Abstract
Bacterial symbioses are widespread among insects, probably being one of the key factors of their evolutionary success. We present the complete genome sequence of Blochmannia floridanus, the primary endosymbiont of carpenter ants. Although these ants feed on a complex diet, this symbiosis very likely has a nutritional basis: Blochmannia is able to supply nitrogen and sulfur compounds to the host while it takes advantage of the host metabolic machinery. Remarkably, these bacteria lack all known genes involved in replication initiation (dnaA, priA, and recA). The phylogenetic analysis of a set of conserved protein-coding genes shows that Bl. floridanus is phylogenetically related to Buchnera aphidicola and Wigglesworthia glossinidia, the other endosymbiotic bacteria whose complete genomes have been sequenced so far. Comparative analysis of the five known genomes from insect endosymbiotic bacteria reveals they share only 313 genes, a number that may be close to the minimum gene set necessary to sustain endosymbiotic life.
Many bacteria live in close association with higher organisms in a symbiotic or parasitic relationship. Whereas much has been learned about pathogenic interactions in the past, little is known about the mechanisms enabling bacteria to have a symbiotic lifestyle. However, symbioses between unicellular and multicellular organisms have contributed significantly to the evolution of life on Earth (1).
Bacterial symbioses are widespread among insects, and it has been estimated that at least 15–20% of all insects live in such symbiotic relationships (2). The early establishment of symbiotic associations among insects and bacteria, ≈300 million years ago (3), has probably been one of the key factors for the evolutionary success of insects, because it may have allowed access to novel ecological niches and to new imbalanced food resources, such as plant sap or blood (4). This is the case for the mutualistic and obligate symbiosis of Buchnera aphidicola with aphids and of Wigglesworthia glossinidia with tsetse flies. These symbiotic bacteria reside in specialized host cells called bacteriocytes, which form symbiotic organs called bacteriomes. The bacterial transmission occurs vertically: the eggs or young embryos are infected by microorganisms derived from the mother.
Most parasitic and symbiotic obligate intracellular bacteria share several genomic features, i.e., bias toward a high A+T content, accelerated sequence evolution (5), and massive genome size reduction with respect to their free-living ancestors (6). This reduction has become so extreme that some Bu. aphidicola strains present the smallest genome sizes (≈450 kb) known to date (7), which may represent ≈400 protein-coding genes. Comparative analyses of the small size genomes of symbiotic and parasitic bacteria will provide interesting insights into the evolution of resident genomes and the minimum set of genes necessary for intracellular life.
In addition to aphids and tsetse flies, social insects such as ants are particularly interesting for understanding mutualistic relationships, because they have developed numerous interactions with different species of animals, plants, and microorganisms. Moreover, ants belong to a different insect order than aphids and tsetse flies. The symbiosis of ants of the genus Camponotus with intracellular bacteria (Blochmannia spp.), located in the midgut and ovaries of the insects, was the first bacterocyte endosymbiosis described (8). As in the above-mentioned bacterial endosymbionts of insects, Blochmannia spp. generally display concordant evolution with their host species (9). This symbiosis has been described so far only within the members of the subfamily Formicinae, which has an estimated age of ≈70 million years, although it is not known whether this symbiosis has been established only in the Formicinae or was an original attribute of ants maintained only in this subfamily (9). Until now, the biological function of this symbiosis remained unknown, because a nutritional basis is not evident at first sight. Although it seems to be a general trend within the genus Camponotus to use honeydew from sap-sucking insects as their main food source, they can feed on a complex diet that may also include dead and live insects, bird excrement, and sweet food waste (10). That adult ants are able to live without their bacterial endosymbionts under laboratory conditions, and that these bacteria seem to degenerate naturally in the course of time, as observed in older queens, suggest that the symbiosis may be of relevance mainly during the early life stages of the ants (11).
Here we present the complete genome sequence of Blochmannia floridanus, the primary endosymbiont of the ant Camponotus floridanus, and its comparison with the previously sequenced genomes of four insect endosymbionts and the obligate parasite Mycoplasma genitalium. The comparative genomics of all known endosymbiont genomes reveals that they share 313 genes, a number that may be close to the minimum gene set necessary to sustain endosymbiotic life. Furthermore, they share 179 genes with M. genitalium, which represents a minimum gene set for bacterial cell life.
Materials and Methods
Bl. floridanus DNA Genomic Purification from Carpenter Ants. C. floridanus were maintained in the laboratory at 30°C and fed with honey water and cockroaches.
