Abstract
The natural form of the hairpin ribozyme comprises two major structural elements: a four-way RNA junction and two internal loops carried by adjacent arms of the junction. The ribozyme folds into its active conformation by an intimate association between the loops, and the efficiency of this process is greatly enhanced by the presence of the junction. We have used single-molecule spectroscopy to show that the natural form fluctuates among three distinct states: the folded state and two additional, rapidly interconverting states (proximal and distal) that are inherited from the junction. The proximal state juxtaposes the two loop elements, thereby increasing the probability of their interaction and thus accelerating folding by nearly three orders of magnitude and allowing the ribozyme to fold rapidly in physiological conditions. Therefore, the hairpin ribozyme exploits the dynamics of the junction to facilitate the formation of the active site from its other elements. Dynamic interplay between structural elements, as we demonstrate for the hairpin ribozyme, may be a general theme for other functional RNA molecules.
Ribozymes, cellular RNA molecules that catalyze chemical reactions, have fundamental implications for the evolution of life on the planet and provide insight into biocatalysis in general (1). Like protein enzymes, they must fold into a conformation that provides a local environment in which catalysis can proceed. Branched helical junctions are very common in RNA and frequently are essential for their function (2), but the dynamic mechanisms by which the junctions contribute to RNA folding are not well understood.
The hairpin ribozyme is one of the nucleolytic ribozymes that bring about a site-specific cleavage of the RNA backbone by means of a transesterification reaction in which the 2′ oxygen attacks the 3′ phosphorus, thereby breaking the bond to the 5′ oxygen. The natural form of the hairpin ribozyme comprises two major structural elements: a four-way RNA junction and two internal loops carried by adjacent A and B arms of the junction (Fig. 1a). The loop–loop interaction is essential for the ribozyme activity, generating a 105-fold acceleration of site-specific cleavage or ligation reactions. Although the minimal form without arms C and D can still catalyze the cleavage reaction and has yielded much insight into the catalytic mechanism (3), its folding requires two to three orders of magnitude higher Mg2+ concentration (4–6), and the internal equilibrium between cleavage and ligation is shifted compared with the natural form (7).
Fig. 1.
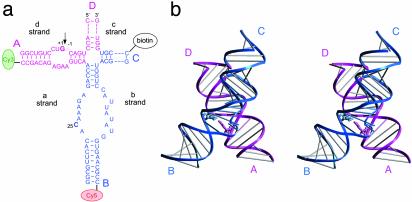
(a) The hairpin ribozyme used in FRET studies. The secondary structure comprises two internal loops in adjacent arms (A and B) of a four-way junction. Ribozyme folding brings the donor and acceptor fluorophores closer to each other, increasing FRET. The cleavage site is marked by an arrow; however, cleavage activity is prevented by substitution of a deoxyribonucleotide at the –1 position. (b) Stereo view of the crystal structure of the hairpin ribozyme (9), with the G+1–C25 interaction highlighted. The helical arms are coaxially stacked in pairs, A on D (magenta) and B on C (blue).
Ensemble fluorescence resonance energy transfer (FRET) studies have shown that the two loops are brought into close proximity at submillimolar Mg2+ concentrations (4, 8), and extensive contacts between them have been identified from crystallographic analysis (9). The folded ribozyme has the neighboring helical arms coaxially stacked in pairs, A on D and B on C, with an antiparallel orientation of the continuous strands (Fig. 1b). The simple junction generated by replacing the loops with Watson–Crick base pairing (which we call the 4H junction to distinguish it from the junction in the context of the ribozyme) also exhibits coaxial stacking of arms with the same stacking preference (10). However, the axes of the 4H junction were found to be approximately perpendicular at submillimolar Mg2+ concentrations. Because the ribozyme folds efficiently under these conditions, it was unclear how the junction could assist the folding if there is substantial separation of arms A and B as indicated by the perpendicular structure.
Single-molecule studies can reveal details of biochemical reactions that are invisible in ensemble measurements (reviewed in refs. 11 and 12). For example, folding and unfolding studies of individual RNA molecules revealed multiple pathways and new intermediates (13, 14). In particular, single-molecule FRET methods (15) provide a powerful means of observing the dynamic structural changes of biomolecules as well as subpopulations in a heterogeneous mixture (13, 16–20). Applying this approach to the hairpin ribozyme and 4H junction, we have obtained evidence that the hairpin ribozyme folds through a discrete intermediate by using the dynamics of the four-way junction.
