Abstract
The facultative intracellular bacterium Francisella tularensis causes the zoonotic disease tularemia. F. tularensis resides within host macrophages in vivo, and this ability is essential for pathogenesis. The transcription factor MglA is required for the expression of several Francisella genes that are necessary for replication in macrophages and for virulence in mice. We hypothesized that the identification of MglA-regulated genes in the Francisella genome by transcriptional profiling of wild-type and mglA mutant bacteria would lead to the discovery of new virulence factors utilized by F. tularensis. A total of 102 MglA-regulated genes were identified, the majority of which were positively regulated, including all of the Francisella pathogenicity island (FPI) genes. We mutated novel MglA-regulated genes and tested the mutants for their ability to replicate and induce cytotoxicity in macrophages and to grow in mice. Mutations in MglA-regulated genes within the FPI (pdpB and cds2) as well as outside the FPI (FTT0989, oppB, and FTT1209c) were either attenuated or hypervirulent in macrophages compared to the wild-type strain. All of these mutants exhibited decreased fitness in vivo in competition experiments with wild-type bacteria. We have identified five new Francisella virulence genes, and our results suggest that characterizations of additional MglA-regulated genes will yield further insights into the pathogenesis of this bacterium.
The facultative intracellular bacterium Francisella tularensis is the etiological agent of tularemia, a zoonotic disease of humans, rabbits, rodents, and hares (16). The clinical manifestations of tularemia vary depending on the virulence of the strain and the route of inoculation. Inhalation of F. tularensis subspecies tularensis, the most virulent subspecies, results in the pneumonic form of tularemia, a fulminating disease in humans (12). F. tularensis can be acquired through the skin, from a tick bite, or from contact with an infected animal, which results in the ulceroglandular form of the disease. Francisella can also be contracted through the conjuctiva, causing the oculoglandular form of tularemia. Less commonly, ingestion of contaminated foods or water may result in clinical symptoms. Once the bacterium enters the body, it travels to the draining lymph nodes and then spreads to the liver, lungs, and spleen of infected humans or animals, where it replicates to high numbers. Francisella novicida rarely causes disease in humans but causes a systemic tularemia-like disease in mice and therefore offers a good model with which to study Francisella pathogenesis (24, 35, 37).
Francisella is found primarily within macrophages in the host (2). Therefore, Francisella-macrophage interactions have been studied in vitro as a model for in vivo interactions. Francisella readily enters unactivated macrophages and is initially contained within a vacuole. The Francisella-containing vacuole has been reported to avoid phagosome-lysosome fusion before the bacteria escape the phagocytic vacuole and replicate to high numbers in the cytosol (10, 17). It has also been reported that Francisella induces apoptosis (26) and abrogates Toll-like receptor signaling (44). However, when Francisella enters activated macrophages, cytosolic Francisella induces a host defense response, and the macrophage undergoes rapid caspase-1-dependent cell death (33). Several mutants that do not induce macrophage death in activated macrophages have been identified (33). Unlike wild-type bacteria, these mutants reside within a vacuole at 6 h postinfection and are attenuated for growth in macrophages and in mice (6, 35). Although it is known that replication in macrophages is important in the pathogenesis of tularemia, very little is known about the genetic and molecular mechanisms that govern Francisella interactions with macrophages.
Baron and Nano previously isolated a spontaneous Francisella mutant with altered colony morphology on plates containing the chromogenic phosphatase substrate XP, which indicated that this strain may have been a spontaneous phosphatase mutant or altered in the expression of an exported phosphatase (6). This mutant was unable to replicate in macrophages, and the mutated locus was identified by a complementation strategy and named mglAB, for macrophage growth locus (6). MglA and MglB are homologs of SspA and SspB from Escherichia coli, which are global regulators of the stringent starvation response. SspA is a transcription factor involved in the regulation of genes contributing to the stringent starvation response, acid resistance, and phage P1 late genes (19, 20, 49, 50). SspB is an accessory factor that determines substrate specificity for the ClpXP protease (31).
Additional studies have shown that the mglA transcript is induced inside macrophages (5), and MglA has subsequently been found to regulate the expression of several genes necessary for replication in macrophages and for virulence, including the intracellular growth locus genes iglA, iglC, and iglD and the pathogenicity determinant protein genes pdpD and pdpA (29). These genes lie within a ∼30-kb stretch of the genome that has characteristics of a pathogenicity island (13), including a lower G+C content in the DNA of this region as well as the presence of virulence genes (35). This region has been designated the Francisella pathogenicity island (FPI).
The sequencing of the Francisella genome (27, 35) allowed us to construct a Francisella microarray to examine global transcriptional responses. Since MglA was identified as a regulator of virulence genes, we began by investigating the transcriptional differences between wild-type Francisella and an mglA mutant during growth in rich media, conditions under which MglA-regulated genes are expressed (5). We identified 102 MglA-regulated genes, the majority of which are positively regulated, including the entire FPI. We mutated eight MglA-regulated genes both within and outside the FPI and show that five of these genes encode novel virulence factors utilized by Francisella during in vitro infections of macrophages and during in vivo infection in mice.
MATERIALS AND METHODS
Bacterial strains and growth.
Wild-type F. novicida strain U112 and an isogenic mglA point mutant (GB2) described previously (6, 35) were obtained from Karen Elkins. Bacteria were grown with aeration overnight in tryptic soy broth (Difco Laboratories, Detroit, MI) supplemented with 0.2% cysteine. Bacteria were plated onto tryptic soy agar (Becton Dickinson, Sparks, MD) supplemented with 0.1% cysteine (Sigma, St. Louis, MO) or modified Mueller-Hinton (MH) agar (Difco) supplemented with 0.025% ferric pyrophosphate (Sigma), 0.02% IsoVitaleX (Becton Dickinson), 0.1% glucose, and 0.025% calf serum (GIBCO, Carlsbad, CA). Kanamycin (15 μg/ml), erythromycin (50 μg/ml), and streptomycin (400 μg/ml) (all from Sigma) were added to the growth media when appropriate. E. coli strain DH12S was used for cloning and was grown in Luria broth (Difco Laboratories, Detroit, Mich.).
Microarray design.
Open reading frames (ORFs) for the Francisella tularensis LVS (live vaccine strain) and SchuS4 genome sequences were predicted by using Glimmer2 (11, 40). Corresponding U112 genes are referred to by the SchuS4 locus tag. Oligonucleotides of 70 bp were selected for each ORF using ArrayOligoSelector (9). Oligonucleotides were designed to represent genes present in (i) SchuS4 and LVS, (ii) LVS and not SchuS4, or (iii) SchuS4 and not LVS based on a cutoff of 70% identity over 80% of the sequence length. Whenever possible, two 70-mer oligonucleotides were selected for each ORF (88%); however, this was not possible in some cases due to predicted cross-hybridization. The specificity of the designed oligonucleotides was verified in silico by an examination of BLAST similarity and the free energies of the oligonucleotides against their second-best matches in the genome. Oligonucleotides were purchased from QIAGEN (Valencia, CA) and resuspended in 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Glass slide preparation, printing, and processing after printing were performed as described previously (14). The final array contains 4,219 independent spots representing 2,242 ORFs.
Time courses and RNA isolation.
Liquid cultures of U112 and mglA mutant strain GB2 were grown overnight at 37°C with shaking and subcultured to an optical density at 600 nm (OD600) of 0.006 in 300 ml. Samples were taken for serial dilutions to verify the OD600 and to determine the CFU for time zero, and the cultures were then placed into a shaking 37°C water bath to minimize changes in temperature. Samples were taken throughout the growth curve for RNA isolation and to determine the OD600 and CFU counts. Ten samples were taken during the first growth curve at 0.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, and 7.5 h. Seven samples were taken during the second growth curve at 1, 3, 5, 6, 7, 8, and 10 h. RNA was harvested as previously described (45). Briefly, bacteria were collected onto 0.45-μm nitrocellulose filters (Millipore, Bedford, Mass.) by vacuum filtration, placed into a 50-ml conical tube, and then immediately frozen in liquid N2 and stored at −80°C. RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) as previously described (45). The aqueous phase was used in the RNeasy Clean-Up protocol (QIAGEN, Chatsford, CA) with an on-column DNase digestion step for 40 min (QIAGEN, Chatsford, CA). The concentration and purity of RNA were assessed by measuring the OD260 and the OD280, and the integrity of the RNA was verified on a 1% agarose gel.
Preparation of cDNA probes, hybridization, and analysis.
For each of the time courses, equal quantities of RNA from each of the U112 and GB2 samples were combined to create a common reference for the experiment. For each time point or reference sample, cDNA was synthesized from 1 μg of total RNA in a standard reverse transcription (RT) reaction using 5 μg of random hexamers (Amersham Biosciences, Piscataway, NJ) and Superscript III (Invitrogen). Amino allyl dUTPs were incorporated at this step (2.5 mM dATP, 2.5 mM dCTP, 2.5 mM dGTP, 1 mM amino allyl dUTP, and 1.5 mM dTTP) (Invitrogen). cDNA was purified using Zymo DNA purification columns as specified by the manufacturer (Zymo Research Corp., Orange, CA), and samples from each time point were labeled with Cy5 (red) fluorophores and the reference was labeled with Cy3 (green) fluorophores as previously described (Amersham Biosciences) (45). Unincorporated fluorophores were quenched using 5 μl of 4 M hydroxylamine and incubated in the dark for 15 min. The samples from each time point and reference samples were then combined, and unincorporated dyes were removed using Zymo DNA purification columns. For hybridization, cDNA was eluted in 19 μl of Tris-EDTA buffer, and 2 μl of 20 mg/ml yeast tRNA (Invitrogen), 4.25 μl of 20× SSC, and 0.75 μl of 10% sodium dodecyl sulfate were added to the probe. Probes were denatured for 2 min at 99°C, spun at 17,900 × g, and cooled at room temperature before they were added to the arrays. The samples and arrays were incubated at 60°C for ∼14 h, and stringency washes were performed as described previously (14). Briefly, the arrays were washed for 2 min each in a series of four washes with increasing stringency: (i) 2× SSC-0.03% sodium dodecyl sulfate, (ii) 2× SSC, (iii) 1× SSC, and (iv) 0.2× SSC. The microarrays were scanned and analyzed using a Gene Pix 4000A scanner and GENEPIX5.1 software (both from Axon Instruments, Redwood City, CA).
