Abstract
4-Diphosphocytidyl-2C-methyl-d-erythritol kinase, an essential enzyme in the nonmevalonate pathway of isopentenyl diphosphate and dimethylallyl diphosphate biosynthesis, catalyzes the single ATP-dependent phosphorylation stage affording 4-diphosphocytidyl-2C-methyl-d-erythritol-2-phosphate. The 2-Å resolution crystal structure of the Escherichia coli enzyme in a ternary complex with substrate and a nonhydrolyzable ATP analogue reveals the molecular determinants of specificity and catalysis. The enzyme subunit displays the α/β fold characteristic of the galactose kinase/homoserine kinase/mevalonate kinase/phosphomevalonate kinase superfamily, arranged into cofactor and substrate-binding domains with the catalytic center positioned in a deep cleft between domains. Comparisons with related members of this superfamily indicate that the core regions of each domain are conserved, whereas there are significant differences in the substrate-binding pockets. The nonmevalonate pathway is essential in many microbial pathogens and distinct from the mevalonate pathway used by mammals. The high degree of sequence conservation of the enzyme across bacterial species suggests similarities in structure, specificity, and mechanism. Our model therefore provides an accurate template to facilitate the structure-based design of broad-spectrum antimicrobial agents.
Keywords: galactose kinase/homoserine kinase/mevalonate kinase/phosphomevalonate kinase, enzyme mechanism, nonmevalonate, phosphorylation
The ATP-dependent 4-diphosphocytidyl-2C-methyl-d-erythritol (CDP-ME) kinase (EC 2.7.1.148) participates in the biosynthesis of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). These isomers are the universal five-carbon precursors of isoprenoids, a diverse and important family of natural products that includes sterols, dolichols, triterpenes, and ubiquinones, and components of macromolecules such as the prenyl groups of prenylated proteins and isopentenylated tRNAs (1–3). Isoprenoids contribute to many biological functions, including electron transport in respiration and photosynthesis, hormone-based signaling, apoptosis, meiosis, protein cleavage, and degradation (4). In addition, they provide important structural components of cell membranes (1).
Two biosynthetic routes to IPP and DMAPP have evolved. In eukaryotes, archaebacteria, and a few eubacteria, the precursor biosynthesis is through the mevalonate pathway (3–7). This begins with the conversion of three molecules of acetyl-CoA to 3-hydroxy-3-methylglutaryl-CoA followed by reduction, phosphorylation, and decarboxylation to generate IPP, some of which is isomerized to DMAPP. The last three steps in this pathway are ATP-dependent and catalyzed by the structurally related mevalonate kinase (MVK), phosphomevalonate kinase, and mevalonate 5-diphosphate decarboxylase.
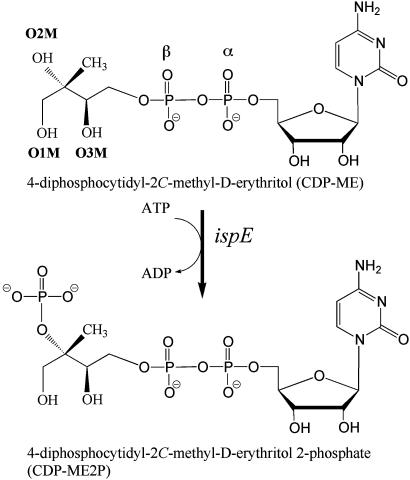
In chloroplasts, algae, cyanobacteria, most eubacteria, and the apicomplexa, IPP and DMAPP synthesis is accomplished by seven enzymes in a pathway named after one of the intermediates, the 1-deoxy-d-xylulose-5-phosphate (DOXP) pathway (5–8). The first stage in the nonmevalonate route is the condensation of pyruvate and d-glyceraldehyde 3-phosphate to produce DOXP (9, 10). The second and third stages convert DOXP to 2C-methyl-d-erythritol-4-phosphate (11, 12) and transfer the erythritol derivative onto a nucleotide, resulting in CDP-ME (13–15). Stage four, the only ATP-dependent step in the pathway, is catalyzed by CDP-ME kinase (16, 17), the subject of the present study. Here, CDP-ME kinase catalyzes the transfer of the γ-phosphoryl moiety of ATP to CDP-ME, forming 4-diphosphocytidyl-2C-methyl-d-erythritol-2-phosphate (CDP-ME2P) and ADP (Fig. 1). Next, CDP-ME2P is converted to 2C-methyl-d-erythritol-2,4-cyclodiphosphate and CMP (18, 19). The cyclodiphosphate then, in two enzyme-catalyzed stages, undergoes reduction and elimination to 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate and onto IPP and DMAPP (20–23).
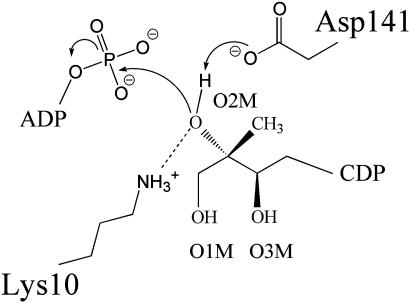
Fig. 1.
The reaction catalyzed by CDP-ME kinase.