Recently, single-molecule fluorescence studies of the minimal form of the hairpin ribozyme (i.e., lacking the junction) dissected the folding and reaction kinetics, finding a single rate of folding of 0.008 s–1, with significant heterogeneity in the rate of unfolding, which could quantitatively explain complex cleavage kinetics (16). In our studies of the natural form, we find that the rate of folding is nearly three orders of magnitude faster, with heterogeneity in the rates of both folding and unfolding processes. We propose that the previously unknown intermediate, absent in the minimal form, is responsible for the large acceleration of the hairpin ribozyme folding in its natural form.
Materials and Methods
RNA Preparation. RNA was synthesized by using tert-butyldimethylsilyl/phosphoramidite chemistry or purchased from Dharmacon (Boulder, CO) and deprotected and purified as described previously (21). The hairpin ribozyme and 4H junction were assembled from oligonucleotides of the sequences given in Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Single-Molecule Spectroscopy. Single-molecule fluorescence measurements were made by using both a wide-field prism-type total internal reflection microscope (with up to 50-ms time resolution) (13) and a confocal scanning microscope (with up to 10-μs time resolution) (22). A detailed description is provided in Supporting Text.
Determination of Rates by Dwell-Time Analysis. After correcting for donor leakage into the acceptor channel and the direct excitation of the acceptor, the apparent FRET efficiency Eapp was calculated by using Eapp = IA/(IA + ID), where IA and ID are the intensities of the sensitized emission of the acceptor and the donor emission, respectively (23). Because the quantum yields and detection efficiencies of Cy3 and Cy5 are similar, Eapp closely parallels the true efficiency of energy transfer. Distances were calculated by using an experimentally determined Ro of 60 Å (under the assumption that orientational factor κ2 = 2/3) for Cy3/Cy5. States were identified from Eapp histograms, and dwell times were measured for each state if the time resolution of the data allowed the clear observation of transitions. To avoid bias toward fast fluctuating molecules that show many more transitions than slowly fluctuating molecules, dwell-time histograms were derived by using a weighting factor inversely proportional to the number of transitions observed from each molecule. Such histograms were fit by an exponential decay function to obtain the average dwell times. In the analysis of heterogeneity, the algebraic average of the dwell times was calculated for each state of individual molecules because of the limited number of transitions observed before photobleaching.
Determination of Rates of Conformational Change by Cross-Correlation. Fast junction dynamics were analyzed by cross-correlating donor and acceptor signals (22). ID and IA were measured from individual junctions with 10-μs time resolution by using the confocal microscope. The cross-correlation function, CC(Δt), was calculated according to ∫ID(t)IA(t – Δt)dt and averaged over at least 20 molecules under each condition. For a two-state fluctuation such as between UD and UP, CC(Δt) takes the form Ae–Δt/τ where τ is the correlation time and is an inverse of the sum of the forward and backward rates (kD→P + kP→D) (22).
Measurement of Cleavage Activity. The substrate strand (d′) was shortened so that the 3′ end of the cleaved product was rapidly released (7). The ribozymes were immobilized in a buffer containing no Mg2+, and a flow system employing a syringe pump was used to deliver 10 mM Mg2+ solution while monitoring single-molecule fluorescence signals at a reduced exposure rate to minimize photobleaching (one 100-ms frame per second) by using the wide-field microscope. The same RNA species were used for bulk measurements of the rate of cleavage. After addition of Mg2+ ions, the cleavage reaction was observed as a change in fluorescence intensity. Cleavage activity in bulk solution was also measured by using radiolabels (27) instead of fluorescence labels.
Results and Discussion
We assembled hairpin ribozyme molecules from four RNA strands each. Donor (Cy3) and acceptor (Cy5) fluorophores were attached to the 5′ termini of the A and B arms, respectively (Fig. 1a). The relative proximity of the two arms can be deduced by measuring the FRET efficiency between the fluorophores. The molecules were attached to a streptavidin-coated glass or quartz surface by means of biotin conjugated to the 5′ terminus of the C arm. Both the cleavage reaction and folding kinetics of single molecules showed highly heterogeneous behavior with similar rates to those measured in bulk solution (Fig. 4; see also Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 4.