Normalized data were collected using the Stanford Microarray Database (43). Spots were excluded from analysis due to obvious abnormalities, a regression correlation of <0.6, or a Cy3 net mean intensity of <100. Only spots with at least 70% good data across the experiment were included for analysis. The ratios of the red (time point sample) channels to green (reference) channels for each spot were expressed as log2(red/green) and used for hierarchical clustering using the CLUSTER program (15). Results were visualized using the TREEVIEW program (15). Using data from all of the microarrays and both growth curves, significant differences between the wild-type and mutant growth curves were determined using the Significance Analysis for Microarrays (SAM) program, v. 1.21 (46). Using the two-class statistical analysis tool in the SAM program, a list of genes whose transcript levels were significantly increased or decreased between the two groups was calculated. A calculated false discovery rate of <1% was used to assign significance, and a twofold cutoff in the change in expression level was imposed. The genes identified by SAM analysis are included in Table 1.
TABLE 1.
List of MglA-regulated genesaa
| Locus tag | Gene | Gene product | KEGG classification | Scoreb | Fold changec |
|---|---|---|---|---|---|
| 10 genes significantly upregulated in mglA mutant | |||||
| FTT0551 | Conserved hypothetical protein, pseudogene | 1.95 | 2.11 | ||
| FTT0552 | Aldehyde dehydrogenase | Metabolism | 1.82 | 2.01 | |
| FTT0553 | Hypothetical protein | 1.97 | 2.20 | ||
| FTT0747c | Hypothetical protein | 1.70 | 2.00 | ||
| FTT0844 | Rossman fold oxidoreductase, pseudogene | 1.59 | 2.22 | ||
| FTT1195c | Conserved hypothetical protein, pseudogene | 2.64 | 2.09 | ||
| FTT1288 | Conserved hypothetical membrane protein | 1.41 | 2.15 | ||
| FTT1529 | fadE | Acyl-coenzyme A dehydrogenase | Metabolism | 1.20 | 2.37 |
| FTT1532 | Hypothetical protein | 0.98 | 2.07 | ||
| FTT1741c | NAD-dependent formate dehydrogenase, fragment | 1.67 | 2.01 | ||
| 92 genes significantly downregulated in mglA mutant | |||||
| FTT0005 | gabD1 | Succinate-semialdehyde dehydrogenase, fragment | Metabolism | −2.65 | 0.49 |
| FTT0070c | ampG | Major facilitator superfamily transport protein | −3.53 | 0.42 | |
| FTT0101 | Conserved membrane hypothetical protein | −1.61 | 0.37 | ||
| FTT0122 | oppA | Oligopeptide transporter, subunit A, ABC transporter periplasmic protein | −3.05 | 0.45 | |
| FTT0123 | oppB | Oligopeptide transporter, subunit B, ABC transporter, membrane protein, pseudogene | −2.80 | 0.44 | |
| FTT0125 | oppD | Oligopeptide transporter, subunit D, ABC transporter, ATP-binding protein | Environmental information processing | −2.88 | 0.49 |
| FTT0194c | Conserved hypothetical membrane protein | −2.42 | 0.50 | ||
| FTT0218c | Cytochrome b561 family protein, pseudogene | −2.68 | 0.48 | ||
| FTT0242 | Hypothetical protein | −2.25 | 0.50 | ||
| FTT0251 | ilvE | Branched-chain amino acid aminotransferase | Metabolism | −2.68 | 0.47 |
| FTT0252 | leuA | 2-Isopropylmalate synthase | Metabolism | −2.98 | 0.46 |
| FTT0254c | Hypothetical protein | −5.11 | 0.14 | ||
| FTT0267 | Hypothetical protein | −8.35 | 0.18 | ||
| FTT0296 | pcp | Pyrrolidone-carboxylate peptidase | Genetic information and processing | −2.35 | 0.40 |
| FTT0297 | Hypothetical membrane protein | −2.53 | 0.47 | ||
| FTT0298 | holC | DNA polymerase III (CHI subunit) protein | Metabolism, genetic information and processing | −3.08 | 0.40 |
| FTT0310 | Amino acid permease | −3.44 | 0.37 | ||
| FTT0311c | Hypothetical protein | −6.73 | 0.08 | ||
| FTT0383 | Hypothetical protein | −6.05 | 0.27 | ||
| FTT0403 | def1 | Peptide deformylase | −4.79 | 0.28 | |
| FTT0421 | Outer membrane lipoprotein, pseudogene | −5.40 | 0.29 | ||
| FTT0511 | Pyridoxine/pyridoxal 5-phosphate biosynthesis protein | Metabolism | −4.96 | 0.28 | |
| FTT0512 | Glutamine amidotransferase family protein | −4.10 | 0.26 | ||
| FTT0611c | Beta-lactamase | Environmental information processing and metabolism | −6.19 | 0.19 | |
| FTT0612 | ospD2 | Hypothetical protein | −4.56 | 0.39 | |
| FTT0613c | Hypothetical protein | −3.80 | 0.19 | ||
| FTT0651 | Proton-dependent oligopeptide transport family protein | Environmental information processing | −4.42 | 0.34 | |
| FTT0723c | Short-chain dehydrogenase/reductase family | −3.19 | 0.49 | ||
| FTT0724c | dacB1 | d-Alanyl-d-alanine carboxypeptidase (penicillin binding protein) family protein, pseudogene | −4.64 | 0.26 | |
| FTT0726c | Glycerophosphoryl diester phosphodiesterase family protein | Metabolism | −3.74 | 0.36 | |
| FTT0852 | Conserved hypothetical protein, pseudogene | −4.55 | 0.21 | ||
| FTT0878c | msrB | Peptide methionine sulfoxide reductase | Genetic information and processing | −2.18 | 0.50 |
| FTT0883 | Alcohol dehydrogenase class III, pseudogene | Metabolism | −3.44 | 0.44 | |
| FTT0905 | Type IV pilus glycosylation protein | −2.80 | 0.38 | ||
| FTT0960 | Hypothetical protein | −3.58 | 0.45 | ||
| FTT0961 | mdaB | Modulator of drug activity B | −3.07 | 0.47 | |
| FTT0974 | Amidinotransferase family protein, pseudogene | −2.93 | 0.46 | ||
| FTT0975 | Hypothetical protein | −3.48 | 0.40 | ||
| FTT0980 | Aminotransferase, class II | Metabolism | −6.01 | 0.17 | |
| FTT0981 | Hypothetical protein | −6.34 | 0.14 | ||
| FTT0987 | Hypothetical protein | −2.79 | 0.35 | ||
| FTT0988 | Hypothetical protein | −2.90 | 0.29 | ||
| FTT0989 | Hypothetical protein | −4.97 | 0.18 | ||
| FTT0998 | Hypothetical lipoprotein | −1.34 | 0.37 | ||
| FTT1032 | Conserved hypothetical membrane protein, pseudogene | −5.37 | 0.21 | ||
| FTT1033 | yihQ | Glycosyl hydrolase family 31 protein, pseudogene | Metabolism | −6.82 | 0.16 |
| FTT1034c | ndh | NADH dehydrogenase | Metabolism | −1.75 | 0.42 |
| FTT1089 | Isochorismatase hydrolase family protein | −2.52 | 0.26 | ||
| FTT1090 | Conserved hypothetical membrane protein | −3.80 | 0.21 | ||
| FTT1091 | Isochorismatase hydrolase family protein | −2.71 | 0.39 | ||
| FTT1122c | Hypothetical lipoprotein | −2.86 | 0.23 | ||
| FTT1127 | Rhodanese-like family protein | −2.92 | 0.41 | ||
| FTT1141 | Conserved hypothetical protein, pseudogene | −2.29 | 0.42 | ||
| FTT1146c | galM | Aldose 1-epimerase (pseudogene) | Metabolism | −2.71 | 0.46 |
| FTT1147c | dfp | 4′-Phosphopantothenoylcysteine decarboxylase, phosphopantothenoylcysteine synthetase, flavin mononucleotide binding (bifunctional protein) | Metabolism | −2.76 | 0.43 |
| FTT1180 | Conserved hypothetical protein, pseudogene | −4.39 | 0.26 | ||
| FTT1191 | Aminoacylase | Metabolism | −2.67 | 0.46 | |
| FTT1209c | Metallopeptidase family M13 protein, pseudogene | −6.16 | 0.08 | ||
| FTT1242 | Hypothetical protein | −1.75 | 0.25 | ||
| FTT1287 | cbs | Cystathionine beta-synthase (cysteine synthase) | Metabolism | −2.57 | 0.50 |
| FTT1289 | Hypothetical protein | −2.00 | 0.38 | ||
| FTT1344 | pdpA | Hypothetical protein | −4.21 | 0.20 | |
| FTT1345 | pdpB | Hypothetical protein | −4.87 | 0.17 | |
| FTT1346 | Hypothetical protein | −5.15 | 0.19 | ||
| FTT1347 | Hypothetical protein | −3.37 | 0.29 | ||
| FTT1348 | Hypothetical protein | −3.52 | 0.34 | ||
| FTT1349 | Hypothetical protein | −6.54 | 0.17 | ||
| FTT1350 | Hypothetical protein | −5.63 | 0.19 | ||
| FTT1351 | Hypothetical protein | −4.83 | 0.15 | ||
| FTT1352 | Hypothetical protein | −4.94 | 0.16 | ||
| FTT1353 | cds2 | Hypothetical protein | −5.08 | 0.18 | |
| FTT1354 | pdpC | Hypothetical protein | −3.07 | 0.36 | |
| FTT1355 | cds1 | Hypothetical protein | −2.27 | 0.35 | |
| FTT1356c | iglD | Intracellular growth locus, subunit D | −5.54 | 0.28 | |
| FTT1357c | iglC | Intracellular growth locus, subunit C | −5.93 | 0.11 | |
| FTT1358c | iglB | Intracellular growth locus, subunit B | −7.62 | 0.06 | |
| FTT1359c | iglA | Intracellular growth locus, subunit A | −6.28 | 0.09 | |
| FTT1360c | pdpD | Hypothetical protein | −4.44 | 0.19 | |
| FTT1379c | Conserved hypothetical protein, pseudogene | −3.24 | 0.45 | ||
| FTT1388 | Hypothetical protein | −2.26 | 0.42 | ||
| FTT1391 | panD | Aspartate-1-decarboxylase | Metabolism | −2.79 | 0.41 |
| FTT1400c | Hypothetical protein | −3.57 | 0.45 | ||
| FTT1536c | Hypothetical protein | −3.62 | 0.44 | ||
| FTT1541c | Hypothetical protein | −3.80 | 0.29 | ||
| FTT1542c | omp26 | Outer membrane protein 26 | −5.28 | 0.14 | |
| FTT1558c | Conserved hypothetical protein, pseudogene | −4.84 | 0.32 | ||
| FTT1565c | Glycosyl hydrolase, family 3, pseudogene | Metabolism | −3.01 | 0.24 | |
| FTT1650c | Chorismate mutase | Metabolism | −5.36 | 0.11 | |
| FTT1682 | Conserved hypothetical protein, pseudogene | −3.49 | 0.23 | ||
| FTT1734c | fopA1 | Outer-membrane-associated protein, fragment pseudogene | −2.55 | 0.49 | |
| FTT1755 | gabD2 | Succinate-semialdehyde dehydrogenase, pseudogene | −2.69 | 0.49 | |
| FTT1771 | Hypothetical protein | −2.95 | 0.27 |
U112 genes are referred to by the SchuS4 locus tag. FPI genes are in boldface type.
