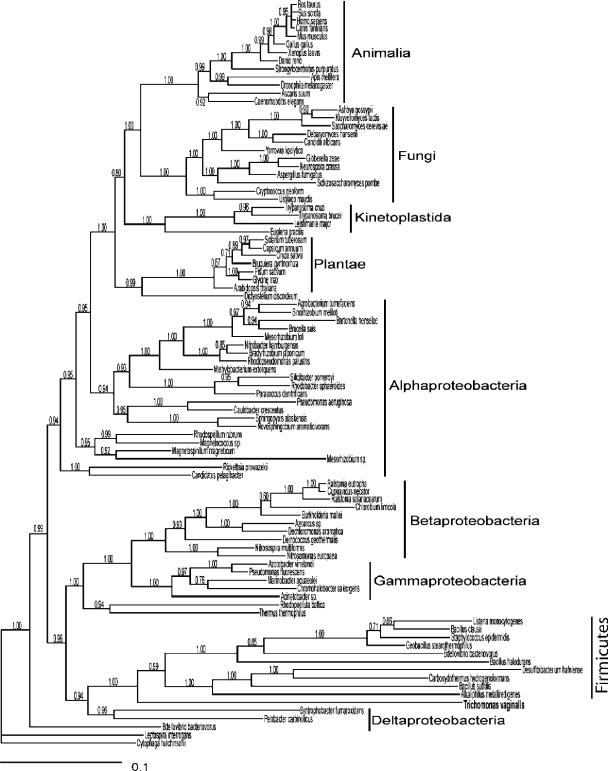
Phylogenetic evaluation of trichomonad H and L proteins.
To elucidate the evolutionary origins of the H and L proteins, we performed a phylogenetic analysis for both in the context of all eukaryotic, bacteria, and archaeal homologues using a Bayesian statistical procedure. The lengths of the MC3 burn-in were 10,000 and 15,000 of 10,000,000 generations for H and L proteins, respectively. There was negligible resolution of phylogenetic relationships of H proteins in this analysis (data not shown) but strong resolution of L protein phylogenetic relationships, as indicated by posterior probabilities (Fig. 7). While phylogenetic analysis strongly supported the common ancestry of eukaryotic L proteins and their origin from within the α-proteobacteria (reflective of mitochondrial origins), the T. vaginalis protein proved an exception by exhibiting putative common ancestry with the firmicute bacteria.
FIG. 7.
Phylogenetic analysis of dihydrolipoamide dehydrogenase (L protein). The phylogeny of L proteins, with Bayesian posterior probabilities superimposed upon the tree, is shown. Horizontal branch lengths are representative of evolutionary change. The phylogeny strongly supports different origins of the T. vaginalis protein from that of other eukaryotic homologues, with common ancestry shared between the firmicute bacteria forms and the T. vaginalis protein.
DISCUSSION
We have identified and characterized two members of the glycine cleavage system, also known as the GDC, from the hydrogenosomes of the anaerobic protozoan T. vaginalis. In other eukaryotes, these proteins are typically found in mitochondria where they play critical roles in amino acid metabolism. Both T. vaginalis H proteins and the L protein were found to be localized in hydrogenosomes (Fig. 2), suggesting a role for this organelle in amino acid metabolism. We found that T. vaginalis has two H proteins, encoded by independent genes, both of which are effective substrates of the L protein (Table 1). The H proteins in plants (2), mammals (6), Escherichia coli (19), and yeast (33) are encoded by a single gene. In some plants, tissue-specific alternative splicing results in two H proteins with or without an N-terminal extension of two amino acids (23, 24). Of the organisms previously examined, only Alkaliphilus metalliredigenes (a member of the firmicutes), was found to possess two H proteins encoded by two different genes (accession numbers ZP_00800556 and ZP_00800478); however, no evidence for their biochemical activity was documented.