The bacteriocytes containing the endosymbiont bacteria were purified by an adaptation of the procedure described by Harrison et al. (12). The abdomens of ≈100 C. floridanus pupae were lightly crushed on isolation buffer (35 mM Tris·Cl, pH 7.6/25 mM KCl/250 mM sucrose) in a glass homogenizer and the insect debris removed by filtration through nylon filters with a pore size from 100 to 28 μm. The bacterial cell pellets were collected and subjected to DNase I digestion on ice for 1 h (1 mg/ml DNase I in isolation buffer supplemented with 10 mM MgCl) to eliminate the remaining ant DNA. EDTA was added to a final concentration of 50 mM. The bacteria were harvested by brief centrifugation and washed three times to remove all traces of DNase I before further treatment.
For the isolation of genomic DNA, the pellets were resuspended in 200 μl of lysis buffer (6 mM Tris·Cl, pH 7.6/10 mM EDTA/1 M NaCl/0.5% Brij35/0.2% deoxycholate/0.2% Nalauroylsarcosine) to which 0.5 mg/ml RNase and 1 mg/ml lysozyme were added. The mixture was incubated for 3–4 h at 37°C before proteinase K was added to a final concentration of 0.2 mg/ml, and incubation was continued overnight. Genomic DNA was finally purified by a standard phenol/chloroform protocol (13).
To evaluate the level of DNA contamination, DNA was analyzed by Southern hybridization using the digoxigenin oligonucleotide labeling kit (Boehringer Mannheim), with probes that recognize the 16S rRNA, the eukaryotic elongation factor EF1-α, and mitochondrial cytochrome oxidase. No host nuclear DNA was detected, and the preparation was estimated to contain 97% Bl. floridanus DNA.
Whole Genome Random Shotgun Sequencing. Shotgun sequence libraries were prepared as described (14). Dye terminator cycle sequence analysis was performed with sequencing kits from Applied Biosystems at the sequencing facility of the Universitat de València.
All trace data were analyzed by using the staden package software program (15) for trimming of vector sequences, data assembly, editing, and finishing processes. Ambiguities were reanalyzed by primer walking, and all polymorphisms were checked manually to exclude false positives. A total of 11,865 sequence reads were generated (average read length: 615 nt). The final assembly contained 11,238 Bl. floridanus-derived sequences. Over 9-fold coverage was achieved.
Gene Prediction and Annotation. ORFs were identified with glimmer and genmark.hmm programs (16, 17), and the putative encoded proteins were compared with sequences present in public databases by using blastp (18). Other putative coding regions, either genes or pseudogenes, were identified by blast, searching protein databases against 1-kb fragments of genome nucleotide sequences. Detected frame shifts in coding sequences were revised in the original sequencing readings to determine the gene or pseudogene status. In general, no ORF smaller than 100 aa was considered a gene, unless similarity with a previously described protein was detected by blast. Genes encoding shorter proteins than those described for Bu. aphidicola or Escherichia coli were studied in detail by comparison to other bacterial species to detect errors in annotation and to resolve the gene or pseudogene status.
Amino acid sequence alignments for each protein with the homologous proteins of complete genome sequences from γ-and β-proteobacteria species were obtained with clustalw (19). These alignments and the nucleotide genome sequence were used to inspect individually each ORF for start codon assignment, alternative putative start codons, and putative Shine–Dalgarno sequences.
On the basis of similarity and other criteria, we identified orthologous genes in several γ-proteobacterial species, including E. coli, Salmonella spp., Vibrio chloreae, and the endosymbionts W. glossinidia, and Bu. aphidicola strains from Acyrtosiphon pisum, Schizaphis graminum, and Baizongia pistaciae. Paralogous and orthologous genes were also identified according to phylogenetic trees and a previous study (6).
tRNAs were identified by using the program trna-scan (www.genetics.wustl.edu/eddy/tRNAscan-SE; ref. 20). rRNAs and other small RNAs were identified by blastn searches of the intergenic regions vs. RNA-specifying genes in Genome Information Broker (GIB; http://gib.genes.nig.ac.jp). Their limits were hand-curated on the basis of the sequence or the secondary structures described for other close bacterial genes, such as the ones from E. coli.
The graphical display of the annotated genome was obtained by using gff2Ps (22).
Functional Analysis of the Predicted ORFs. The possible ORFs were classified on the basis of cluster of orthologous genes classification (23) and the Riley and Labedan classification for E. coli and Salmonella (24), with some modifications. Metabolic pathways were examined by using the on-line service at KEGG (www.genome.ad.jp/kegg; ref. 25).