Relative level of significance assigned by SAM.
Change (n-fold) between the average of all wild-type microarrays and all mglA mutant microarrays.
qRT-PCR.
Quantitative real-time RT-PCR (qRT-PCR) was performed with the rTth enzyme (Applied Biosystems, Foster City, CA) and gene-specific primers (Table 2) using a Bio-Rad iCycler according to the manufacturer's instructions. SYBR green was used to detect signals. To calculate the gene-specific message, a standard curve was plotted for each primer set using a dilution series of RNA from a sample known to contain the message of interest. To compare transcript abundance from the wild-type and that from an mglA mutant, values were normalized to DNA helicase (FTT0121) to obtain relative quantities of message.
TABLE 2.
Primers used in this study
| Primer and use | Sequencea | Primer and use | Sequencea | |
|---|---|---|---|---|
| Mutagenesis | FTT1771r1 | AtatatatTCTAGAaacccctaaagattgtttcg | ||
| KanF | atatatatGCGGCCGCtttatggacagcaagcgaac | FTT1771f2 | gttgtatggttatgcagttg | |
| KanR | atatatatGCGGCCGCtcaattcagaagaactcgtc | FTT1771r2 | gctctggagagtggaat | |
| pdpAf1 | atatatatGTCGACctttagctcctatccgctaa | FTT1032f1 | AtatatatGTCGACtgagatactttacgtgcagaag | |
| pdpAinv1 | atatatatGCGGCCGCgctttgagagtctttatagtt | FTT1032inv1 | AtatatatGCGGCCGCgctacttatacatacagtgat | |
| pdpAinv2 | atatatatGCGGCCGCcgagacattctcactaactc | FTT1032inv2 | AtatatatGCGGCCGCgtactctttgtgagatattta | |
| pdpAr1 | atatatatTCTAGAtttgtatccgcatcagaagt | FTT1032r1 | AtatatatTCTAGAatggatcttacaccatttag | |
| pdpAf2 | ctgactctaactactgattac | FTT1032f2 | agaccaattagtggtaagac | |
| pdpAr2 | catcatctttagagctaaga | FTT1032r2 | ctatcacggtattgctataa | |
| pdpBf1 | atatatatGTCGACttagagtcgctttcatgtag | |||
| pdpBinv1 | atatatatGCGGCCGCaaagataaagcctcaagctc | Complementation | ||
| pdpBinv2 | atatatatGCGGCCGCttttatggtctttgaggcag | CmF | ataATCGATatgatggagaaaaaaatcactggatatacc | |
| pdpBr2 | atatatatTCTAGAactcctgaatatcctaagtac | CmR | ataGCGGCCGCttacgccccgccctgccactc | |
| pdpBf2 | aggagagtatcagcatattg | omp26f | ataGCGGCCGCggttgtcactcatcgtatttgg | |
| pdpBr2 | aagtgattctgaatcagacg | omp26r | ataCATCGAtccccttaagattgtcg | |
| cds2f1 | atatatatGTCGACatcatccataaattctggag | cds2compf1 | atatatatGTCGACatcatccataaattctggag | |
| cds2inv1 | atatatatGCGGCCGCtctcccatttaagatagaac | cds2compinv1 | atatatatGCGGCCGCtctcccatttaagatagaac | |
| cds2inv2 | atatatatGCGGCCGCtaatatggtaggagatgagg | cds2compinv2 | atatatatGCGGCCGCtttatcagatagtgattcgg | |
| cds2r1 | atatatatTCTAGAaaacatacaaggctctgaga | cds2compr1 | atatatatTCTAGAaaacatacaaggctctgaga | |
| cds2f2 | tttagtatcagctcttaccc | oppBcompF | atatatatGTCGACtgtatacatgtgatagtgac | |
| cds2r2 | agttgaagcaagtatcacag | oppBcompinv1 | atatatatGCGGCCGCtttgccattatttgaccctc | |
| FTT0612f1 | AtatatatGTCGACagtaatagcctttggagttg | oppBcompinv2 | atatatatGCGGCCGCaaacgcagcaacaaatagag | |
| FTT0612inv1 | AtatatatGCGGCCGCctatctctctttatacgaca | oppBcompR | atatatatTCTAGAaagtgcaatagtttgccca | |
| FTT0612inv2 | AtatatatGCGGCCGCttaggtatgattgtcaatgc | FTT1209compf1 | AtatatatGTCGACtatttgcagctactatagg | |
| FTT0612r1 | AtatatatTCTAGAggtatgtcattagtttggct | FTT1209compinv1 | atatatatGCGGCCGCctgctaaataagtgtcattattg | |
| FTT0612f2 | aggcatactaaaagctatcc | FTT1209compinv2 | AtatatatGCGGCCGCacaaagagttaatatctggtg | |
| FTT0612r2 | cgacacttgagcaaacgat | FTT1209compr1 | AtatatatTCTAGActgtagtatcaagctcggaa | |
| FTT0989f1 | AtatatatGTCGACatcccagttaacaactgagt | omp26F_pvuII | atatatCAGCTGggttgtcactcatcgtatttgg | |
| FTT0989inv1 | AtatatatGCGGCCGCtgaagatagacaatcagaac | CmR_pvuII | atatatCAGCTGttacgccccgccctgccactc | |
| FTT0989inv2 | AtatatatGCGGCCGCctttcacagcataaattaactt | FTT0989compf1 | atatatACTAGTttaaaatctcacttacaaaaaatg | |
| FTT0989r1 | AtatatatTCTAGAatgcaggtatcatatgcttg | FTT0989compr1 | atatatACTAGTgcctagtattaaaaataacctt | |
| FTT0989f2 | tctaccatcaacaacttgttc | |||
| FTT0989r2 | caacttgctgaatagtatcat | qRT-PCR | ||
| oppBf1 | AtatatatGTCGACaagtgcaatagtttgtccac | hel F4 | gggatgtcgccttttgattttc | |
| oppBinv1 | AtatatatGCGGCCGCaaacgcagcaacaaatagag | hel R4 | ctcttttgtcccttgtgcttgc | |
| oppBinv2 | AtatatatGCGGCCGCaaccaaaaaccactatctcc | mglA R1 | gctccttttgctttgatagt | |
| oppBr1 | AtatatatTCTAGAtgtatacatgtgatagtgac | mglA F1 | agaaagtaatccagatgctc | |
| oppBf2 | accactaactttagccgctt | pepO F3 | ggatgggttcagagtctaag | |
| oppBr2 | tggcttggattgcagacttt | pepO R3 | ccaagtgtatcgagatcttg | |
| FTT1209cf1 | AtatatatGTCGACtatttgcagctactataggg | oppB F2 | gtagaaacgcttaactctcc | |
| FTT1209cinv1 | AtatatatGCGGCCGCcctcaacaaaagattcaaca | oppB R2 | atagtacccgtcagaatacc | |
| FTT1209cinv2 | AtatatatGCGGCCGCacaaagagttaatatctggtg | ospD2 F1 | acaccagcattgtgtatggc | |
| FTT1209cr1 | AtatatatTCTAGActgtagtatcaagctcggaa | ospD2 R1 | cttgtgaatgaccctgttgc | |
| FTT1209cf2 | gcttgtcttagttatggttg | pdpB F6 | tactggacttattagtgggagc | |
| FTT1209cr2 | caggaagcactgttggattt | pdpB R6 | cgcagatgatggtaaagaac | |
| FTT1771f1 | AtatatatGTCGACacaatattagcagctaaaggc | 1326 F7 | tctgccttgtatttgtgaagc | |
| FTT1771inv1 | AtatatatGCGGCCGCctaaatcctcttaaggagtt | 1326 R7 | ttgattggactgtggaggag | |
| FTT1771inv2 | AtatatatGCGGCCGCgttctgttagtttgttttgta |
Capital letters indicate restriction site.