The species that use the DOXP pathway include the causal agents for diverse and serious human diseases including leprosy, malaria, bacterial meningitis, various gastrointestinal and sexually transmitted infections, tuberculosis, and certain types of pneumonia (5). The enzymes of the DOXP pathway have no orthologs in humans and have attracted interest as potential targets for structure-based antimicrobial drug development. DOXP reductoisomerase, the enzyme that catalyzes the second reaction in the pathway, is inhibited by the antibiotic fosmidomycin (24, 25), a compound that has performed well in recent clinical trials against malaria (26). Such chemical validation of the reductoisomerase is complemented by genetic studies, which have proven that the enzymes responsible for stages three to five of the pathway are essential in eubacteria (27–31). The crystallographic studies necessary to support structure-based drug discovery have produced models for DOXP reductoisomerase (32–34), 2C-methyl-d-erythritol-4-phosphate cytidylyltransferase (15, 35), and 2C-methyl-d-erythritol-2,4-cyclodiphosphate synthase (36–39). We now turn our attention to CDP-ME kinase.
On the basis of sequence comparisons, CDP-ME kinase is recognized as a member of the galactose kinase/homoserine kinase (HSK)/mevalonate kinase/phosphomevalonate kinase (GHMP) kinase superfamily (40), so-called after the four founding members, namely galacto, homoserine, mevalonate, and phosphomevalonate kinases (41). The GHMP family now also includes mevalonate 5-diphosphate decarboxylase (42) and archaeal shikimate kinase (43). Crystal structures are available for several GHMP kinases (40, 42, 44–47) although only HSK (38) has provided experimental details for a ternary complex. In other cases modeling has been used to support mechanistic studies (45, 46).
To elucidate details of the enzyme structure, specificity, and mechanism we have determined the high-resolution crystal structure of recombinant Escherichia coli CDP-ME kinase in complex with substrate and the hydrolysis-resistant ATP analogue adenosine 5′-(β,γ-imino)triphosphate (AMP-PNP). Sequence and structural comparisons with functional and fold-homologues are also presented.
Experimental Procedures
Sample Preparation and Crystallization. The E. coli ispE gene that encodes CDP-ME kinase was amplified from genomic DNA by PCR and cloned into the pET15b expression vector (Novagen). The resulting plasmid was heat shock-transformed into E. coli strain BL21 (DE3) (Stratagene) for protein expression. Cells were grown in M9 medium supplemented with the usual amino acids except that l-selenomethionine (100 mg/liter) replaced l-methionine. The culture was incubated at 37°C for 3 h to an OD600 of 0.5, and protein expression was induced overnight at room temperature with 0.5 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested by centrifugation and disrupted in a French press. After centrifugation, the supernatant was filtered and applied to a metal-chelating HiTrap column (Amersham Pharmacia) previously equilibrated with nickel chloride (50 mM) followed by 50 mM Tris hydrochloride, pH 7.7, containing 50 mM NaCl (buffer A). The column was washed with 25 mM bis-Tris propane, pH 7.7, containing 10 mM imidazole. CDP-ME kinase was eluted by 90 mM imidazole and dialyzed overnight against buffer A. The protein solution was applied to a Q-Sepharose anion exchange column (Amersham Pharmacia), washed with 50 mM Tris hydrochloride, pH 8.5, and 50 mM NaCl, and developed with 100 mM NaCl. Fractions were analyzed on SDS/PAGE, combined, dialyzed against buffer A, and then concentrated to 25 mg/ml. Sample purity was assessed by SDS/PAGE and matrix-assisted laser desorption time-of-flight MS; the latter technique also confirmed full incorporation of l-selenomethionine.
The enzyme was incubated with 2 mM AMP-PNP (Sigma) and 3 mM CDP-ME (48) on ice for 2 h to provide a stock solution for crystallization. Monoclinic plate-like crystals grew in hanging drops consisting of 1 μl of stock, 1 μl of reservoir solution (20% polyethylene glycol 8000, 0.2 M magnesium acetate, 0.1 M sodium cacodylate, pH 6.5), and 0.2 μl of 0.25 M sulfo-betaine.
Data Collection, Structure Solution, and Refinement. Crystals were maintained at 100 K in the presence of 20% glycerol as a cryoprotectant, and measurements were made on beamline ID29 at the European Synchrotron Radiation Facility with an ADSC-Q210 charge-coupled device detector. A single crystal of l-selenomethionine derivilized protein was used to measure 180° of data at a wavelength (λ 0.9791 Å), determined from a Se K-edge x-ray absorption near-edge scan, to maximize the dispersive f″ signal. A single-wavelength anomalous dispersion approach provided initial phases.
Data were processed and scaled with denzo/scalepack (49). Twelve Se positions were identified, and phases were calculated to 2-Å resolution with a figure of merit of 0.33 by using the program solve (50). Density modification (51) by solvent flattening and 2-fold noncrystallographic symmetry (NCS) averaging raised the figure of merit to 0.59 and resulted in an electron density map of excellent quality. The map was interpreted automatically (51), and a model of 242 residues for each subunit was constructed. The remaining amino acids were included by using the program o (52) after rounds of refinement (53–55) interspersed with map inspection. NCS restraints were used in the early stages of refinement then completely released. The careful placement of solvent molecules and active site ligands concluded the analysis. The refinement process was monitored with the use of Rfree (56); further details are in Table 1. Figures were prepared with molscript (57), raster3d (58), promotif (59), and aline (C. S. Bond, personal communication).