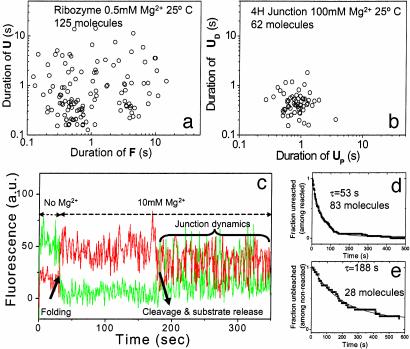
Heterogeneity in ribozyme folding and cleavage reaction kinetics. (a) Average duration of U vs. average duration of F for 125 ribozyme molecules showing up to 50-fold heterogeneity. (b) Average duration of UD vs. average duration of UP for 62 4H junction molecules showing much narrower distribution than in a. (c) Analysis of substrate cleavage in a single ribozyme molecule with a shortened substrate so that the 3′ product is rapidly released. Upon addition of Mg2+, the molecule folds, resulting in increased FRET. After 180 s, the molecule exhibits frequent fluctuations in FRET characteristic of the 4H junction; we deduce that the substrate has now been cleaved and released, leading to loss of loop–loop interaction. (d) Fraction unreacted vs. time since Mg2+ addition for the 83 molecules that showed the ribozyme reaction before photobleaching, fitted to a single exponential decay. (e) Fraction not photobleached vs. time since Mg2+ addition for the 28 molecules that underwent photobleaching before cleavage could occur, fitted to a single exponential decay.
Folding and Unfolding Transitions of the Hairpin Ribozyme. Single-molecule time records of the ribozyme display clear transitions between states with high and low FRET efficiency (corresponding to significant differences in the separation of the loop-carrying A and B arms), but the relative durations of these states exhibits a strong dependence on Mg2+ concentration (Fig. 2a). At 0.1 mM Mg2+, ribozymes remain mostly in a low FRET state (Eapp ≈ 0.25), with brief excursions into a high FRET state (Eapp ≈ 0.9). With an increase in Mg2+ concentration, the dwell times increase for the high FRET state and decrease for the low FRET state. Above 1 mM Mg2+, the ribozyme molecules remain mostly in the high FRET state until the fluorophores photobleach. The ribozyme is folded and active under these conditions, and the Eapp corresponds to a distance of ≈40 Å, consistent with the separation of the A and B helices in the folded ribozyme (Fig. 1b). Therefore, we assign the high FRET state to the folded ribozyme (termed F) as observed in the crystal (9). The low FRET state is termed U for unfolded ribozyme, although measurements with better time resolution show that U is actually a mixture of at least two different states (see below). Whereas the time traces shown in Fig. 2a illustrate the stabilization of the F state over the U state with increasing Mg2+ concentration, it should be noted that these rates exhibit significant heterogeneity between molecules, as discussed below. Fig. 6, which appears as supporting information on the PNAS web site, shows additional traces at 0.5 mM Mg2+.
Fig. 2.
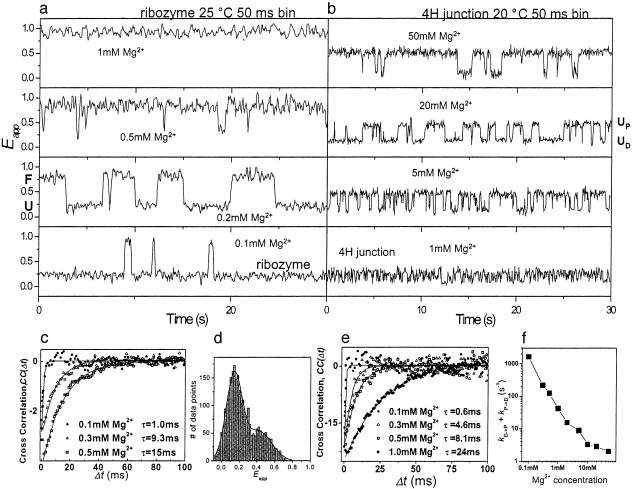
Structural transitions in single hairpin ribozyme and 4H junctions. Shown are single-molecule time records of Eapp for the hairpin ribozyme (a) and 4H junction (b). (c) ID and IA were measured from individual hairpin ribozymes in the unfolded state with 10-μs time resolution. Cross-correlation was averaged over at least 20 molecules under each condition. Negative cross-correlations indicate FRET changes due to conformational changes between UD and UP. Single exponential decay fits are shown. Data for 0.5 mM Mg2+ were obtained by using a G+1A variant, which does not fold at this Mg2+ concentration. (d) Eapp histogram of a single ribozyme in the unfolded state at 0.2 mM Mg2+ with two Gaussian fits. The proximal (≈0.43) and distal (≈0.15) states comprise 40% and 60% of the total population, respectively. (e) Cross-correlation between ID and IA for the 4H junction with single exponential decay fits. (f) The rate of 4H junction fluctuations (kD→P + kP→D) obtained from the data shown in e as a function of Mg2+ concentration. The data points at 2, 5, 10, 20, and 50 mM Mg2+ were obtained from the dwell-time analysis.