Mutagenesis.
To create Francisella deletion mutants, the regions of the chromosome 5′ and 3′ to the gene of interest were amplified by PCR, introducing NotI and SalI or NotI and XbaI sites, respectively, and cloned into a Topo-TA vector (Invitrogen) (primers are listed in Table 2). The Topo plasmid was purified, and the insert was cut out with the appropriate restriction enzymes. We amplified the pFNLTP7 (32) kanamycin resistance cassette by PCR, introducing NotI restriction sites on both sides of the gene. The 5′ and 3′ homologous regions and the NotI-digested kanamycin cassette were then ligated into the pACYC184 backbone digested with XbaI and SalI. The resulting plasmid was then transformed into chemically competent strain U112 as previously described (3). Mutants were selected on modified Mueller-Hinton agar (Difco Laboratories) supplemented with kanamycin (15 μg/ml). Allelic replacement of the gene of interest was assessed by PCR to verify that U112 had undergone double homologous recombination, and these primers are listed in Table 2 as primers F2 and R2. Clones whose genomic DNA was amplified by PCR to the correct size were then sequenced and tested for their ability to grow in tryptic soy broth plus 0.2% cysteine by determining the OD600 and counting CFU.
To select for a spontaneous streptomycin-resistant strain of GB2 for competition experiments, 100 μl of a culture grown overnight was plated onto modified MH agar supplemented with streptomycin (400 μg/ml). Individual streptomycin-resistant clones were passaged on modified MH agar with streptomycin to ensure that the clones were streptomycin resistant and tested for in vitro growth defects in rich media.
Complementation of mutants.
The chloramphenicol cassette from pACYC184 (NEB) was amplified with primers CmF and CmR (Table 2), introducing ClaI and NotI restriction sites, respectively. The omp26 promoter was amplified from pLG62a (L. Gallagher and C. Manoil, unpublished data) with primers omp26f and omp26r (Table 2), introducing NotI and ClaI sites, respectively. The resulting PCR products were digested with ClaI and ligated to create a chloramphenicol resistance cassette driven by the omp26 promoter. This cassette was cloned into the NotI site of pMTM-T3 (34). Additionally, a chloramphenicol version of pDSK519 (23) was made by excising the kanamycin cassette using PvuII. The omp26 promoter-driven chloramphenicol cassette was amplified using omp26F_PvuII and cmR_PvuII, cut with PvuII, and ligated to the pDSK519 backbone. The resulting plasmid was called pDSC.
oppB, FTT1209c, and cds2 mutants were complemented in cis using a strategy similar to the mutagenesis strategy described above. In this strategy, the PCR of the 5′ region included a full-length version of the gene of interest and sufficient upstream and downstream regions to allow for homologous recombination. The omp26 promoter-driven chloramphenicol cassette described above was cut with NotI and, along with the 5′ and 3′ digested regions, was ligated to SalI- and XbaI-digested pMPM-T6 omega (34). The plasmid was then transformed into the corresponding mutant, and allelic replacement was confirmed by screening for chloramphenicol resistance and kanamycin sensitivity. The FTT0989 mutant was complemented in trans by amplifying the FTT0989 gene and promoter using FTT0989compF1 and FTT0989compR1 (Table 2), introducing SpeI sites. pDSC was cut using XbaI and ligated to SpeI-cut FTT0989. Colonies were screened for the insertion of FTT0989 by PCR. Plasmid DNA was isolated from colonies with insertions and transformed into a chemically competent FTT0989 mutant as described previously (3). Complemented mutants were selected on Mueller-Hinton agar (Difco Laboratories) supplemented with chloramphenicol (7.5 μg/ml). All complemented clones were then sequenced and tested for their ability to grow in tryptic soy broth plus 0.2% cysteine by measuring the OD600 and counting CFU.
Bone-marrow-derived macrophages.
Bone-marrow-derived macrophages were prepared as described elsewhere previously (41). Briefly, bone marrow was collected from the femurs of mice. Bone marrow cells were plated onto sterile petri dishes and incubated in Dulbecco's modified Eagle medium with high glucose, l-glutamine, 110 mg/liter sodium pyruvate, and pyroxidine hydrochloride (Invitrogen Life Technologies) and supplemented with 10% fetal calf serum (Invitrogen Life Technologies), 10 mM HEPES (Invitrogen Life Technologies), and 30% macrophage colony-stimulating factor (M-CSF)-conditioned medium. M-CSF-conditioned medium was collected from an L929 M-CSF cell line. Bone marrow cells were incubated at 37°C under 5% CO2, and macrophages were harvested after 6 days. All assays were performed using standard tissue culture plates at 37°C under 5% CO2 in similar media, excluding M-CSF-conditioned medium.
Macrophage replication assay.
Bone-marrow-derived macrophages were seeded in 24-well plates at a density of 2.5 × 105 macrophages per well and incubated at 37°C and 5% CO2 overnight. The macrophages were then infected at a multiplicity of infection (MOI) of 25:1 (bacteria-to-macrophage ratio) and centrifuged at 730 × g for 15 min to mediate attachment. The macrophages were incubated (time zero) for 1 h and washed three times with warm media. Macrophages were lysed at 2 and 10 h postinfection with 1% saponin in water for 5 min and then diluted in phosphate-buffered saline and plated onto tryptic soy agar with 0.1% cysteine. Plates were incubated at 37°C and 5% CO2 for 2 days, and colonies were enumerated. Statistical analysis was performed using Student's two-tailed t test for independent means.
Cytotoxicity.
Bone-marrow-derived macrophages were seeded in 96-well plates at 5 × 104 cells per well and incubated overnight with heat-killed F. novicida, with approximately 100 heat-killed bacteria per macrophage. Cells were infected at a multiplicity of infection of 100:1 or 10:1 (bacteria-to-macrophage ratio) and centrifuged at 730 × g for 15 min to mediate attachment. Medium was removed after 1 h and replaced with phenol red-free Dulbecco's modified Eagle medium with high glucose, l-glutamine, 110 mg/liter sodium pyruvate, and pyroxidine hydrochloride (Invitrogen Life Technologies) supplemented with 10% fetal calf serum. Cell death was quantified colorimetrically using the CytoTox96 lactate dehydrogenase (LDH) release kit according to the manufacturer's instructions (Promega, Madison, WI).
Mouse infections.
Six- to eight-week-old female wild-type C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) were kept under specific-pathogen-free conditions in filter-top cages. Mice were provided with sterile water and food ad libitum. In competitive index (CI) experiments, mice were infected subcutaneously with equal amounts of the mutant strain (5 × 104 cells) and wild-type strain U112 (5 × 104 cells) for a total of 105 bacteria in a 0.05-ml volume. The mice were monitored for signs of morbidity and mortality twice daily for the entirety of the study. Spleens were harvested at 2 days postinfection and homogenized, and dilutions were plated to determine CFU/g tissue on both MH agar and MH agar supplemented with the appropriate antibiotic to select for mutant bacteria. Competitive indices were calculated as the ratio of mutant to wild type of the output, normalized for the input, and significance was calculated by comparing the log of the CI to 0. Standard errors were calculated and significance of results was determined by the application of the Mann-Whitney statistical test to the log10 value of the CI. All animal infection experiments were approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of Stanford University.
Microarray accession numbers.
Results of the SAM analyses are available at the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE 5468.
RESULTS
Transcriptional profiles of wild-type and mglA mutant Francisella strains in rich media.
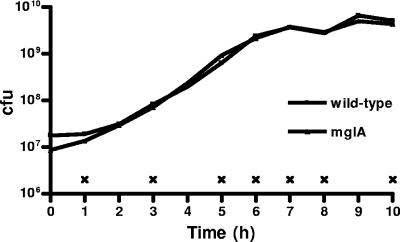
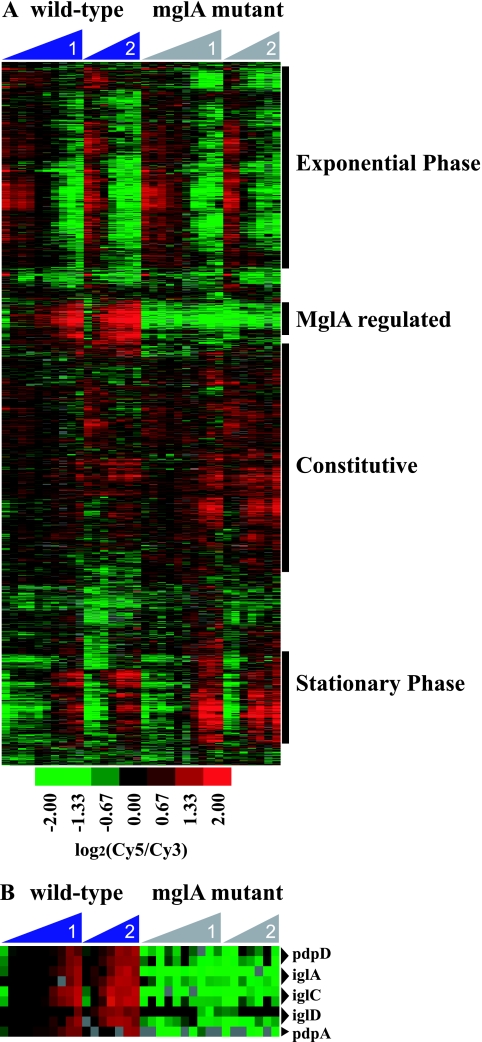
To identify novel MglA-regulated genes, we examined the global transcriptional profiles of genes expressed by wild-type Francisella and an mglA mutant (6) during growth in rich media at 37°C, conditions that induce MglA expression but do not impair the growth of the mglA point mutant (Fig. 1). RNA was collected from the two strains at 10 (experiment 1) or 7 (experiment 2) time points during the growth curves to represent lag, exponential, and stationary phases (Fig. 1). RNA was used to generate cDNA probes that were hybridized to a Francisella microarray (see Materials and Methods for a description of the microarray). In a hierarchical cluster, the majority of the genes separated into distinct gene expression patterns representing those expressed during lag and exponential phases, stationary phase, and constitutive expression (Fig. 2A). One cluster, however, contained genes that were differentially regulated in the mglA mutant compared to the wild-type. This cluster contained all of the genes (pdpD, iglA, iglC, iglD, and pdpA) known to be transcriptionally regulated by MglA (29) (Fig. 2B), thereby validating our results. The cluster also contained ∼100 genes not previously known to be regulated by MglA, suggesting that MglA regulates the expression of roughly 100 genes.