Table 1. Data collection and refinement statistics.
| Resolution, Å/completeness | 2.0/94.3 (92.3) |
| No. of measurements/unique reflections | 364, 197/39,761 |
| I/σ(I)/Rmerger, % | 12.9 (5.6)/4.8 (14.7) |
| Protein residues/water molecules | 566/383 |
| AMP-PNP/CDP-ME/Cl- | 2/2/1 |
| Rwork, %/no. of reflections | 16.6/37,773 |
| Rfree, %/no. of reflections | 20.2/1,988 |
| Wilson B, Å2 | 17.8 |
| Average isotropic thermal parameters, Å2 | |
| Main chain/side chain | 15.3/18.4 |
| Solvents/AMP-PNP/CDP-ME | 28.9/44.0/29.1 |
| rms deviation for bond lengths, Å/bond angles, ° | 0.010/1.33 |
| Ramachandran analysis, % | |
| Favored regions/additionally allowed regions | 92.9/7.1 |
Numbers in parentheses refer to a high-resolution bin of approximate width 0.1 Å.
Results and Discussion
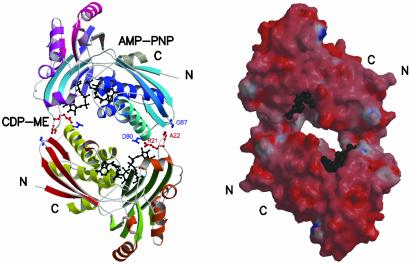
Subunit Architecture and Quaternary Structure. Recombinant E. coli CDP-ME kinase has been expressed, purified, and crystallized. The crystals display space group P21 with unit cell dimensions of a = 61.0, b = 76.3, c = 68.6 Å, and β = 107.8°. The asymmetric unit consists of two subunits, A and B, of approximate total mass 62 kDa. They assemble with C2 symmetry to form an extended homodimer, of approximate dimensions 90 × 50 × 33 Å (Fig. 2), positioned in the unit cell with the NCS axis near perpendicular to the ab plane. The surface-accessible area of the dimer is 25,150 Å2. The surface areas for subunits A and B are ≈13,210 and 12,980 Å2, respectively so ≈1,040 Å2, i.e., only 4% of the total surface area of two monomers, are buried on dimer formation. Despite such a small area of interaction, a dimer is observed in gel filtration experiments and persists even in matrix-assisted laser desorption time-of-flight MS.
Fig. 2.
The CDP-ME kinase dimer. (Left) Ribbon diagram viewed parallel to the NCS 2-fold. The residues shown as red or blue balls and sticks (Arg-21, Ala-22, Asp-80, and Gly-87) contribute to dimer formation through electrostatic interactions. Substrate and cofactor are shown as black balls and sticks. (Right) Surface representation is colored blue for basic and red for acidic residues. Ligands are depicted as black spheres.
The homodimer encloses a solvent-filled channel, ≈23 Å in length and 10 Å wide, with two active sites positioned at either end of this channel (Fig. 2). A single type of subunit–subunit interface, adjacent to the substrate-binding site, creates the dimer and involves the placement of a type-I turn between β2 and β3 of one subunit into a depression formed between the C-terminal section of α1 and β6 of the partner. Salt bridges between Arg-21 on one subunit with Asp-80 of the partner, hydrogen bonds donated from the amide of Gly-87 to the carbonyl of Ala-22, a number of solvent-mediated hydrogen bonding networks, and the hydrophobic interactions between the side chains of Tyr-25 of one subunit and Lys-76 of the partner contribute to the stability of the dimer.
The NCS is well conserved in the asymmetric unit. For example, the rms deviation between 278 Cα atoms is 0.6 Å. The only difference of note is that in subunit A, the γ-phosphate of AMP-PNP is in two positions, each assigned an occupancy of 0.5. One site corresponds to that fully occupied in subunit B and is placed to suggest how catalysis occurs, whereas the other is directed out toward solvent. We only detail the active site of subunit B (see below).
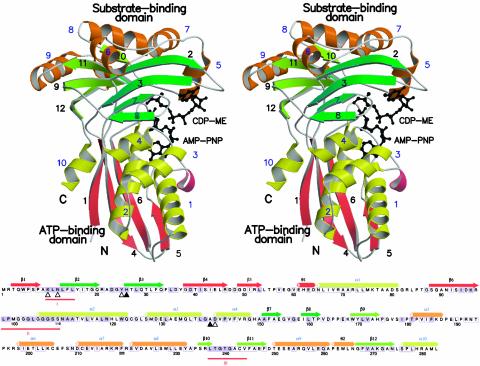
The CDP-ME kinase subunit comprises 10 α-helices (≈40% of residues), 12 β-strands (≈28% of residues), and 2 short 310 sections (Fig. 3). The subunit displays the characteristic two-domain fold of the GHMP kinase superfamily (60). One domain, the cofactor or ATP-binding domain, comprises residues 1–10, 34–150, and 275–283, which form a four-stranded β-sheet (in order β1-β4-β6-β5, with β4 antiparallel to the others) on one side with a five-helix bundle (α1-α4, α10) on the other. The second domain is constructed from residues 11–33 and 151–274 with, at its core, two four-stranded twisted antiparallel β-sheets (β2-β3-β7-β8 and β10-β11-β9-β12) and five helices at the periphery of the domain. This is the CDP-ME or substrate-binding domain.