Dynamics of the 4H Junction. Single-molecule time records of the 4H junction at various Mg2+ concentrations exhibit fundamentally different structural dynamics. The 4H junction has been proposed to convert from a perpendicular to an antiparallel structure with increasing Mg2+ concentration (10). Therefore, we expected that the 4H junction would exhibit anti-correlated changes in the dwell times of the two states with increasing Mg2+ concentrations. Strikingly, single-molecule studies revealed a qualitatively different behavior. The 4H junction underwent rapid fluctuations between two states with Eapp ≈ 0.5 and Eapp ≈ 0.15 that we refer to as UP (proximal) and UD (distal), respectively, and the rates of fluctuations increased for decreasing Mg2+ concentration (Fig. 2b). Clearly the FRET efficiency of the higher FRET state is significantly lower than that of the ribozyme. The relative populations of the two states remained approximately constant as Mg2+ concentration was varied (≤10 mM) (Fig. 7, which is published as supporting information on the PNAS web site), in stark contrast to the behavior of the ribozyme. Therefore, the dynamics cannot be synchronized by changing the Mg2+ concentration, and stopped-flow studies in this Mg2+ concentration range do not reveal changes in FRET (data not shown). It is very likely that the proposed perpendicular structure of the 4H junction is in fact due to time-averaging of the two conformations. At Mg2+ concentrations ≥20 mM, a bias toward UP is observed (Fig. 7), consistent with the previous ensemble studies that indicated the formation of an antiparallel structure (10). The estimated distance of ≈60 Å between the fluorophores is also consistent with helices A and B subtending an acute angle. Therefore, we assign the UP state to an antiparallel structure with A on D stacking analogous to that observed in the complete ribozyme (4, 9). Previous time-resolved FRET studies of the 4H junction (in 12 mM Mg2+) also detected two populations (5) but assigned the higher FRET state to the folded (docked) state instead of a junction-specific state, and it was not possible to determine whether and how fast the two populations interconvert.
At concentrations <1 mM Mg2+, the fluctuations of the 4H junction are too fast to be clearly resolved, so a cross-correlation analysis was used to determine their rates (22). ID and IA were measured with 10-μs time resolution, and the cross-correlation function, CC(Δt), was calculated (Fig. 2e). In all cases, CC(Δt) showed negative values at Δt = 0 and gradually decayed to zero at larger Δt, indicative of anti-correlated fluctuations of ID and IA. The correlation time, τ, obtained via single exponential decay fit, became longer with increasing Mg2+ concentration, merging with τ measured via dwell-time analysis at higher Mg2+ concentrations (Fig. 2f). We conclude that the 4H junction fluctuates between the UD and UP states over a wide range of Mg2+ concentrations (0.1–50 mM).
The Ribozyme Inherits and Exploits the Junction Dynamics. Because the UP state brings the A and B arms of the 4H junction (loop-carrying arms in the ribozyme) into close proximity, we hypothesized that the unfolded state of the ribozyme exhibits fluctuations quantitatively similar to the 4H junction and that the UP state is an intermediate that precedes the folding of the ribozyme. To test whether the dynamics of the 4H junction are preserved for the ribozyme, we required conditions where the unfolded state is substantially populated (<1 mM Mg2+). Because the junction dynamics are too fast for dwell-time analysis under these conditions (Fig. 8, which is published as supporting information on the PNAS web site), we performed a cross-correlation analysis of ID and IA for the unfolded data segments only. As for the 4H junction, CC(Δt) was negative near zero time, and τ became shorter at lower Mg2+ concentrations and was within a factor of two of τ for the 4H junction under the same conditions (Fig. 2c). Furthermore, the Eapp histogram of state U of a single molecule binned at 3 ms can be fitted by two Gaussian distributions, centered at 0.15 and 0.4 (Fig. 3d), closely corresponding to those of UD and UP for the 4H junction. We conclude that the unfolded ribozyme fluctuates between two states very similar to UD and UP of the 4H junction, henceforth called UD and UP for the ribozyme as well.