FIG. 1.
Wild-type and mglA mutant Francisella bacteria replicate with similar kinetics in rich media at 37°C. Shown is a representative growth curve for wild-type and mglA mutant Francisella strains. ×, time points when RNA was isolated from each strain.
FIG. 2.
Transcriptional profiles of wild-type and mglA mutant Francisella strains in rich media. (A) Hierarchical gene cluster of microarrays from two independent wild-type and mglA mutant growth curves. Columns represent individual time points, increasing from left to right during the growth curve. Rows represent individual genes. Genes were clustered into groups that were expressed in lag/exponential phase or stationary phase or that were constitutively expressed. One distinct cluster was expressed in wild-type bacteria but not in the mglA mutant. (B) Expression patterns of the mglAB operon and the known MglA-regulated genes pdpD, iglA, iglC, iglD, and pdpA. Duplicate gene spots that passed the filtering criteria are included.
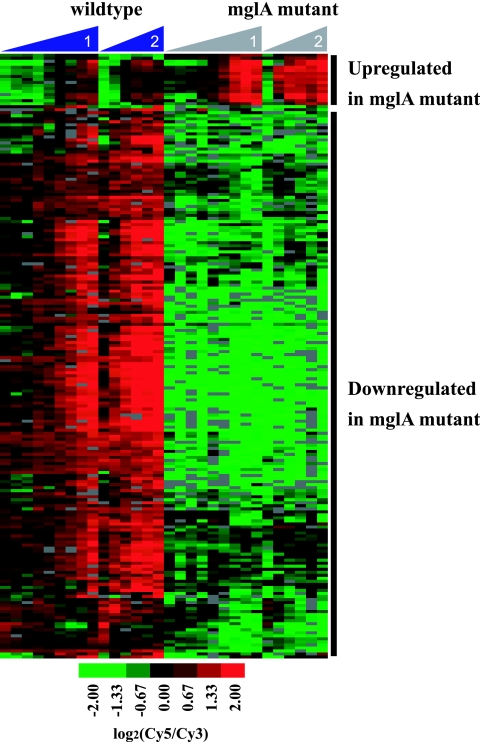
To identify genes whose expression was significantly different in the mglA mutant compared to the wild-type, we performed two-class statistical analysis of the data from experiments 1 and 2 using SAM (46). Genes whose expression varied by ≥2-fold between the wild type and mutant with a false discovery rate of <1 are included in Table 1. Ten genes were expressed at higher levels in the mglA mutant than in the wild type, suggesting negative regulation of these genes by an MglA-dependent mechanism. However, the majority of the genes (92 genes) were downregulated in the mglA mutant compared to the wild type, suggesting positive regulation of these genes by an MglA-dependent mechanism (Fig. 3). These genes include the previously identified MglA-regulated genes in the FPI: pdpD, pdpA, iglA, iglC, and iglD (29). Gene FTT1091, which encodes a protein with homology to an amidase from Nostoc punctiforme, has previously been shown by proteomic analysis to be regulated by MglA (29). This gene was also transcriptionally regulated in an MglA-dependent manner in our experiments, further validating our results. We also identified many genes that lie within the FPI that were not previously known to be MglA regulated. Indeed, pdpB, pdpC, and all of the genes located between these open reading frames, CDS1 through CDS9, are also positively regulated by MglA (Table 1), showing that the entire FPI is under the control of MglA.
FIG. 3.
Clustering of the 102 MglA-regulated genes as determined by SAM. Seventeen spots representing 10 genes were expressed at higher levels in the mutant than in the wild type, and 183 spots representing 92 genes were downregulated in the mutant compared to the wild-type. Columns represent individual time points, increasing from left to right during the growth curve, and rows represent individual genes.
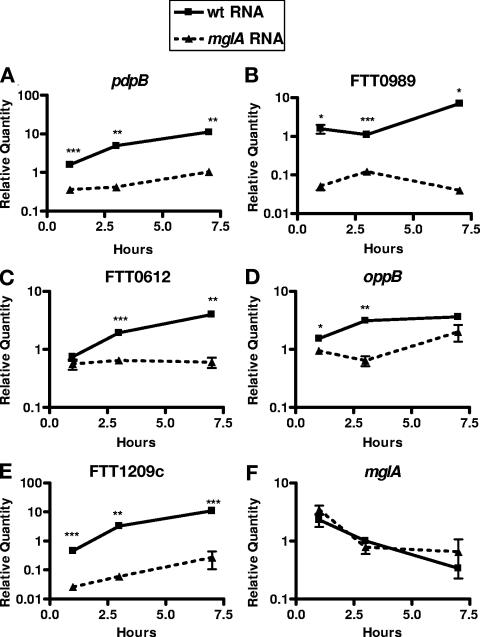
We chose five MglA-regulated genes to verify our microarray results by real-time RT-PCR. We included mglA as a control because the microarray results suggested that the gene was not differentially regulated between the wild-type and mutant bacteria. Using gene-specific primers, we quantified the transcript abundance in RNA from either the wild type or the mglA mutant at three time points representing the lag, exponential, and stationary phases of the growth curve (Fig. 4). The values were normalized to FTT0121, a DNA helicase whose expression changed minimally during the growth curve, as indicated by microarray analysis. Similar to our microarray data, transcripts of the putative MglA-regulated genes were significantly higher in RNA collected from the wild-type strain than in RNA collected from the mglA mutant, while the mglA transcript displayed similar quantities and kinetics between the two strains. These data show that the microarray results strongly correlate with qRT-PCR data and that the five genes tested are indeed regulated by MglA.
FIG. 4.
Quantification of selected MglA-regulated genes by real-time reverse transcription-PCR. Differences in transcript abundance of MglA-regulated genes were confirmed by real-time RT-PCR using RNA from the second growth curve. Gene-specific primers for (A) pdpB, (B) FTT0989, (C) FTT0612, (D) oppB, (E) FTT1209c, and (F) mglA were used to amplify either wild-type (wt) RNA, represented by a solid line, or mglA mutant RNA, represented by a dashed line. Testing of samples was performed in triplicate. Experiments were performed at least three times. Means and standard deviations from a representative experiment are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with the wild type).
A diverse repertoire of genes are regulated by MglA.
MglA-regulated genes can be separated into several different categories. First, 43 of the MglA-regulated genes are either hypothetical or conserved hypothetical genes according to the published Francisella tularensis genome sequence (35). This means that the largest category of MglA-regulated genes encodes proteins with unknown functions, suggesting that these proteins may have novel functions utilized by Francisella under MglA-activating conditions, such as intracellular growth. Twenty-four genes have been annotated as pseudogenes, representing another large class of MglA-regulated genes. The microarray was constructed using sequence data from F. tularensis subspecies tularensis strain SchuS4 and Francisella tularensis subspecies holarctica strain LVS but probed with cDNA from F. novicida. Since the F. novicida genome sequence has not been released yet, we do not know whether genes within this category are also pseudogenes in F. novicida. Additionally, it is possible that MglA regulates some F. novicida-specific genes that are not represented on our microarray and that are therefore not present in our data set.
The F. tularensis SchuS4 genome sequence has recently been added to the Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/dbget-bin/www_bfind?F.tularensis). KEGG is a bioinformatics database that uses information about protein interaction networks in well-studied organisms to predict the cellular pathway(s) and function of proteins in other organisms (7, 22). The majority of the MglA-regulated genes were not assigned to a pathway (76 genes), which is consistent with the large number of hypothetical genes and pseudogenes. However, 26 genes were grouped into the categories of metabolism (20 genes), environmental information processing (3 genes), or genetic information and processing (3 genes). Within the category of metabolism, amino acid (10 genes) and carbohydrate (8 genes) metabolism genes appeared to be widely represented. For example, ivlE is involved in the biosynthesis and degradation of leucine, isoleucine, and valine, and galM is involved in glycolysis. The variety of categories identified by our analysis suggests that MglA regulates several different cellular processes during the growth conditions described here.
Construction of mutants in MglA-regulated genes.