Fig. 3.
Sequence and structure of E. coli CDP-ME kinase. (Upper) Stereoview ribbon diagram of a monomer (β-strand numbers are black, and α-helix numbers blue). Strands and helices in the cofactor-binding domain are red and yellow, respectively. Strands associated with the two β-sheets in the substrate-binding domain are light and dark green, and the helices are orange. Substrate and AMP-PNP are depicted as black ball-and-stick models. N and C identify the amino and carboxyl termini. (Lower) Amino acid sequence of E. coli CDP-ME kinase with elements of secondary structure assigned and colored according to Upper. Residues strictly conserved or highly homologous in enzymes from S. typhi, Vibrio cholera, Haemophilus influenzae, Pseudomonas aeruginosa, Neisseria meningitidis, M. tuberculosis, Chlamydia trachomatis, and Clostridium perfringens are black on a purple background; when conserved in at least six of the sequences they are enclosed in a purple box. Sequences were retrieved from expasy (www.expasy.ch) and aligned (clustalw, ref. 63). ▵ identify residues interacting with the substrate using side chains; ▴ are for those using main chain. His-26 uses both. θ1 and θ2 identify 310-helices, and three red lines mark motifs I, II, and III.
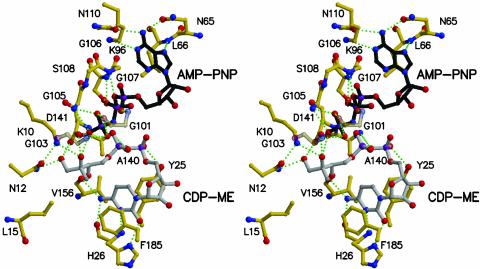
Binding of AMP-PNP and CDP-ME: Specificity and Mechanism. The ATP-binding site is constructed by two sections of the protein structure. One segment comprises the C-terminal section of β6, the N-terminal section of α2, and a glycine-rich phosphate-binding loop linking these elements of secondary structure. The helix dipole of α2 may contribute to phosphate binding. The other segment lies over the purine-binding pocket and comprises the loop, with a turn of 310-helix, linking β5 to α1 (Fig. 3 Upper). The adenine binds in an aliphatic cleft formed by the side chains of Val-57, Val-60, Leu-66, Ile-67, Lys-96, and Met-100, near Cα of Gly-107. The purine forms hydrogen-bonding interactions that stabilize the less common syn orientation of the base with respect to the glycosidic bond. The side-chain carbonyl groups of two asparagines (Asn-65, Asn-110), on either side of Ade N6, accept hydrogen bonds from the amino group (Fig. 4). The asparagines ND2 groups participate in a network of water-mediated hydrogen-bonding interactions to the amide and carbonyl groups of Val-57 and Val-60, respectively. Asn-110 ND2 also interacts with Ser-7 OG and Asp-39 OD2, the latter positioned by hydrogen bonds donated from the main-chain amides of Arg-97 and Leu-98 and a salt bridge with Lys-96. Ade N1 and N7 accept hydrogen bonds from Lys-96 NZ and the amide of Leu-66, respectively.
Fig. 4.
Stereoview of the active site. Bonds in AMP-PNP are black, bonds in the protein are yellow, and bonds in CDP-ME are gray. Selected hydrogen-bonding interactions are shown as green dashed lines. Water molecules have been omitted; Gly-101 and Gly-103 are semitransparent for the purpose of clarity.
The ribose and phosphates of AMP-PNP are solvent accessible. The α-phosphate binds a water molecule, which also interacts with Lys-96 NZ, the Pro-99 carbonyl, and Gly-101 amide. The α–β linking oxygen accepts a hydrogen bond from the amide of Gly-107 whereas the β-phosphate accepts hydrogen bonds donated from the hydroxyl and main-chain amide of Ser-108. A well-ordered water molecule interacts with this phosphate and participates in hydrogen-bonding interactions with Asp-141 OD2, the γ-phosphate of AMP-PNP, and the β-phosphate of the CDP-ME substrate. A γ-phosphate oxygen of AMP-PNP accepts a hydrogen bond donated from the amide group of Gly-105. Another interacts with the carbonyl of Gly-101 and is likely protonated given the close contact of 2.7 Å. The proton may participate in a bifurcated hydrogen bond to the α-phosphate oxygen nearby; alternatively, the α-phosphate itself may also be protonated. Two water molecules interact with the terminal phosphate, one links to the β-phosphate (see above), whereas the other forms hydrogen bonds with the amide group of Thr-240 and the O2M hydroxyl of CDP-ME, the phosphorylation site.