Fig. 3.
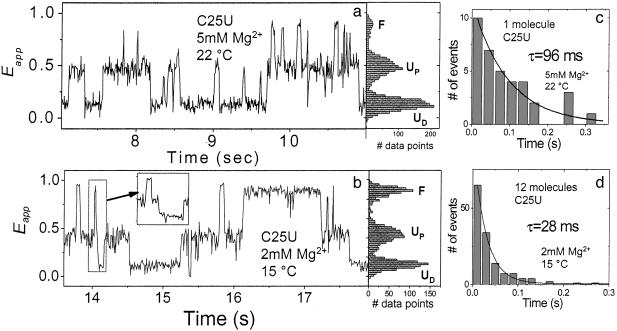
An intermediate folding state in a C25U variant hairpin ribozyme. (a and b) Examples of time records for two variant ribozyme molecules, with corresponding Eapp histogram [10-ms (a) and 9-ms (b) bin times]. b Inset shows an expanded plot (3-ms bin time) of the data segment marked by a dashed rectangle. (c and d) Dwell-time histograms of UP during transitions between UD and F and single exponential decay fits. If the UP was not resolved within the bin time used, zero was assigned as the dwell time.
The existence of three distinct states in the hairpin ribozyme (UD, UP, and F) is a new finding afforded by the sensitivity of single-molecule measurements. Previous time-resolved ensemble FRET measurements of the natural form of the hairpin ribozyme did not detect three populations and have been interpreted by using a two-state folding model, docked (folded) and undocked (unfolded) (5, 24). Because these studies could not distinguish between the FRET values of the higher FRET states of the 4H junction (UP) and the docked ribozyme (F), the docked state in their analysis may have included the proximal state (hence UP and F) instead of the pure docked state (F) only. We also note that such a proximal state is absent in the minimal form of the hairpin ribozyme (16), which displayed two-state fluctuations.
These results provide new insight into the mechanism of the hairpin ribozyme folding. The rapid conformational exchange within the unfolded state (UD ↔ UP) repeatedly brings the two loop elements into proximity, increasing the probability of interaction between them and thus folding. This is consistent with thermodynamic studies that found a reduction in the entropic cost of folding for the natural form of the ribozyme compared with the minimal form (24). Loop–loop interaction involves numerous specific contacts (9), and the local conformation of the loops in the folded ribozyme is significantly altered from that in the isolated loops (25, 26). Therefore, multiple conformational adjustments will be required to achieve the active state. The UD ↔ UP transition occurs at a rate of ≈100 s–1 in 0.5 mM Mg2+, whereas we show below that the rate of formation of the F state is significantly slower, ≤3 s–1 (Fig. 4a). The folding under these conditions must be limited by the conformational changes involved in loop–loop interaction rather than by the junction dynamics.
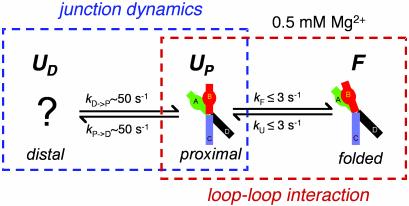
The Intermediate State Observed in a Folding-Impaired Sequence Variant of the Ribozyme. Our model predicts that the UP state is an obligatory intermediate in a sequential folding pathway (UD ↔ UP ↔ F, see Fig. 5). The large rate differences between UD ↔ UP and UP ↔ F make it difficult to test the obligatory nature of UP because below 1 mM Mg2+ UP is too short-lived for such a test, whereas at or above 1 mM Mg2+ the ribozyme spends most of the time in the F state. Therefore, we studied a sequence variant (C25U) that is impaired in folding (27). The crystal structure reveals that G+1 is extruded from loop A and is inserted into a pocket within loop B, where it forms a Watson–Crick base pair with C25 of loop B (Fig. 1b). This pairing is perturbed in C25U, destabilizing the F state and allowing us to observe unfolded ribozymes in millimolar Mg2+ concentrations where the fluctuations between UD and UP are slow enough to be directly resolved. Indeed, as shown in Fig. 3 a and b, C25U clearly exists in three states corresponding to UD, UP, and F. These assignments are justified because (i) the lifetime of UD for the 4H junction and the C25U remain within a factor of 1.2 of each other for three different Mg2+ concentrations (Fig. 9, which is published as supporting information on the PNAS web site), (ii) FRET values are consistent with the assignments, and (iii) previous studies have shown that C25U possesses activity, albeit 200-fold lower (27), and therefore state F must be accessible. This is an equilibrium observation of three-state structural transitions by single-molecule fluorescence measurements.