We selected eight MglA-regulated genes for mutagenesis and further study. Two genes within the FPI, pdpB and cds2, were chosen because this region has already been shown to contain virulence factors (35). We chose four genes outside the FPI based upon homology to known virulence factors as well as whether they encode either possible secretion signals or conserved catalytic domains (Table 3). These mutated MglA-regulated genes include FTT0612, which encodes a protein similar to OspD3 (SenA), a putative enterotoxin from Shigella flexneri (36); FTT0989, which encodes a protein with similarity to transglutaminase-like enzymes; FTT0123 (oppB), which encodes a protein similar to oligopeptide permease subunit B (OppB) from Pasteurella multocida; and FTT1209c, which encodes a protein with similarity to endothelin-converting enzyme (ece-1) from Shewanella putrefaciens. Lastly, two mutants were made using the hypothetical genes FTT1032 and FTT1771. Isogenic mutants were created for each gene by allelic exchange and replacement of the majority of the open reading frame with a kanamycin resistance cassette. All the mutants and complemented mutants grew similarly to the wild-type strain at 37°C in rich media (data not shown).
TABLE 3.
Genes chosen for further studies
| Locus tag | Gene | Product | Homolog according to phi-psi BLAST | E value |
|---|---|---|---|---|
| FTT1345 | pdpB | Hypothetical protein | None | |
| FTT1353 | Hypothetical protein | None | ||
| FTT0612 | Hypothetical protein | OspD3, Shigella dysenteriae | 3.E-35 | |
| FTT0989 | Hypothetical protein | COG1305, transglutaminase-like enzymes, putative cysteine proteases, Pseudomonas aeruginosa | 9.E-22 | |
| FTT0123 | oppB | Oligopeptide transporter, subunit B, ABC transporter, membrane protein, pseudogene | oppB, Pasteurella multocida | 4.E-70 |
| FTT1209c | Metallopeptidase family M13 protein, pseudogene | Endothelin-converting enzyme 1, Shewanella putrefaciens | 1.E-165 | |
| FTT1032 | Conserved hypothetical membrane protein, pseudogene | Hypothetical protein TDE2706, Treponema denticola | 4.E-34 | |
| FTT1771 | Hypothetical protein | None |
MglA-regulated genes contribute to replication in macrophages.
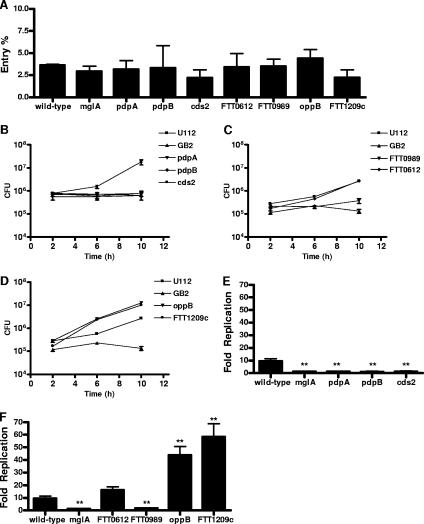
Each of the mutants was tested for the ability to enter and replicate in unactivated bone-marrow-derived macrophages. None of the mutants were defective for macrophage entry, as indicated by levels of intracellular mutant bacteria that were similar to those of wild-type bacteria at 2 h postinfection (Fig. 5A). By 10 h, the macrophages infected with wild-type Francisella contained approximately 10-fold-higher numbers of intracellular bacteria compared to the levels of bacteria at 2 h. Similar to the mglA and pdpA mutants, which are known to have a growth defect in macrophages, the pdpB and cds2 mutants had similar levels of intracellular bacteria at 2 h and 10 h postinfection (Fig. 5E), indicating that there was no death, no replication, or a balance between the two. A mutant in FTT0989, which is located outside of the pathogenicity island, also contained similar levels of intracellular bacteria at 2 h and 10 h postinfection (Fig. 5F). The FTT0612, FTT1032, and FTT1771 mutants replicated to the same extent as the wild type, showing that these genes are not required for growth in macrophages (Fig. 5F and data not shown). Surprisingly, two mutants, oppB and FTT1209c, replicated to higher levels than the wild type by 10 h postinfection (Fig. 5F). We repeated the replication assay in the presence of gentamicin and obtained similar results, suggesting that the increased replication was not due to mutant growth in the extracellular medium (data not shown). Taken together, these results show that genes regulated by an MglA-dependent mechanism are involved in facilitating, as well as limiting, intracellular replication.
FIG. 5.
MglA-regulated genes contribute to Francisella replication in macrophages. Bone-marrow-derived macrophages were infected at an MOI of 25:1. Samples were taken at 2 h, 6 h, and 10 h postinfection. (A) Percent bacterial entry into macrophages was calculated by dividing the 2-h counts by the input. Macrophages were infected with (B) pdpB and cds2 mutants, (C) FTT0612 and FTT0989 mutants, and (D) oppB and FTT1209c mutants. Bacterial replication (n-fold) was calculated by dividing the 10-h bacterial counts by the averages of the 2-h bacterial counts for (E) pdpB and cds2 mutants and (F) FTT0612, FTT0989, oppB, and FTT1209c mutants. The wild type, the mglA mutant, and pdpA mutants were included as controls. Samples were performed in triplicate. Experiments were performed at least three times. Means and standard deviations from a representative experiment are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with the wild type).
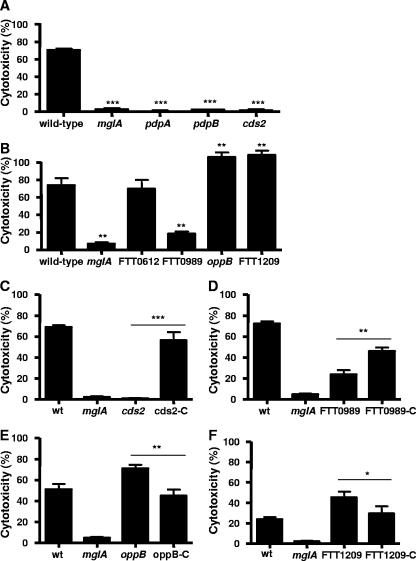
MglA-regulated genes are involved in macrophage cytotoxicity.
We showed previously that mutants known to have growth defects in macrophages were also defective in inducing cytotoxicity of activated macrophages (33). Therefore, we assessed the ability of each of our mutants to kill activated macrophages, as measured by an LDH release assay. As previously shown, the mglA and pdpA mutants did not kill macrophages, while 73% of wild-type Francisella-infected macrophages died by 8 h postinfection (33) (Fig. 6A). The pdpB and cds2 mutants, which were unable to replicate intracellularly, were severely defective in their ability to kill activated macrophages. We were unable to obtain an intact pdpB allele for complementation of the pdpB mutant. However, the cds2 mutant was complemented in cis by recombining a wild-type cds2 gene into the chromosome. Using this strategy, we achieved full complementation (Fig. 6C) where cds2 was under the control of its native promoter and in a single copy.
FIG. 6.
MglA-regulated genes are involved in macrophage cytotoxicity. Macrophages were prestimulated with heat-killed, wild-type Francisella cells ∼18 h prior to infection. Cytotoxicity was measured by LDH release at 8 h postinfection. The mglA and pdpA mutants were included as controls. Macrophages were infected at an MOI of 100:1 with (A) pdpB and cds2 mutants and (B) FTT0612, FTT0989, oppB, and FTT1209c mutants. Mutants were complemented, and cytotoxicity was measured at (C) 8 h postinfection with an MOI of 100:1; (D) 10 h postinfection with an MOI of 20:1; (E) 8 h postinfection with an MOI of 100:1; and (F) 10 h postinfection with an MOI of 10:1. Testing of samples was performed in triplicate in each experiment. Experiments were performed at least three times. Means and standard deviations from a representative experiment are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with wild-type for A and B or mutant compared to the complemented mutant by the bar in C to F). wt, wild type.
The FTT0989 (transglutaminase homolog) mutant also induced a significantly lower level of cell death than the wild-type strain (Fig. 6B). We found that the FTT0989 mutant was partially complemented by the addition of a plasmid expressing FTT0989 in trans under the control of its native promoter (Fig. 6D). The partial trans complementation could be due to the overexpression of the FTT0989 gene product. Unfortunately, we were unable to obtain the cis complementation construct to address the issue of copy number. The FTT0612 (ospD3 homolog), FTT1032, and FTT1771 mutants, which replicated similarly to the wild-type bacteria in macrophages, induced levels of cytotoxicity that were similar to that of the wild-type strain (Fig. 6B and data not shown).
The two mutants that had a higher rate of replication in macrophages than wild-type Francisella, oppB and FTT1209c (ece-1 homolog), induced significantly higher levels of cell death than the wild-type at 8 h (Fig. 6B). The oppB and FTT1209c mutants were complemented in cis, and both constructs were under the control of their native promoters and in single copy to avoid possible problems of overexpression. These complemented mutants displayed lower levels of macrophage cytotoxicity that were similar to the levels induced by wild-type bacteria (Fig. 6E and F). These data show that we have identified both attenuated and hypervirulent mutants and, together with the replication data, highlight the striking correlation between the ability of bacteria to replicate in unactivated macrophages and the induction of cytotoxicity in activated macrophages.
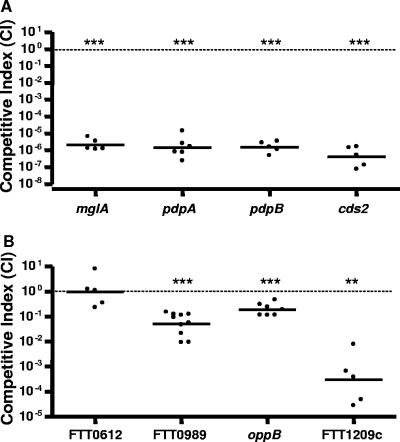
MglA-regulated genes contribute to Francisella virulence in mice.