The CDP-ME binding site is formed by contributions from β3, α5, and three turns that link α3 to α4, β7 to β8, and β10 to β11. The cytosine is tucked in toward β3, sandwiched between the aromatic side chains of Tyr-25 and Phe-185 with the ribose directed out of the cleft and placed between Tyr-25 and Pro-182. A water molecule interacts with the ribose O2′ and also Cyt O2. His-26 is a key residue providing three hydrogen-bonding interactions with the pyrimidine. Cyt O2 and N3 accept hydrogen bonds donated by His-26 ND1 and amide, respectively, whereas Cyt N4 donates to the His-26 carbonyl. The His-26 side chain is oriented by NE2 accepting a hydrogen bond from the side chain of Arg-192 (data not shown). The substrate α-phosphate interacts with the hydroxyl group of Tyr-25 and a water molecule. The O link between α- and β-phosphate accepts a hydrogen bond from the amide of Ala-140 and the β-phosphate has solvent-mediated associations with Asp-141 OD2, the β-phosphate of AMP-PNP, and Thr-181 OG1. The CDP-ME tail, with α- and β-phosphates in a staggered conformation, is directed over toward the γ-phosphate of AMP-PNP, and the O1M and O2M hydroxyls form hydrogen bonds with Asp-141 OD1 and OD2, respectively. The CDP-ME O2M group also interacts with the carbonyl of Val-156 via a water bridge whereas O3M accepts hydrogen bonds from Lys-10 NZ and interacts with a water molecule that bridges over to the AMP-PNP γ-phosphate. The CDP-ME O4M hydroxyl accepts a hydrogen bond from Asn-12 ND2 and donates one to Asp-141. Asn-12 is held in place by OD1 accepting hydrogen bonds from Lys-10 NZ and the Gly-239 amide.
The catalytic center is located in a deep cavity near the interface of the cofactor and substrate-binding domains. Lys-10 and Asp-141 are positioned here to participate in hydrogen-bonding interactions with CDP-ME O2M (Fig. 4). This combination of residues would polarize the hydroxyl group and facilitate proton abstraction with Asp-141 acting as a general base to generate the O2M nucleophile and Lys-10 also contributing to the stabilization of the transition state (Fig. 5). The binding of the cofactor places the γ-phosphate to accept nucleophilic attack from CDP-ME O2M. This will likely involve an associative in-line mechanism to form a pentacovalent intermediate, and subsequently the production of 4-diphosphocytidyl-2C-methyl-d-erythritol-2-phosphate and ADP. Such a mechanism is widely recognized as the most likely for enzyme-catalyzed phospho-group transfer (60, 61).
Fig. 5.
The proposed mechanism for CDP-ME kinase.
In contrast to structures of rat MVK (45) and Methanococcus jannaschii HSK (40), we see no evidence for a metal ion binding to the cofactor phosphates despite the presence of magnesium ions in the crystallization buffer. Well-defined peaks in electron and difference density maps in the active sites were assigned as water molecules on the basis of interatomic distances and types of contacts with functional groups. In rat MVK and M. jannaschii HSK, Mg2+ is bound to the ATP triphosphate and coordinated by a conserved glutamic acid residue (Glu-130 and Glu-193, respectively). This residue is not conserved in CDP-ME kinase, which in a structural overlay is occupied by Gly-139 (data not shown).
GHMP kinase superfamily members carry three conserved sequence motifs (40), which in E. coli CDP-ME kinase occur at Lys-11–Tyr-15 (I), Pro-99–Ser-109 (II), and Leu-237–Val-244 (III). They are positioned beside each other at the domain–domain interface and help to create the active site. Motif I forms a bulge after β1 and leading into β2; it is adjacent to motif III, which forms a type II turn between β10 and β11, and together these sections of polypeptide help to create the catalytic center. Motif II forms a loop that closes over then extends into the N-terminal region of α2 and binds the ATP phosphates.
Structural Homologues. An architectural comparison using dali (62) identified a close structural relationship between CDP-ME kinase and four members of the GHMP kinase superfamily, HSK (40, 44), MVK (45), phosphomevalonate kinase (46), and mevalonate 5-diphosphate decarboxylase (42). The overall structure of the ATP-binding domains and the core of the substrate-binding domains are well conserved. However, there are noteworthy differences in the catalytic centers and purine-binding pockets. The structural variation on the surface of these domains contributes to different modes of oligomerization in the GHMP family of enzymes (40), whereas the differences in the catalytic centers reflect the different molecular features required for distinct specificities. In particular, we note that the pyrimidine-binding site in CDP-ME kinase is constructed with an extension to the first β-turn-β unit, a feature lacking in the other enzymes, which process smaller substrates lacking a nucleotide moiety.
The most relevant structures for a detailed comparison are HSK and MVK. These are the best characterized of the GHMP family with structures of ligand complexes having been determined (40, 44, 45) and they are also most similar to CDP-ME kinase. A single subunit of CDP-ME kinase aligned with HSK (Protein Data Bank ID code 1FWK) with a Z-score of 21 and an rms deviation of 3.0 Å for 254 Cα atoms. The alignment with MVK (Protein Data Bank ID code 1KKH) is comparable with a Z-score of 20 and rms deviation of 3.0 Å for 251 Cα atoms. The Z-score is a measure of the statistical significance of the best alignment. Typically, dissimilar proteins will have a Z-score <2.0.