Fig. 5.
Proposed model for the stepwise folding of the hairpin ribozyme. The ribozyme undergoes folding into the active conformation in two stages, corresponding to the properties of the junction and the interactions between the loops. The structural dynamics of the junction promote rapid active site formation due to the spontaneous fluctuations between the two states of the junction, UD and UP. State UD comprises a number of distinct conformations that cannot be distinguished on the basis of our current data. The ribozyme exploits the frequent encounters between the two loops in UP to achieve the F state. Single-molecule spectroscopy provides information on the previous and subsequent history of a given state, allowing us to conclude that the UP state is an obligatory intermediate in the formation of the ultimate F state. The rates shown are measured in the presence of 0.5 mM Mg2+ ions.
Single-molecule time records clearly demonstrate that direct transitions between UD and F are rare. Even when a transition from F to UD seems abrupt (Fig. 3b, dashed rectangle), a close inspection with a shorter bin time (3 ms vs. 9 ms) usually reveals that the molecule passes through UP (Fig. 3b Inset). Of 36 transitions from UD to F of from F to UD for the molecule shown in Fig. 3a (5 mM Mg2+, 22°C), four seemed to be bypassing UP within 10-ms. time resolution, whereas the 96-ms average lifetime of UP (Fig. 3c) predicts that 10% of them will be shorterlived than 10 ms. Similarly, among 140 transitions observed from 12 molecules at 2 mM Mg2+ and 15°C, 25 (18%) showed a <6-ms duration, whereas the 28-ms lifetime of UP predicts 19% (Fig. 3d). Although this analysis does not rigorously rule out the possibility that a small fraction of transitions between UD and F may occur without passing through UP, the proportion of apparent direct conversions between UD and F is fully explicable in terms of the lifetime of the UP state and the time resolution of the data. Therefore, it is highly probable that UP is an obligatory intermediate in the ribozyme folding.
Persistent Heterogeneity in both Folding and Unfolding Rates Originates from the Loops. The relative populations of the three states vary significantly among C25U molecules, indicating the molecular heterogeneity in the stability of the F state (16). To further our understanding of the folding kinetics, we characterized the statistically significant heterogeneities in the folding and unfolding rates among individual ribozymes without the C25U modification. Average dwell times of the F and U states were obtained from each molecule, and a scatter plot (Fig. 4a) shows a 50-fold variation for both. We have observed large heterogeneities in ribozymes immobilized using two alternative strategies (BSA-biotin vs. PEG-biotin; ref. 28; see Fig. 10, which is published as supporting information on the PNAS web site), for ribozymes synthesized by using either tert-butyldimethylsilyl or ACE chemistries and in 1 M Na+ without Mg2+ (Fig. 10). The heterogeneity seems to be due mainly to the loops, because the average dwell times of the UD and UP states of the 4H junction exhibit a much narrower distribution (Fig. 4b). Stopped-flow kinetic studies of folding and unfolding also displayed evidence for heterogeneous kinetics with rates that are within a factor of 1.5 of those deduced from single-molecule measurements (Fig. 11, which is published as supporting information on the PNAS web site). Therefore, it seems probable that the heterogeneity arises from the fundamental nature of the ribozyme structure and is unlikely to be an artifact. This is probably because of variations in loop substructures that persist over multiple folding and unfolding events, as have been proposed for the minimal form (16).
Interestingly, unlike the minimal form that showed heterogeneity only in the F state dwell times (16), we observe heterogeneity in the dwell times of both the F and U states (for example, two ribozymes can have very different dwell times of the U state; Fig. 6). It is likely that this difference is due to a change in the identity of the rate-limiting step of folding. The junction efficiently juxtaposes the loops such that they are primed for interaction, and we have shown that the formation of the F state is rate-limiting. Nevertheless, the dominant component in the folding rate of the natural ribozyme is ≈3 s–1 at 0.5 mM Mg2+ (Fig. 4a), which is nearly three orders of magnitude faster than the folding rate of 0.008 s–1 measured for the minimal form in the presence of a much higher Mg2+ concentration (12 mM) (16). We suggest that, in the absence of the junction, the encounter between the loops is rate-limiting for the folding, and thus the effects of the heterogeneity are observable only in the reverse reaction.