We identified mutations in MglA-regulated genes that were either avirulent or hypervirulent in in vitro assays and sought to further investigate the fitness of these mutants in Francisella infection in vivo. We performed competitive index experiments where the relative fitness of an individual Francisella mutant could be compared to that of the wild-type strain within a single mouse. The CI is expressed as the ratio of the number of mutant bacteria recovered from the tissue divided by that of the wild type and is normalized to the ratio of the mutant to the wild type from the input. Therefore, a competitive index value that is less than 1 indicates that the mutant is less fit than the wild-type strain in vivo. Mice were infected subcutaneously with an equal mixture of wild-type and mutant bacteria, and spleens were harvested at 2 days postinfection to determine the levels of bacterial colonization of each strain. Consistent with our in vitro results, we found that the pdpB and cds2 mutants showed dramatic decreases in fitness (CI of <1 × 10−6 [lower limit of detection]) (Fig. 7). In fact, the mglA mutant and all the FPI mutants tested here had similar CI values (Fig. 7). The FTT0989 (transglutaminase homolog) mutant, which showed a defect in intramacrophage replication and cytotoxicity, also showed a decrease in fitness in vivo (CI of 0.051). The FTT0612 (ospD3 homolog), FTT1032, and FTT1771 mutants, which behaved similarly to the wild-type strain in the macrophage assays, were not attenuated in vivo (CI of 0.997) (data not shown). The oppB and FTT1209c (ece-1 homolog) mutants, which replicated better than the wild-type strain in macrophages and induced higher levels of host cell death, were less fit in vivo than the wild type, with CI values of 0.19 and 0.0003, respectively. These results suggest that higher rates of replication seen in vitro are not necessarily beneficial in vivo. Taken together, these data highlight the role of MglA-regulated genes in Francisella pathogenesis in vivo.
FIG. 7.
MglA-regulated genes contribute to Francisella virulence in mice. Mice were infected subcutaneously with equal amounts of mutant and wild-type Francisella cells. At 2 days postinfection, spleens were taken for counts, and the competitive index was calculated. (A) Competitive indices of pdpB and cds2 FPI mutants. A spontaneous streptomycin-resistant mglA mutant (see Materials and Methods for a description) and the pdpA mutants were included as controls. (B) Competitive indices of FTT0612, FTT0989, oppB, and FTT1209c mutants. At least five mice were used to calculate the CI value for each mutant strain. Each spot represents the CI value from an individual mouse, and bars represent the geometric means. Significance was calculated by comparing the log of the CI value to 0. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (compared with 0).
DISCUSSION
We performed a transcriptional comparison of genes expressed in wild-type Francisella and those expressed by an isogenic mglA mutant during growth in rich media (Fig. 1 to 3). Intriguingly, the entire FPI was regulated by MglA. We targeted several MglA-regulated genes and created eight mutant strains, including two uncharacterized genes in the FPI. By testing mutant strains for several of the MglA-regulated genes in virulence assays both in vitro and in vivo, we identified novel Francisella virulence factors (Fig. 4 to 6).
We mutated pdpB and cds2, two FPI genes that have not yet been characterized. Similar to the pdpA, pdpD, and iglC FPI mutants, the pdpB and cds2 mutants were avirulent, as determined by bacterial replication, macrophage cytotoxicity, and virulence in mice (Fig. 5, 6, and 7) (17, 28, 29, 33, 35). It is unclear how the FPI contributes to Francisella replication and virulence; however, all the FPI mutants tested here exhibited similar phenotypes in every assay. This is strongly suggestive of a system comprised of multiple components whereby mutating one component results in the inactivation of the entire system. Interestingly, PdpB has limited similarity to a domain in IcmF, a protein that may be involved in type IV secretion in Legionella pneumophila (42, 47). The FPI proteins IglA and IglB share homology with Edwardsiella tarda EvpA (22% identity) and EvpB (29% identity), respectively, which are involved in the secretion of virulence proteins (39). The fact that mutations in several FPI genes have similarly dramatic phenotypes, together with the homology of some of the FPI genes to secretion system genes in other bacteria, suggests that the FPI may encode a secretion system that is essential for Francisella intracellular replication, macrophage cytotoxicity, and virulence in vivo.
In addition to the FPI genes, we identified several novel genes that appear to be important for intracellular replication, macrophage cytotoxicity, and virulence in mice. For example, similar to FPI mutants, an FTT0989 mutant is attenuated for replication inside macrophages, does not induce macrophage death, and is outcompeted by wild-type bacteria in mice. The protein encoded by FTT0989 has a transglutaminase domain containing the catalytic triad characteristic of this domain, suggesting that the protein may have transglutaminase activity. The protein also contains a putative secretion signal and may therefore be secreted by Francisella, making it available to interact with host cells and thereby contribute to virulence. Indeed, FTT0989 plays an important role in Francisella pathogenesis, since it was attenuated for growth in macrophages and in mice (Fig. 5 and 7). Cytotoxic necrotizing factor type 1 from Escherichia coli and dermonecrotic toxin from Bordetella pertussis are two characterized transglutaminases that activate Rho GTPases of the host cells and are involved in virulence (25).
We identified two additional mutants, FTT1209c and oppB (FTT0123), that had similar phenotypes to one another in macrophages. Both of these mutants replicated to higher levels in macrophages than wild-type Francisella and induced higher levels of cytotoxicity in activated macrophages (Fig. 5 and 6). However, these mutants were attenuated in vivo (Fig. 7). FTT1209c encodes a predicted M13 metallopeptidase with striking similarity to mammalian endothelin-converting enzyme (ECE-1). FTT1209c has several homologs, including PepO from Porphyromonas gingivalis, which plays a role in host cell invasion (1, 4). FTT1209c has been annotated as a pseudogene in the sequenced F. tularensis strain SchuS4 (35). This gene does not appear to be a pseudogene in F. novicida strain U112, the strain chosen for our experiments (unpublished data). Both of the genes from the two species contain full-length N-terminal and C-terminal M13 peptidase domains as well as the HEXXH metal binding motif, suggesting that both gene products may act as metallopeptidases. However, FTT1209c in U112 has a predicted signal peptide that is absent in SchuS4. It is unknown if the signal sequence is necessary for the phenotypes described here.
FTT0123 (oppB) encodes a protein similar to OppB from Pasteurella multocida. The Opp complex has been identified as a virulence factor for several pathogens, including L. monocytogenes, group A Streptococcus, Bacillus cereus, and Bacillus thuringiensis (8, 18, 38, 48). The opp operon (oppABCDF) encodes an ATP-dependent transport system involved in the uptake of small peptides, which can serve as carbon and nitrogen sources and may have signaling properties (30). For group A Streptococcus and B. cereus, mutations in the opp operon result in an inability to express virulence factors, suggesting a regulatory role for Opp in these two species. oppB has also been annotated as a pseudogene in SchuS4 due to the presence of a nonsense mutation that results in a truncated protein. This mutation is not present in oppB from U112, where it appears to encode a full-length protein (unpublished data). Perhaps SchuS4 has evolved other ways to make it competitive in nature despite the alteration of this locus. Future experiments will be designed to investigate this possibility. Taken together, the phenotypes of the FTT1209c and OppB mutants suggest that limiting intracellular growth is a critical aspect of Francisella pathogenesis.
To our knowledge, this is the first description of Francisella mutants that replicate to higher levels than wild-type bacteria within macrophages, and these data suggest that oppB and FTT1209c may be negative regulators of Francisella growth. This suggests that Francisella encodes factors that limit its growth in macrophages and that controlling its growth rate is important for pathogenesis. Controlling replication within a macrophage may allow Francisella to grow intracellularly without triggering macrophage cell death and exposing the bacteria to complement, antibodies, etc. In addition, Francisella may use the macrophage for dissemination from the skin to the draining lymph nodes, and uncontrolled replication of Francisella could result in rapid macrophage death and elimination of the ability to disseminate efficiently.
Not all MglA-regulated genes are expected to play a role in virulence. In keeping with this assumption, we have presented data for FTT0612 (OspD3 homolog), FTT1032, and FTT1771 mutants that behaved similar to the wild type in the macrophage assays and did not contribute to virulence in a mouse model of infection. This suggests that these genes are dispensable for virulence in Francisella.
While the present work describes many new MglA-regulated genes, the mechanism of MglA regulation remains unknown. However, it is interesting that while MglA-regulated genes are maximally expressed during the stationary phase, the mglA transcript is maximally expressed in the lag and early exponential phases (our work and see reference 5). MglA is closely related to E. coli SspA, a stringent starvation protein. The SspA regulon overlaps that of another transcription factor (20), and SspA works with an additional cofactor to transcribe phase P1 late genes (19). It is possible that the MglA regulon presented here represents the combined action of MglA and an additional cofactor and/or multiple layers of regulation. Additional studies will be required to investigate this possibility.
We have identified five novel MglA-regulated virulence genes that either facilitate or limit replication within macrophages and play a role in virulence in vivo. This suggests that in addition to the bacterium's ability to grow intracellularly, the bacterium's ability to keep its own intracellular replication in check is also essential to Francisella pathogenesis. The growing number of MglA-regulated virulence factors underscores the idea that MglA is an important regulator of virulence factors. Nonetheless, the majority of MglA-regulated genes remain uncharacterized. Twenty out of 102 MglA-regulated genes are predicted to play a role in metabolism, particularly in amino acid metabolism (Table 1). Work with Listeria monocytogenes has shown that amino acid metabolism genes are essential for intracellular growth (21). This is particularly relevant, since L. monocytogenes escapes the vacuole and replicates in the cytosol of infected cells, similar to Francisella. However, the most interesting class of MglA-regulated genes may be the 43 hypothetical genes with no known or characterized homologs. Characterization of these genes may lead to the elucidation of new processes in bacterial pathogenesis that are shared with other pathogens as well as some that may be unique to Francisella.