Both HSK and CDP-ME kinase bind the cofactor purine moiety in syn orientation with respect to the ribose. There is strong conservation of secondary structure around the whole ATP-binding site and this extends to conservation of side chains in the purine-binding pocket. In E. coli CDP-ME kinase the aliphatic side chains that contribute to the purine-binding pocket (Leu-66, Ile-67, Leu-53, Ile-94) are replaced by Val-63, Ala-64, Ile-48, and Ile-85 in M. jannaschii HSK. Strikingly, four of the hydrogen-bonding interactions between the base and the enzyme, only possible with the adenine in syn conformation, are also conserved. These involve the side chains of Asn-62, Lys-87, and Asp-34 strictly conserved as Asn-65, Lys-96, and Asp-39. Adjacent to and orientating the lysine side chains are the conserved aspartate residues, Asp-34 and Asp-39 in CDP-ME kinase and HSK, respectively. The remaining hydrogen-bonding interaction involves Asn-110 in CDP-ME kinase that in HSK is Ser-101, positioned such that the amine is replaced by the hydroxyl, thereby retaining the hydrogen bond donating capacity.
By way of contrast, in the binary complex of rat MVK-ATP the nucleotide adopts the more common antiorientation (43). Although much of the MVK ATP-binding domain overlays well with the corresponding domain of CDP-ME kinase we note that MVK carries an additional β-strand, producing an extended β-sheet, and a completely different loop leading into helix α1. In addition, the adenine-binding pocket of MVK carries specific amino acid side chains (e.g., Tyr-80 and Ile-120, which correspond to Leu-66 and Asn-110 in E. coli CDP-ME kinase), which on steric considerations would preclude a syn conformation for the base.
As described, the ternary complex structure suggests a likely mechanism based on the combination of Lys-10 and Asp-141. The mechanism postulated for HSK is different because there is no obvious catalytic base in that active site (40), and we note that the corresponding positions of Lys-10 and Asp-141 in HSK are Thr-14 and Asn-141. MVK, on the other hand, has the same lysine-aspartate combination (Lys-13 and Asp-160 in the M. jannaschii enzyme) and is likely to catalyze phosphoryl transfer in similar fashion to CDP-ME kinase.
Functional Homologues. The sequences of CDP-ME kinase from eight species, selected to be representative of bacterial pathogens, were aligned with the sequence of the E. coli enzyme (Fig. 3 Lower). Pair-wise alignments indicate that identity and homology range from 90% to 95% between E. coli and Salmonella typhi enzymes to 21% and 38% between E. coli and Mycobacterium tuberculosis enzymes, respectively. The averages are 45% and 60% over all species. Amino acid conservation extends throughout the sequence and is particularly well maintained in elements of secondary structure and in the active sites. Some 29 residues have been discussed above in the context of direct or solvent-mediated interactions involving CDP-ME kinase with ligands and also in organizing the active site. Of these, 16 are absolutely conserved in all nine sequences and an additional 9 are strictly conserved in at least six of the sequences.
In conclusion, our crystallographic analysis of a ternary complex of E. coli CDP-ME kinase has delineated the structure, specificity, and mechanism of the enzyme responsible for phosphorylation in the nonmevalonate route of isoprenoid biosynthesis and one shown to be essential for cell viability. The high level of sequence conservation indicates a well-conserved structure of CDP-ME kinase in microbial pathogens, and our model provides a high-resolution template for a structure-based approach to aid the search for inhibitors of the enzyme and subsequently of isoprenoid biosynthesis. Such inhibitors could be lead compounds to support the development of novel broad-spectrum antimicrobial drugs.
Acknowledgments
We thank the European Synchrotron Radiation Facility for access and support and Charles Bond for advice. This work was funded by the Wellcome Trust, the Engineering and Physical Sciences Research Council–Biotechnology and Biological Sciences Research Council, and the European Synchrotron Radiation Facility (to W.N.H.), and the Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie, and Hans-Fischer Gesellschaft (to A.B.).
Abbreviations: AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate; CDP-ME, 4-diphosphocytidyl-2C-methyl-d-erythritol; DMAPP, dimethylallyl diphosphate; DOXP, 1-deoxy-d-xylulose-5-phosphate; GHMP, galactose kinase/homoserine kinase/mevalonate kinase/phosphomevalonate kinase; HSK, homoserine kinase; IPP, isopentenyl diphosphate; MVK, mevalonate kinase; NCS, noncrystallographic symmetry.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 10J4).
Note Added in Proof. Further refinement has resolved the detail of the cofactor γ-phosphate in active site A. This group now occupies a single position similar to that in active site B.