Biochemical analysis using the recombinant H1, H2, and L proteins demonstrates that they interact in vitro (Fig. 5). The experimental Kms for the lipoylated H proteins were 2.6 μM and 3.7 μM, which suggest an affinity of the proteins for the dihydrolipoamide dehydrogenase (L protein). These values were much lower than that found for the free lipoamide (Table 1). H and L proteins were also found to exist as monomers and homodimers, respectively, in the hydrogenosomes, similar to their counterparts in other organisms (Fig. 3). However, sucrose density gradient analysis (Fig. 6) and BN-PAGE of the organellar fractions of T. vaginalis suggest that H and L do not form a stable complex in the hydrogenosomes and hence do not support a strong physical interaction between the two. The interactions between H and L proteins in pea leaf mitochondria are well studied, and the experimental Km for this H protein is 27 μM (31), significantly higher than that for rTvH proteins. In green leaves, GDC can be present in concentrations of up to 200 mg/ml (8). Although the ratio of the protein subunits in the pea has been roughly estimated as 4P:27H:9T:2L (9), it is not well understood how they interact. They may be able to assemble spontaneously within the mitochondrial matrix based on their behavior in vitro at protein concentrations above 0.25 mg/ml, with the H protein predicted to be the central core or the “structural and mechanistic heart” of the complex (31). Recent crystallographic data and the analysis of their interaction by nuclear magnetic resonance studies provide a clearer view, indicating structure-function relationships of and between the individual GDC subunits. Several lines of evidence strongly suggest that, except for the catalytic interaction with the lipoyl arm, there is no apparent molecular recognition and interaction between L protein and the reduced H protein. It is assumed that the main role of H protein could be to maintain the hydrophobic lipoate in a state that is fully accessible to the catalytic site of L protein (15).
In addition to GDC, SHMT plays a vital role in the interconversion of serine and glycine. It catalyzes the reversible transfer of a methylene group from serine to tetrahydrofolate with the formation of glycine and 5,10-methylenetetrahydrofolate. As this gene would be expected to be present to support a functional GDC, we searched the ∼7× coverage of Trichomonas vaginalis genome and found a single open reading frame homologous to SHMT (TVAG_109540). Moreover, the predicted protein contains a putative hydrogenosomal presequence consistent with the presence of SHMT in the hydrogenosome. These data further support a probable role for this unusual organelle in amino acid metabolism.
Earlier reports reveal that the L protein is not only a component of GDC but also a component of the E3 subunit of the α-ketoacid dehydrogenase complexes, comprised of pyruvate dehydrogenase, α-ketoglutarate dehydrogenase, and the branched chain α-ketoacid dehydrogenase. Our results indicate that T. vaginalis possessess a single L protein encoded by an independent gene. Furthermore, the in vitro interaction of the recombinant T. vaginalis glycine cleavage H and L proteins indicated the possibility of the presence of the GDC multienzyme complex in the hydrogenosome. To date, members of α-ketoacid dehydrogenase complexes have not been identified in T. vaginalis; however, the possibility of the L protein playing an important role as a component of the E3 subunit of the α-ketoacid dehydrogenase complexes in the hydrogenosomes cannot be excluded. In the pea (P. sativum), the mitochondrial L protein was found to be encoded by a single gene and shared between α-ketoacid dehydrogenase complexes and GDC (10). In Arabidopsis thaliana, two genes encoding mitochondrial dihydrolipoamide dehydrogenase (mtLPD1 and mtLPD2) have been reported. mtLPD seems to provide L protein for GDC, whereas the mtLPD2 gene product mainly interacts with α-ketoacid dehydrogenases (26, 27). However, from the high sequence identity between mtLPD1 and mtLPD2 (92%), the authors concluded that both L proteins can work in either multienzyme complex. Plasmodium falciparum has two dihydrolipoamide dehydrogenase isoforms encoded by two different genes. It has been demonstrated that both genes are functional, with one of them being part of a pyruvate dehydrogenase complex that is present exclusively in the apicoplast of Plasmodium and the other probably involved in both the α-ketoacid dehydrogenase complex and the GDC in the mitochondria (29, 37).