Rates of Cleavage Reactions in Single-Ribozyme Molecules. The heterogeneity between the rates of loop–loop interaction in different ribozyme molecules does not seem to be an artifact of the RNA chemistry or surface attachment. Moreover, we have demonstrated that there is good agreement between the rates measured in the single molecules and those measured in bulk solution. To explore this question further, we have examined the activity of the surface-attached ribozyme molecules. A shortened substrate strand (d′) was used so that the 3′ product of cleavage rapidly dissociates (7), and a flow system was used to deliver a 10 mM Mg2+ solution while monitoring single-molecule fluorescence signals. A representative fluorescence time record of a single ribozyme is shown in Fig. 4c. Upon addition of Mg2+, the molecule folds and FRET increases. Approximately 150 s later, rapid fluctuations in FRET values begin. We interpret them as the four-way junction dynamics after 3′ product release because (i) no further changes in the behavior are observed, even after 1 h, ruling out product release slower than 1 min but faster than 1 h; (ii) high time-resolution data obtained afterward show that the fluctuations occur between two states and the rate of fluctuation increases if the Mg2+ concentration is lowered (Fig. 12, which is published as supporting information on the PNAS web site), consistent with the junction dynamics we report above; and (iii) molecules assembled with the 5′ product d strand exhibited similar fluctuation, which also showed increased rates for decreasing Mg2+ concentration (Fig. 12). FRET values are different from those of the 4H junction, presumably because helix A contains a single-stranded region.
Of 111 molecules that showed folding upon Mg2+ addition, 83 reacted before photobleaching, and 28 did not show reaction before photobleaching. The reaction kinetics of the 83 molecules was fit by using a single exponential decay with a decay time of 53 s (Fig. 4d). The photobleaching kinetics of the 28 molecules was substantially slower with an average time of 188 s (Fig. 4e), indicating that there are at least two populations, the slower of which undergoes photobleaching before cleavage. Assuming the photobleaching lifetime is 188 s, we estimate the reaction time of fast-reacting population to be 75 s, similar to the fast rate obtained by fitting bulk solution data using two exponential decays (70 s for fluorescence measurements and 120 s for radiolabel measurements; data not shown). The heterogeneity may reside in the cleavage rate or in the unfolding rate of the cleaved ribozyme (16).
Conclusion
The importance of the junction for the hairpin ribozyme has already been well established (4–7); our work provides the most direct evidence yet for the actual mechanism. The structural dynamics of the junction, rather than its static structure, promote rapid active site formation due to the spontaneous fluctuations between the two states of the junction, UD and UP. The ribozyme exploits the frequent encounters between the two loops in UP to achieve the folded state (Fig. 5). Single-molecule spectroscopy provides information on the previous and subsequent history of a given state, allowing us to conclude that the UP state is an obligatory intermediate in the formation of the ultimate F state.
The presence of the junction accelerates the folding of the hairpin ribozyme by nearly three orders of magnitude, as well as substantially reducing the required Mg2+ ion concentration for folding into the physiological range. The junction should therefore be regarded as an integral part of the ribozyme that ensures efficient folding, a kind of folding enhancer. A similar situation has been found in the hammerhead ribozyme, where the Mg2+ ion concentration required for activity is greatly reduced (again into the physiological range) by inclusion of the loops on helices I and II (A. Khvorova, personal communications); it is likely that interaction between the loops promotes folding into the active conformation. Such auxiliary folding elements may be common in small autonomously folding RNA species.
Supplementary Material
Acknowledgments
Funding was provided by National Institutes of Health Grant GM65367, National Science Foundation Career Award 0134916, National Science Foundation MRI Grant 0215869, a Cottrell Scholar award, a Sloan fellowship, and a Searle Scholars award (to T.H.) and by Cancer Research UK (D.M.J.L.). M.K.N. was partially supported by a National Institutes of Health molecular biophysics training grant.
Abbreviation: FRET, fluorescence resonance energy transfer.