References
- 1.Sacchettini, J. C. & Poulter, C. D. (1997) Science 277, 1788–1789. [DOI] [PubMed] [Google Scholar]
- 2.Rohmer, M. (1999) in Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Sterols, ed. Cane, D. (Elsevier, Amsterdam), Vol. 2, pp. 45–67. [Google Scholar]
- 3.Beytia, E. D. & Porter, J. W. (1976) Annu. Rev. Biochem. 45, 113–142. [DOI] [PubMed] [Google Scholar]
- 4.Edwards, P. A. & Ericsson, J. (1999) Annu. Rev. Biochem. 68, 157–185. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, Y. & Doolittle, W. F. (2000) Mol. Microbiol. 37, 703–716. [DOI] [PubMed] [Google Scholar]
- 6.Kuzuyama, T. (2002) Biosci. Biotechnol. Biochem. 66, 1619–1627. [DOI] [PubMed] [Google Scholar]
- 7.Lange, B. M., Rujan, T., Martin, W. & Croteau, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13172–13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rohdich, F., Kis, K., Bacher, A. & Eisenreich, W. (2001) Curr. Opin. Chem. Biol. 5, 535–540. [DOI] [PubMed] [Google Scholar]
- 9.Sprenger, G. A., Schorken, U., Weigert, T., Grolle, S., de Graaf, A. A., Taylor, S. V., Begley, T. P., Bringer-Meyer, S. & Sahm, H. (1997) Proc. Natl. Acad. Sci. USA 25, 12857–12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lois, L. M., Campos, N., Putra, S. R., Danielsen, K., Rohmer, M. & Boronat, A. (1998) Proc. Natl. Acad. Sci. USA 95, 2105–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, S., Kuzuyama, T., Watanabe, H. & Seto, H. (1998) Proc. Natl. Acad. Sci. USA 95, 9879–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzuyama, T., Takahashi, S., Takagi, M. & Seto, H. (2000) J. Biol. Chem. 30, 19928–19932. [DOI] [PubMed] [Google Scholar]
- 13.Rohdich, F., Wungsintaweekul, J., Fellermeier, M., Sagner, S., Herz, S., Kis, K., Eisenreich, W., Bacher, A. & Zenk, M. H. (1999) Proc. Natl. Acad. Sci. USA 96, 11758–11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuzuyama, T., Takagi, M., Kaneda, K., Dairi, T. & Seto, H. (2000) Tetrahedron Lett. 41, 703–706. [Google Scholar]
- 15.Richard, S. B., Bowman, M. E., Kwiatkowski, W., Kang, I., Chow, C., Lillo, A. M., Cane, D. E. & Noel, J. P. (2001) Nat. Struct. Biol. 8, 641–647. [DOI] [PubMed] [Google Scholar]
- 16.Lüttgen, H., Rohdich, F., Herz, S., Wungsintaweekul, J., Hecht, S., Schuhr, C. A., Fellermeier, M., Sagner, S., Zenk, M. H., Bacher, A. & Eisenreich, W. (1999) Proc. Natl. Acad. Sci. USA 97, 1062–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuzuyama, T., Takagi, M., Kaneda, K., Watanabe, H., Dairi, T. & Seto, H. (2000) Tetrahedron Lett. 41, 2925–2928. [Google Scholar]
- 18.Takagi, M., Kuzuyama, T., Kaneda, K., Watanabe, H., Dairi, T. & Seto, H. (2000) Tetrahedron Lett. 41, 3395–3398. [Google Scholar]
- 19.Herz, S., Wungsintaweekul, J., Schuhr, C. A., Hecht, S., Lüttgen, H., Sagner, S., Fellermeier, M., Eisenreich, W., Zenk, M. H., Bacher, A. & Rohdich, F. (2000) Proc. Natl. Acad. Sci. USA 97, 2486–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altincicek, B., Duin, E. C., Reichenberg, A., Hedderich, R., Kollas, A. K., Hintz, M., Wagner, S., Wiesner, J., Beck, E. & Jomaa, H. (2002) FEBS Lett. 532, 437–440. [DOI] [PubMed] [Google Scholar]
- 21.Kollas, A. K., Duin, E. C., Eberl, M., Altincicek, B., Hintz, M., Reichenberg, A., Henschker, D., Henne, A., Steinbrecher, I., Ostrovsky, D. N., et al. (2002) FEBS Lett. 532, 432–436. [DOI] [PubMed] [Google Scholar]
- 22.Hecht, S., Eisenreich, W., Adam, P., Amslinger, S., Kis, K., Bacher, A., Arigoni, D. & Rohdich, F. (2001) Proc. Natl. Acad. Sci. USA 98, 14837–14842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rohdich, F., Hecht, S., Gärtner, K., Adam, P., Krieger, C., Amslinger, D., Arigoni, D., Bacher, A. & Eisenreich, W. (2002) Proc. Natl. Acad. Sci. USA 99, 1158–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzuyama, T., Shimizu, T., Takahashi, S. & Seto, H. (1998) Tetrahedron Lett. 39, 7913–7916. [Google Scholar]
- 25.Jomaa, H., Weisner, J., Sanderbrand, S., Altincicek, B., Weidemeyer, C., Hintz, M., Turbachova, I., Eberl, M., Zeidler, J., Lichtenthaler, H. K., et al. (1999) Science 285, 1573–1576. [DOI] [PubMed] [Google Scholar]
- 26.Missinou, M. A., Borrmann, S., Schindler, A., Issifou, S., Adegnika, A. A., Matsiegui, P. B., Binder, R., Lell B., Wiesner, J., Baranek, T., et al. (2002) Lancet 360, 1941–1942. [DOI] [PubMed] [Google Scholar]
- 27.Freiberg, C., Wieland, B., Spaltmann, F., Ehlert, K., Brotz, H. & Labischinski, H. (2001) J. Mol. Microbiol. Biotechnol. 3, 483–489. [PubMed] [Google Scholar]