Phylogenetic analyses of the H and L proteins from T. vaginalis were performed to address the origin of the hydrogenosomes. While we could not resolve the evolutionary origin of the T. vaginalis H proteins, phylogenetic analyses clearly illustrated common ancestry of the T. vaginalis L protein with L proteins of the firmicute bacteria, strongly suggestive of its acquisition by T. vaginalis via a gene transfer event (1) independent of the origins of the mitochondria. Salcedo et al. (37) previously performed a distance-based phylogenetic analysis of L proteins to determine the evolutionary origins of several Plasmodium falciparum homologues, with origins of the Plasmodium proteins from within a plastidic-cyanobacterial clade. Our sampling of sequences for phylogenetic analysis was based on determining the 200 sequences most similar to the T. vaginalis protein followed by removal of all but one sequence within clades of very similar sequences, resulting in a phylogenetic analysis of ∼100 sequences. The cyanobacterial, Plasmodium, and plastidic sequences were considerably more distant from the T. vaginalis protein, so they were not included in our analysis. Overall, our analysis and that of Salcedo et al. (37) illustrate independent origins of the L proteins of mitochondria, plastids, and hydrogenosomes.
Upon obtaining phylogenetic analyses indicating that the L protein was derived by horizontal gene transfer from a firmicute, we specifically used the P and T proteins from the firmicute Bacillus subtilis, and still no genes with significant homology were uncovered in the T. vaginalis genome. Likewise, a broader search using a hidden Markov model (16) did not reveal likely P or T proteins in the genome. Given the conserved nature of these proteins in different organisms and the paucity of introns in T. vaginalis, it is highly unlikely that their genes were not detected due to a high level of fragmentation by introns (42). Whether T. vaginalis contains highly divergent P and T proteins or lacks these proteins entirely or the genes are simply missing from the genome database remains to be determined.
Supplementary Material
Acknowledgments
We thank our colleagues in the lab for helpful discussions.
This work was supported by National Institutes of Health (NIH) grants to P.J.J., a Burroughs-Wellcome Molecular Parasitology Award to P.J.J., and an NIH Microbial Pathogenesis Training Grant (2-T32-AI-07323) to M.T.B. A.G.M. was supported by the Marine Biological Laboratory's Program in Global Infectious Disease, funded by the Ellison Medical Foundation. Computational resources were provided by the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution (Marine Biological Laboratory) through funds provided by the W. M. Keck Foundation and the G. Unger Vetlesen Foundation.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Andersson, J. O., R. P. Hirt, P. G. Foster, and A. J. Roger. 2006. Evolution of four gene families with patchy phylogenetic distributions: influx of genes into protist genomes. BMC. Evol. Biol. 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauwe, H., and U. Kolukisaoglu. 2003. Genetic manipulation of glycine decarboxylation. J. Exp. Bot. 54:1523-1535. [DOI] [PubMed] [Google Scholar]
- 3.Boxma, B., R. M. de Graaf, G. W. van der Staay, T. A. van Alen, G. Ricard, T. Gabaldon, A. H. van Hoek, S. Y. Moon-van der Staay, W. J. Koopman, J. J. van Hellemond, A. G. Tielens, T. Friedrich, M. Veenhuis, M. A. Huynen, and J. H. Hackstein. 2005. An anaerobic mitochondrion that produces hydrogen. Nature 434:74-79. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, P. J., C. J. Lahti, E. Plumper, and P. J. Johnson. 1997. Targeting and translocation of proteins into the hydrogenosome of the protist Trichomonas: similarities with mitochondrial protein import. EMBO J. 16:3484-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui, E. T., P. J. Bradley, and P. J. Johnson. 1996. A common evolutionary origin for mitochondria and hydrogenosomes. Proc. Natl. Acad. Sci. USA 93:9651-9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choy, F., L. Sharp, and D. A. Applegarth. 2000. Glycine cleavage enzyme complex: rabbit H-protein cDNA sequence analysis and comparison to human, cow, and chicken. Biochem. Cell Biol. 78:725-730. [PubMed] [Google Scholar]