Identification of the Origin of Replication and Gene Order Analysis. The putative origin of replication, in the absence of a diagnostic cluster of DnaA boxes, was determined by GC-skew (G-C/G+C) analysis and gene GC-skew by using the program oriloc (http://pbil.univ-lyon1.fr/software/oriloc.html; ref. 26). We refined the location of the origin by subsequent analysis based on the observation that chromosomal rearrangements centered on the origin and terminus of replication are predominant (27).
Isoelectric Point Analysis. The isoelectric points of Bl. floridanus predicted proteins were compared with those of the corresponding orthologous proteins of E. coli K-12. They were estimated with the program iep implemented in the emboss package (european molecular biology open software suite, www.emboss.org).
Phylogenetic Analysis. The phylogenetic relationship of Bl. floridanus with other γ-proteobacteria, including insect endosymbionts Bu. aphidicola and W. glossinidia, was evaluated by means of maximum likelihood and Bayesian methods. An initial alignment of 61 concatenated conserved protein-coding genes involved in translation from selected bacterial genomes (see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org) was obtained with clustalw (19) and trimmed by using gblocks (29), resulting in 8,713 amino acid positions. The maximum likelihood tree was obtained by the quartet-puzzling method (30). The Mueller–Vingron matrix of amino acid substitution (31), along with a γ model (α = 0.99) for rate variation among sites and a proportion P = 0.14 for invariant sites, was used with 4,000 puzzling steps. The Bayesian analysis (32) proceeded with the JTT (33) model for amino acid substitution, complemented with a γ+ invariant model for rate heterogeneity among sites. Four chains were used with 1,000,000 generations, and trees were sampled every 100 generations. The last 9,000 trees were used for obtaining a consensus tree, although no significant changes were observed when all of the sampled trees were used for obtaining the consensus.
Results and Discussion
General Features of the Genome. The genome of Bl. floridanus consists of a circular chromosome of 705,557 bp with an average G+C content of 27.38%, similar to most analyzed endosymbiotic bacteria. No plasmids were found. Table 1 summarizes the general features of this genome, compared with the other four sequenced genomes of insect endosymbionts: W. glossinidia and three strains of Bu. aphidicola (14, 34–36).
Table 1. Comparison of genome features for all known bacterial endosymbionts from insects.
|
Bu. aphidicola
|
|||||
|---|---|---|---|---|---|
| Features | Bl. floridanus | W. glossinidia | BAp | BSg | BBp |
| Chromosome, bp | 705,557 | 697,724 | 640,681 | 641,454 | 615,980 |
| Plasmids, total length, bp* | 0 | 1 (5,280) | 2 (7,805) | 2 (7,967) | 1 (2,399) |
| G+C content, % | 27.38 | 22 | 26.2 | 26.3 | 25.3 |
| Total gene number | 625 | 661 | 608 | 596 | 545 |
| CDS† | 583 | 619 | 571 | 559 | 508 |
| rRNAs | 3 | 6 | 3 | 3 | 3 |
| tRNAs | 37 | 34 | 32 | 32 | 32 |
| Small RNA genes | 2 | 2 | 2 | 2 | 2 |
| Pseudogenes | 6 | 8 | 12 | 33 | 9 |
| Protein-coding regions, % | 83.2 | 89 | 86.8 | 84.5 | 81.4 |
| Average length ORF, bp | 1,007 | 988 | 991 | 985 | 992 |
Data were taken from original papers, with some minor modifications according to Tamas et al. (14) and our own revision. The status of the previously described Bu. aphidicola BSg pseudogenes lig, infC, endA, mfd, and prfB was changed on the basis of several criteria. BAp, Bu. aphidicola from the aphid Acyrthosiphon pisum; BSg, Bu. aphidicola from the aphid Schizaphis graminum; BBp, Bu. aphidicola from the aphid Baizongia pistacea.
Bu. aphidicola strains BAp and BSg contain two plasmids, one including the leucine operon and another containing the genes involved in tryptophan biosynthesis. This second plasmid has not been completely characterized by sequencing, so we have included only the size of the leucine plasmids. Bu. aphidicola BBp plasmid is a subset of the leucine plasmid. The W. glossinidia plasmid is apparently unrelated to the Bu. aphidicola plasmids.
Total number of protein-coding genes present in the chromosome plus the plasmids. For the tryptophan plasmid in Bu. aphidicola BAp and BSg, only one copy of the trpE and trpG genes has been considered.