- 28.Campos, N., Rodriguez-Concepcion, M., Sauret-Gueto, S., Gallego, F., Lois, L. M. & Boronat, A. (2001) Biochem. J. 353, 59–67. [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell, T. L. & Brown, E. D. (2002) J. Bacteriol. 184, 5609–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campos, N., Rodrguez-Concepcion, M., Seemann, M., Rohmer, M. & Boronat A. (2001) FEBS Lett. 488, 170–173. [DOI] [PubMed] [Google Scholar]
- 31.Altincicek, B., Kollas, A., Eberl, M., Wiesner, J., Sanderbrand, S., Hintz, M., Beck, E. & Jomaa, H. (2001) FEBS Lett. 499, 37–40. [DOI] [PubMed] [Google Scholar]
- 32.Reuter, K., Sanderbrand, S., Jomaa, H., Wiesner, J., Steinbrecher, I., Beck, E., Hintz, M., Klebe, G. & Stubbs, M. T. (2002) J. Biol. Chem. 277, 5378–5384. [DOI] [PubMed] [Google Scholar]
- 33.Yajima, S., Nonaka, T., Kuzuyama, T., Seto, H. & Ohsawa, K. (2002) J. Biochem. (Tokyo) 131, 313–317. [DOI] [PubMed] [Google Scholar]
- 34.Steinbacher, S., Kaiser, J., Eisenreich, W., Huber, R., Bacher, A. & Rohdich, F. (2003) J. Biol. Chem. 278, 18401–18407. [DOI] [PubMed] [Google Scholar]
- 35.Kemp, L. E., Bond, C. S. & Hunter, W. N. (2003) Acta Crystallogr. D 59, 607–610. [DOI] [PubMed] [Google Scholar]
- 36.Steinbacher, S., Kaiser, J., Wungsintaweekul, J., Hecht, S., Eisenreich, W., Gerhardt, S., Bacher, A. & Rohdich, F. (2002) J. Mol. Biol. 316, 79–88. [DOI] [PubMed] [Google Scholar]
- 37.Richard, S. B., Ferrer, J.-L., Bowman, M. E., Lillo, A. M., Tetzlaff, C. N., Cane, D. E. & Noel, J. P. (2002) J. Biol. Chem. 277, 8667–8672. [DOI] [PubMed] [Google Scholar]
- 38.Kemp, L. E., Bond, C. S. & Hunter, W. N. (2002) Proc. Natl. Acad. Sci. USA 99, 6591–6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kishida, H., Wada, T., Unzai, S., Kuzuyama, T., Takagi, M., Terada, T., Shirouzu, M., Yokoyama, S., Tame, J. R. H. & Park, S.-Y. (2003) Acta Crystallogr. D 59, 23–31. [DOI] [PubMed] [Google Scholar]
- 40.Krishna, S. S., Zhou, T., Daugherty, M., Osterman, A. L. & Zhang, H. (2001) Biochemistry 40, 10810–10818. [DOI] [PubMed] [Google Scholar]
- 41.Bork, P., Sander, C. & Valencia, A. (1993) Protein Sci. 2, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonanno, J. B., Edo, C., Eswar, N., Pieper, U., Romanowski, M. J., Ilyin, V., Gerchman, S. E., Kycia, H., Studier, F. W., Sali, A. & Burley, S. K. (2001) Proc. Natl. Acad. Sci. USA 98, 12896–12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daugherty, M., Vonstein V., Overbeek, R. & Osterman, A. (2001) J. Bacteriol. 183, 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, T., Daugherty, M., Grishin, N. V., Osterman, A. L. & Zhang, H. (2000) Structure (London) 8, 1247–1257. [DOI] [PubMed] [Google Scholar]
- 45.Fu, Z., Wang, M., Potter, D., Miziorko, H. M. & Kim, J.-J. (2002) J. Biol. Chem. 277, 18134–18142. [DOI] [PubMed] [Google Scholar]
- 46.Yang, D., Shipman, L. W., Roessner, C. A., Scott, A. I. & Sacchettini J. C. (2002) J. Biol. Chem. 277, 9462–9467. [DOI] [PubMed] [Google Scholar]
- 47.Romanowski, M. J., Bonanno, J. B. & Burley, S. K. (2002) Proteins 47, 568–571. [DOI] [PubMed] [Google Scholar]
- 48.Rohdich, F., Schuhr, C. A., Hecht, S., Herz, S., Wungsintaweekul, J., Eisenreich, W., Zenk, M. H. & Bacher, A. (2000) J. Am. Chem. Soc. 122, 9571–9594. [Google Scholar]
- 49.Otwinowski, Z. & Minor, W. (1996) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 50.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terwilliger, T. C. (2000) Acta Crystallogr. D 56, 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 53.Perrakis, A., Morris, R. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 54.Murshodov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 55.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–764.15299374 [Google Scholar]
- 56.Kleywegt, G. J. & Brünger, A. T. (1996) Structure (London) 4, 897–904. [DOI] [PubMed] [Google Scholar]
- 57.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 58.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 59.Hutchinson, E. G. & Thornton, J. M. (1996) Protein Sci. 5, 212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheek, S., Zhang, H. & Grishin, N. V. (2002) J. Mol. Biol. 320, 855–881. [DOI] [PubMed] [Google Scholar]
- 61.Knowles, J. (2003) Science 299, 2002–2003. [DOI] [PubMed] [Google Scholar]
- 62.Holm, L. & Sander, C. (1995) Trends Biochem. Sci. 20, 478–480. [DOI] [PubMed] [Google Scholar]
- 63.Thomson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]