- 7.Delgadillo, M. G., D. R. Liston, K. Niazi, and P. J. Johnson. 1997. Transient and selectable transformation of the parasitic protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 94:4716-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douce, R., J. Bourguignon, D. Macherel, and M. Neuburger. 1994. The glycine decarboxylase system in higher plant mitochondria: structure, function and biogenesis. Biochem. Soc. Trans. 22:184-188. [DOI] [PubMed] [Google Scholar]
- 9.Douce, R., J. Bourguignon, M. Neuburger, and F. Rebeille. 2001. The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6:167-176. [DOI] [PubMed] [Google Scholar]
- 10.Drea, S. C., R. M. Mould, J. M. Hibberd, J. C. Gray, and T. A. Kavanagh. 2001. Tissue-specific and developmental-specific expression of an Arabidopsis thaliana gene encoding the lipoamide dehydrogenase component of the plastid pyruvate dehydrogenase complex. Plant Mol. Biol. 46:705-715. [DOI] [PubMed] [Google Scholar]
- 11.Dyall, S. D., M. T. Brown, and P. J. Johnson. 2004. Ancient invasions: from endosymbionts to organelles. Science 304:253-257. [DOI] [PubMed] [Google Scholar]
- 12.Dyall, S. D., C. M. Koehler, M. G. Delgadillo-Correa, P. J. Bradley, E. Plumper, D. Leuenberger, C. W. Turck, and P. J. Johnson. 2000. Presence of a member of the mitochondrial carrier family in hydrogenosomes: conservation of membrane-targeting pathways between hydrogenosomes and mitochondria. Mol. Cell. Biol. 20:2488-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Embley, T. M., and W. Martin. 2006. Eukaryotic evolution, changes and challenges. Nature 440:623-630. [DOI] [PubMed] [Google Scholar]
- 15.Faure, M., J. Bourguignon, M. Neuburger, D. MacHerel, L. Sieker, R. Ober, R. Kahn, C. Cohen-Addad, and R. Douce. 2000. Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the glycine decarboxylase multienzyme system 2. Crystal structures of H- and L-proteins. Eur. J. Biochem. 267:2890-2898. [DOI] [PubMed] [Google Scholar]
- 16.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray, M. W. 2005. Evolutionary biology: the hydrogenosome's murky past. Nature 434:29-31. [DOI] [PubMed] [Google Scholar]
- 18.Gray, M. W., G. Burger, and B. F. Lang. 1999. Mitochondrial evolution. Science 283:1476-1481. [DOI] [PubMed] [Google Scholar]
- 19.Heil, G., L. T. Stauffer, and G. V. Stauffer. 2002. Glycine binds the transcriptional accessory protein GcvR to disrupt a GcvA/GcvR interaction and allow GcvA-mediated activation of the Escherichia coli gcvTHP operon. Microbiology 148:2203-2214. [DOI] [PubMed] [Google Scholar]
- 20.Huelsenbeck, J. P., B. Larget, R. E. Miller, and F. Ronquist. 2002. Potential applications and pitfalls of Bayesian inference of phylogeny. Syst. Biol. 51:673-688. [DOI] [PubMed] [Google Scholar]
- 21.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 23.Kopriva, S., C. C. Chu, and H. Bauwe. 1996. H-protein of the glycine cleavage system in Flaveria: alternative splicing of the pre-mRNA occurs exclusively in advanced C4 species of the genus. Plant J. 10:369-373. [DOI] [PubMed] [Google Scholar]
- 24.Kopriva, S., R. Cossu, and H. Bauwe. 1995. Alternative splicing results in two different transcripts for H-protein of the glycine cleavage system in the C4 species Flaveria trinervia Plant J. 8:435-441. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, P. O., and D. L. Swofford. 2001. Back to the future: Bayesian inference arrives in phylogenetics. Trends Ecol. Evol. 16:600-601. [Google Scholar]
- 26.Lutziger, I., and D. J. Oliver. 2000. Molecular evidence of a unique lipoamide dehydrogenase in plastids: analysis of plastidic lipoamide dehydrogenase from Arabidopsis thaliana. FEBS Lett. 484:12-16. [DOI] [PubMed] [Google Scholar]