The genetic map of Bl. floridanus reveals the presence of 625 putative genes and six pseudogenes, all with significant database matches, 555 (88%) of which were assigned a biological function (see Table 3 and Fig. 5, which are published as supporting information on the PNAS web site). The genome contains 583 protein-coding genes, with an average size of 1,007 nucleotides per gene and 42 RNA-specifying genes (three ribosomal RNAs, two small RNAs, and 37 tRNAs specifying all 20 amino acids). The average predicted isoelectric point of the products of the coding sequences is 8.9 (see Fig. 6, which is published as supporting information on the PNAS web site), similar to what has been described for other endosymbionts (34, 36).
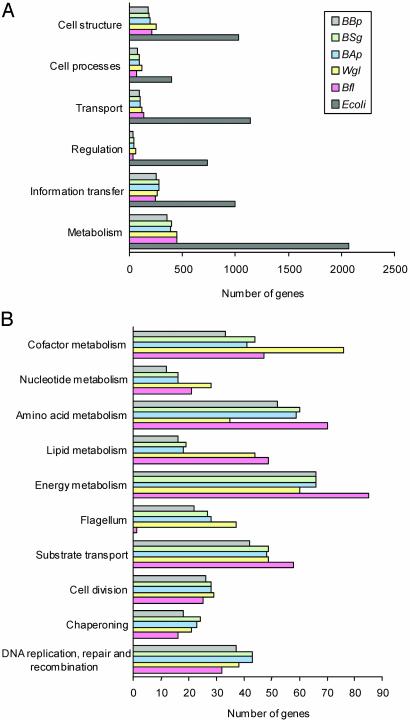
Interestingly, no orphan genes were found in this genome. Generally, the most similar counterparts of Bl. floridanus proteins are among the Enterobacteriaceae. Almost all of the coding sequences (99.3%) have a homologue in E. coli. Fig. 1 represents the gene repertoire of the Bl. floridanus genome by functional categories (24), compared with the previously known genomes of E. coli and other insect endosymbionts (14, 34–37).
Fig. 1.
Comparative analysis by functional categories of the gene repertoires of Bl. floridanus, W. glossinidia, Bu. aphidicola, and E. coli K-12. (A) General comparison among E. coli K-12 and the five insect endosymbionts under study. (B) Comparison among the insect endosymbionts of the number of genes present in some relevant functional categories in which the general comparison is subdivided.
The putative origin of replication was determined by GC-skew analysis, because no diagnostic cluster of DnaA boxes could be identified. Subsequent analysis refined the location of the origin (oriC) to the intergenic region next to gidA. Considering this as the most plausible position for the origin of replication, the genomic alignment of orthologous gene position between Bl. floridanus and E. coli generates an X pattern (see Fig. 7, which is published as supporting information on the PNAS web site), similar to that described for other endosymbionts (6), showing that many chromosomal rearrangements have occurred since the divergence of these two species.
Functional Analysis of the Predicted Protein-Coding Genes. One of the most surprising features of this bacterium is the absence of all known mechanisms for replication initiation (38). Bl. floridanus lacks dnaA, which explains the above-mentioned difficulty to define the oriC of its genome. It should be noted that both Bl. floridanus and W. glossinidia, the only two bacteria in which no dnaA has been found, are located free in the cytosol of bacteriocytes (39, 40), whereas Bu. aphidicola resides in vacuole-like organelles (4) and still retains dnaA to initiate replication. Bacteria in the cytosol might be a potential danger for the host cell and much more difficult to control. Thus, it could be that the development of a stable symbiosis with cytosolic bacteria might have required more direct control of DNA replication of the symbionts by the host, involving loss of dnaA. Furthermore, the other alternative mechanisms reported so far for DNA replication initiation from noncanonic oriC sites are also absent from Bl. floridanus, because it has also lost priA and recA. An explanation for the lack of a DNA replication initiation protein could be that another protein (maybe HplA, the only HU-like nucleoprotein present in Bl. floridanus) is able to recruit DnaB to the right position in DNA to start replication. The loss of most histone-like proteins, which play an auxiliary role during replication, may also imply chromosomal and replicative instability. Nevertheless, Bl. floridanus retains the main functions in DNA replication such as the helicase (dnaB), primase (dnaG), gyrase (gyrA and gyrB), and an almost complete DNA polymerase III (except for the τ and Θ subunits).
A general feature of all sequenced genomes of endosymbionts is the loss of most DNA repair and transcriptional regulation mechanisms. However, a few transcriptional regulators have been identified in this genome, which may indicate they play an important role in the ant–bacterial symbiotic relationship. Its genome encodes Zur, which in E. coli is a repressor of the znu gene cluster encoding the membrane components of the zinc import system (41). Bl. floridanus also codes for SlyA, a transcription factor involved in induction of stress response proteins, including several molecular chaperones (42), which may compensate for the smaller number of chaperones present in Bl. floridanus compared with other endosymbionts. Two more genes that encode putative transcription factors are present: yidZ, encoding a regulatory protein with similarities to the LysR family (43), and Bfl615, a MarR-like regulator (44).