Acknowledgments
We thank Lucy Thompson and Kaman Chan for technical advice. We thank Lucy Thompson, Thomas Henry, and Stanley Falkow for critical reading of the manuscript. We also thank Larry Gallagher and Colin Manoil for the kind gift of pLG62a as well as Karin Elkins for the U112 and GB2 strains and Francis Nano for pVIC314. Thank you to David Schneider for the use of the iCycler.
This work was supported by grants RO1 AI063302 and AI-65359 from the NIAID, NIH, to D.M.M. A.B. was supported by a Cellular and Molecular Biology Training Program grant (NIH 5 T32 GM007276). D.S.W. was supported by a postdoctoral fellowship from the Giannini Family Foundation.
Editor: D. L. Burns
Footnotes
Published ahead of print on 25 September 2006.
REFERENCES
- 1.Ansai, T., W. Yu, S. Urnowey, S. Barik, and T. Takehara. 2003. Construction of a pepO gene-deficient mutant of Porphyromonas gingivalis: potential role of endopeptidase O in the invasion of host cells. Oral Microbiol. Immunol. 18:398-400. [DOI] [PubMed] [Google Scholar]
- 2.Anthony, L. D., R. D. Burke, and F. E. Nano. 1991. Growth of Francisella spp. in rodent macrophages. Infect. Immun. 59:3291-3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anthony, L. S., M. Z. Gu, S. C. Cowley, W. W. Leung, and F. E. Nano. 1991. Transformation and allelic replacement in Francisella spp. J. Gen. Microbiol. 137:2697-2703. [DOI] [PubMed] [Google Scholar]
- 4.Awano, S., T. Ansai, H. Mochizuki, W. Yu, K. Tanzawa, A. J. Turner, and T. Takehara. 1999. Sequencing, expression and biochemical characterization of the Porphyromonas gingivalis pepO gene encoding a protein homologous to human endothelin-converting enzyme. FEBS Lett. 460:139-144. [DOI] [PubMed] [Google Scholar]
- 5.Baron, G. S., and F. E. Nano. 1999. An erythromycin resistance cassette and mini-transposon for constructing transcriptional fusions to cat. Gene 229:59-65. [DOI] [PubMed] [Google Scholar]
- 6.Baron, G. S., and F. E. Nano. 1998. MglA and MglB are required for the intramacrophage growth of Francisella novicida. Mol. Microbiol. 29:247-259. [DOI] [PubMed] [Google Scholar]
- 7.Bono, H., H. Ogata, S. Goto, and M. Kanehisa. 1998. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 8:203-210. [DOI] [PubMed] [Google Scholar]
- 8.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, D. L., B. Y. Lee, and M. A. Horwitz. 2004. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72:3204-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis, D. T., T. V. Inglesby, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. D. Fine, A. M. Friedlander, J. Hauer, M. Layton, S. R. Lillibridge, J. E. McDade, M. T. Osterholm, T. O'Toole, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2001. Tularemia as a biological weapon: medical and public health management. JAMA 285:2763-2773. [DOI] [PubMed] [Google Scholar]
- 13.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 14.Eisen, M. B., and P. O. Brown. 1999. DNA arrays for analysis of gene expression. Methods Enzymol. 303:179-205. [DOI] [PubMed] [Google Scholar]
- 15.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golovliov, I., V. Baranov, Z. Krocova, H. Kovarova, and A. Sjostedt. 2003. An attenuated strain of the facultative intracellular bacterium Francisella tularensis can escape the phagosome of monocytic cells. Infect. Immun. 71:5940-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gominet, M., L. Slamti, N. Gilois, M. Rose, and D. Lereclus. 2001. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40:963-975. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, A. M., H. Lehnherr, X. Wang, V. Mobley, and D. J. Jin. 2003. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol. Microbiol. 48:1621-1631. [DOI] [PubMed] [Google Scholar]
- 20.Hansen, A. M., Y. Qiu, N. Yeh, F. R. Blattner, T. Durfee, and D. J. Jin. 2005. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol. Microbiol. 56:719-734. [DOI] [PubMed] [Google Scholar]
- 21.Joseph, B., K. Przybilla, C. Stuhler, K. Schauer, J. Slaghuis, T. M. Fuchs, and W. Goebel. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188:556-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa, M. 2002. The KEGG database. Novartis Found. Symp. 247:91-101, 101-103, 119-128, 244-252. [PubMed] [Google Scholar]
- 23.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 24.Kieffer, T. L., S. Cowley, F. E. Nano, and K. L. Elkins. 2003. Francisella novicida LPS has greater immunobiological activity in mice than F. tularensis LPS, and contributes to F. novicida murine pathogenesis. Microbes Infect. 5:397-403. [DOI] [PubMed] [Google Scholar]
- 25.Lacerda, H. M., G. D. Pullinger, A. J. Lax, and E. Rozengurt. 1997. Cytotoxic necrotizing factor 1 from Escherichia coli and dermonecrotic toxin from Bordetella bronchiseptica induce p21(rho)-dependent tyrosine phosphorylation of focal adhesion kinase and paxillin in Swiss 3T3 cells. J. Biol. Chem. 272:9587-9596. [DOI] [PubMed] [Google Scholar]
- 26.Lai, X. H., I. Golovliov, and A. Sjostedt. 2001. Francisella tularensis induces cytopathogenicity and apoptosis in murine macrophages via a mechanism that requires intracellular bacterial multiplication. Infect. Immun. 69:4691-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 28.Lauriano, C. M., J. R. Barker, F. E. Nano, B. P. Arulanandam, and K. E. Klose. 2003. Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229:195-202. [DOI] [PubMed] [Google Scholar]
- 29.Lauriano, C. M., J. R. Barker, S. S. Yoon, F. E. Nano, B. P. Arulanandam, D. J. Hassett, and K. E. Klose. 2004. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. USA 101:4246-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazazzera, B. A. 2001. The intracellular function of extracellular signaling peptides. Peptides 22:1519-1527. [DOI] [PubMed] [Google Scholar]
- 31.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354-2356. [DOI] [PubMed] [Google Scholar]
- 32.Maier, T. M., A. Havig, M. Casey, F. E. Nano, D. W. Frank, and T. C. Zahrt. 2004. Construction and characterization of a highly efficient Francisella shuttle plasmid. Appl. Environ. Microbiol. 70:7511-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mariathasan, S., D. S. Weiss, V. M. Dixit, and D. M. Monack. 2005. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J. Exp. Med. 202:1043-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer, M. P. 1995. A new set of useful cloning and expression vectors derived from pBlueScript. Gene 163:41-46. [DOI] [PubMed] [Google Scholar]
- 35.Nano, F. E., N. Zhang, S. C. Cowley, K. E. Klose, K. K. Cheung, M. J. Roberts, J. S. Ludu, G. W. Letendre, A. I. Meierovics, G. Stephens, and K. L. Elkins. 2004. A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186:6430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nataro, J. P., J. Seriwatana, A. Fasano, D. R. Maneval, L. D. Guers, F. Noriega, F. Dubovsky, M. M. Levine, and J. G. Morris, Jr. 1995. Identification and cloning of a novel plasmid-encoded enterotoxin of enteroinvasive Escherichia coli and Shigella strains. Infect. Immun. 63:4721-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Owen, C. R., E. O. Buker, W. L. Jellison, D. B. Lackman, and J. F. Bell. 1964. Comparative studies of Francisella tularensis and Francisella novicida. J. Bacteriol. 87:676-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podbielski, A., B. Pohl, M. Woischnik, C. Korner, K. H. Schmidt, E. Rozdzinski, and B. A. Leonard. 1996. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol. Microbiol. 21:1087-1099. [DOI] [PubMed] [Google Scholar]
- 39.Rao, P. S., Y. Yamada, Y. P. Tan, and K. Y. Leung. 2004. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 53:573-586. [DOI] [PubMed] [Google Scholar]
- 40.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schaible, U. E., and S. H. E. Kaufmann. 2002. Studying trafficking of intracellular pathogens in antigen-presenting cells. Methods Microbiol. 31:343-360. [Google Scholar]
- 42.Sexton, J. A., J. L. Miller, A. Yoneda, T. E. Kehl-Fie, and J. P. Vogel. 2004. Legionella pneumophila DotU and IcmF are required for stability of the Dot/Icm complex. Infect. Immun. 72:5983-5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telepnev, M., I. Golovliov, T. Grundstrom, A. Tarnvik, and A. Sjostedt. 2003. Francisella tularensis inhibits Toll-like receptor-mediated activation of intracellular signalling and secretion of TNF-alpha and IL-1 from murine macrophages. Cell. Microbiol. 5:41-51. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VanRheenen, S. M., G. Duménil, and R. R. Isberg. 2004. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect. Immun. 72:5972-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, C.-H., C.-Y. Lin, Y.-H. Luo, P.-J. Tsai, Y.-S. Lin, M. T. Lin, W.-J. Chuang, C.-C. Liu, and J.-J. Wu. 2005. Effects of oligopeptide permease in group A streptococcal infection. Infect. Immun. 73:2881-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, M. D., T. X. Ouyang, and M. C. Flickinger. 1994. Glutathione S-transferase-sspA fusion binds to E. coli RNA polymerase and complements delta sspA mutation allowing phage P1 replication. Biochem. Biophys. Res. Commun. 201:123-127. [DOI] [PubMed] [Google Scholar]
- 50.Williams, M. D., T. X. Ouyang, and M. C. Flickinger. 1994. Starvation-induced expression of SspA and SspB: the effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol. Microbiol. 11:1029-1043. [DOI] [PubMed] [Google Scholar]