- 27.Lutziger, I., and D. J. Oliver. 2001. Characterization of two cDNAs encoding mitochondrial lipoamide dehydrogenase from Arabidopsis. Plant Physiol. 127:615-623. [PMC free article] [PubMed] [Google Scholar]
- 28.Macherel, D., J. Bourguignon, E. Forest, M. Faure, C. Cohen-Addad, and R. Douce. 1996. Expression, lipoylation and structure determination of recombinant pea H-protein in Escherichia coli. Eur. J. Biochem. 236:27-33. [DOI] [PubMed] [Google Scholar]
- 29.McMillan, P. J., L. M. Stimmler, B. J. Foth, G. I. McFadden, and S. Muller. 2005. The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases. Mol. Microbiol. 55:27-38. [DOI] [PubMed] [Google Scholar]
- 30.Muller, M. 1993. The hydrogenosome. J. Gen. Microbiol. 139:2879-2889. [DOI] [PubMed] [Google Scholar]
- 31.Neuburger, M., A. M. Polidori, E. Pietre, M. Faure, A. Jourdain, J. Bourguignon, B. Pucci, and R. Douce. 2000. Interaction between the lipoamide-containing H-protein and the lipoamide dehydrogenase (L-protein) of the glycine decarboxylase multienzyme system. 1. Biochemical studies. Eur. J. Biochem. 267:2882-2889. [DOI] [PubMed] [Google Scholar]
- 32.Petrin, D., K. Delgaty, R. Bhatt, and G. Garber. 1998. Clinical and microbiological aspects of Trichomonas vaginalis. Clin. Microbiol. Rev. 11:300-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piper, M. D., S. P. Hong, T. Eissing, P. Sealey, and I. W. Dawes. 2002. Regulation of the yeast glycine cleavage genes is responsive to the availability of multiple nutrients. FEMS Yeast Res. 2:59-71. [DOI] [PubMed] [Google Scholar]
- 34.Pütz, S., P. Dolezal, G. Gelius-Dietrich, L. Bohacova, J. Tachezy, and K. Henze. 2006. Fe-hydrogenase maturases in the hydrogenosomes of Trichomonas vaginalis. Eukaryot. Cell 5:579-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quon, D. V., M. G. Delgadillo, A. Khachi, S. T. Smale, and P. J. Johnson. 1994. Similarity between a ubiquitous promoter element in an ancient eukaryote and mammalian initiator elements. Proc. Natl. Acad. Sci. USA 91:4579-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roger, A. J., C. G. Clark, and W. F. Doolittle. 1996. A possible mitochondrial gene in the early-branching amitochondriate protist Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 93:14618-14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salcedo, E., P. F. Sims, and J. E. Hyde. 2005. A glycine-cleavage complex as part of the folate one-carbon metabolism of Plasmodium falciparum. Trends Parasitol. 21:406-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoemaker, J. S., I. S. Painter, and B. S. Weir. 1999. Bayesian statistics in genetics: a guide for the uninitiated. Trends Genet. 15:354-358. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, A. G., and A. J. Roger. 2004. The real ‘kingdoms’ of eukaryotes. Curr. Biol. 14:R693-R696. [DOI] [PubMed] [Google Scholar]
- 40.Sutak, R., P. Dolezal, H. L. Fiumera, I. Hrdy, A. Dancis, M. Delgadillo-Correa, P. J. Johnson, M. Muller, and J. Tachezy. 2004. Mitochondrial-type assembly of FeS centers in the hydrogenosomes of the amitochondriate eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 101:10368-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tachezy, J., L. B. Sanchez, and M. Muller. 2001. Mitochondrial type iron-sulfur cluster assembly in the amitochondriate eukaryotes Trichomonas vaginalis and Giardia intestinalis, as indicated by the phylogeny of IscS. Mol. Biol. Evol. 18:1919-1928. [DOI] [PubMed] [Google Scholar]
- 42.Vanacova, S., W. Yan, J. M. Carlton, and P. J. Johnson. 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. USA 102:4430-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Giezen, M., D. J. Slotboom, D. S. Horner, P. L. Dyal, M. Harding, G. P. Xue, T. M. Embley, and E. R. Kunji. 2002. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 21:572-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson, R. L., L. T. Stauffer, and G. V. Stauffer. 1993. Roles of the GcvA and PurR proteins in negative regulation of the Escherichia coli glycine cleavage enzyme system. J. Bacteriol. 175:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.