References
- 1.Doudna, J. A. & Cech, T. R. (2002) Nature 418, 222–228. [DOI] [PubMed] [Google Scholar]
- 2.Lilley, D. M. J. (1998) Biopolymers 48, 101–112. [DOI] [PubMed] [Google Scholar]
- 3.Berzal-Herranz, A., Joseph, S., Chowrira, B. M., Butcher, S. E. & Burke, J. M. (1993) EMBO J. 12, 2567–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murchie, A. I. H., Thomson, J. B., Walter, F. & Lilley, D. M. J. (1998) Mol. Cell 1, 873–881. [DOI] [PubMed] [Google Scholar]
- 5.Walter, N. G., Burke, J. M. & Millar, D. P. (1999) Nat. Struct. Biol. 6, 544–549. [DOI] [PubMed] [Google Scholar]
- 6.Zhao, Z. Y., Wilson, T. J., Maxwell, K. & Lilley, D. M. J. (2000) RNA 6, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedor, M. J. (1999) Biochemistry 38, 11040–11050. [DOI] [PubMed] [Google Scholar]
- 8.Walter, N. G., Hampel, K. J., Brown, K. M. & Burke, J. M. (1998) EMBO J. 17, 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupert, P. B. & Ferre-D'Amare, A. R. (2001) Nature 410, 780–786. [DOI] [PubMed] [Google Scholar]
- 10.Walter, F., Murchie, A. I. H. & Lilley, D. M. J. (1998) Biochemistry 37, 17629–17636. [DOI] [PubMed] [Google Scholar]
- 11.Mehta, A. D., Rief, M., Spudich, J. A., Smith, D. A. & Simmons, R. M. (1999) Science 283, 1689–1695. [DOI] [PubMed] [Google Scholar]
- 12.Weiss, S. (2000) Nat. Struct. Biol. 7, 724–729. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang, X. W., Bartley, L. E., Babcock, H. P., Russell, R., Ha, T. J., Herschlag, D. & Chu, S. (2000) Science 288, 2048–2051. [DOI] [PubMed] [Google Scholar]
- 14.Liphardt, J., Onoa, B., Smith, S. B., Tinoco, I. & Bustamante, C. (2001) Science 292, 733–737. [DOI] [PubMed] [Google Scholar]
- 15.Ha, T., Enderle, T., Ogletree, D. F., Chemla, D. S., Selvin, P. R. & Weiss, S. (1996) Proc. Natl. Acad. Sci. USA 93, 6264–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhuang, X. W., Kim, H., Pereira, M. J. B., Babcock, H. P., Walter, N. G. & Chu, S. (2002) Science 296, 1473–1476. [DOI] [PubMed] [Google Scholar]
- 17.McKinney, S. A., Declais, A. C., Lilley, D. M. J. & Ha, T. (2003) Nat. Struct. Biol. 10, 93–97. [DOI] [PubMed] [Google Scholar]
- 18.Deniz, A. A., Laurence, T. A., Beligere, G. S., Dahan, M., Martin, A. B., Chemla, D. S., Dawson, P. E., Schultz, P. G. & Weiss, S. (2000) Proc. Natl. Acad. Sci. USA 97, 5179–5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuler, B., Lipman, E. A. & Eaton, W. A. (2002) Nature 419, 743–747. [DOI] [PubMed] [Google Scholar]
- 20.Rothwell, P. J., Berger, S., Kensch, O., Felekyan, S., Antonik, M., Wohrl, B. M., Restle, T., Goody, R. S. & Seidel, C. A. (2003) Proc. Natl. Acad. Sci. USA 100, 1655–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson, T. J. & Lilley, D. M. (2002) RNA 8, 587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, H. D., Nienhaus, G. U., Ha, T., Orr, J. W., Williamson, J. R. & Chu, S. (2002) Proc. Natl. Acad. Sci. USA 99, 4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha, T. J., Ting, A. Y., Liang, J., Caldwell, W. B., Deniz, A. A., Chemla, D. S., Schultz, P. G. & Weiss, S. (1999) Proc. Natl. Acad. Sci. USA 96, 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klostermeier, D. & Millar, D. P. (2000) Biochemistry 39, 12970–12978. [DOI] [PubMed] [Google Scholar]
- 25.Cai, Z. & Tinoco, I., Jr. (1996) Biochemistry 35, 6026–6036. [DOI] [PubMed] [Google Scholar]
- 26.Butcher, S. E., Allain, F. H. & Feigon, J. (1999) Nat. Struct. Biol. 6, 212–216. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, T. J., Zhao, Z. Y., Maxwell, K., Kontogiannis, L. & Lilley, D. M. J. (2001) Biochemistry 40, 2291–2302. [DOI] [PubMed] [Google Scholar]
- 28.Ha, T., Rasnik, I., Cheng, W., Babcock, H. P., Gauss, G., Lohman, T. M. & Chu, S. (2002) Nature 419, 638–641. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.