Similarly to W. glossinidia, but unlike Bu. aphidicola, Bl. floridanus encodes most genes necessary for the synthesis of a normal Gram-negative cell wall, including the lipopolysaccharide components of the outer membrane. The complete lipoprotein transport system lolACDE (45) is also present, which confirms the assumption of a well structured cell wall that possibly renders these bacteria more resistant to a hostile environment. This is even further substantiated by the presence of the tol-pal gene cluster, involved in the uptake of biomolecules and outer membrane stability (46), which is absent in Bu. aphidicola and in many obligate intracellular parasites (47). Because the cytosol of a eukaryotic host cell may not be as benign an environment as previously thought (48), the need for protection from the host environment and/or the relatively recent symbiotic association may help to explain the maintenance of the genes responsible for cell wall integrity in Bl. floridanus and W. glossinidia. However, unlike other analyzed endosymbionts, Bl. floridanus has completely lost the flagellar apparatus that has been suggested to be involved in transport functions, but also in the invasion of bacteriocytes, ovaries, or embryos by the bacteria (49).
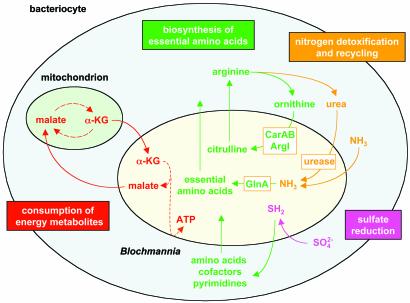
The analysis of the gene content of Bl. floridanus supports that, as in previously studied endosymbionts, its symbiosis with carpenter ants has a nutritional basis and is mutualistic (Fig. 2).
Fig. 2.
Relevant metabolic interactions between Bl. floridanus and its host cell, as deduced from its genome sequence. For clarity, the metabolic pathways shown are simplified. α-KG, α-ketoglutarate; ArgI, ornithine carbamoyltransferase chain I; CarAB, carbamoyl-phosphate synthetase; GlnA, glutamine synthetase.
Similarly to Bu. aphidicola, Bl. floridanus encodes most biosynthetic pathways required for the production of amino acids essential to the insect hosts (50) (see Table 4, which is published as supporting information on the PNAS web site). Most interestingly, Bl. floridanus also contains a urease gene cluster encoding the structural genes of urease and its accessory factors required for the assembly of the nickel-containing enzyme. Urease hydrolyzes urea to CO2 and ammonia, the latter of which is a potent cell poison. Because Bl. floridanus codes for a glutamine synthetase that is missing in Bu. aphidicola, ammonia can be recycled before toxic concentrations are accumulated. On the other hand, Bl. floridanus has lost a major part of the arginine biosynthesis pathway, retaining only those enzymes that catalyze the synthesis of citrulline from ornithine. Therefore, Bl. floridanus resembles a mammalian mitochondrion, which is involved in the urea cycle, providing citrulline to the host cytoplasm where it is transformed to arginine. This suggests that Bl. floridanus contributes to the biosynthesis and degradation of arginine, and that the different reactions of the arginine metabolism are running in separate compartments. On the basis of these findings, we propose that arginine plays a central role in this symbiotic relationship. Ants may use arginine as a nitrogen storage compound for times when high anabolic activities are supported by little or no uptake of substrates, e.g., during metamorphosis. When required, the stored nitrogen may be mobilized by the action of host cell-derived or bacterial arginases and the Bl. floridanus-encoded urease. Because in several bacterial and fungal pathogens urease is a virulence factor (51), the urease gene cluster might represent a remnant of a formerly pathogenic relationship, which has been transformed into a useful or even essential symbiotic factor.
Insects lack the assimilatory sulfur reductive pathway and can, therefore, synthesize cysteine only if reduced sulfur is available (50). Like Bu. aphidicola, and unlike W. glossinidia, Bl. floridanus has retained all enzymes necessary for sulfate reduction. In addition, Bl. floridanus is the only known endosymbiont that also contains the cysUWA operon, encoding an ABC-type sulfate carrier (52). Thus, Bl. floridanus should be able to efficiently incorporate even trace amounts of sulfate and make it available to the host even if feeding on a diet extremely poor in reduced sulfur.
In exchange for the nutritional benefits given to the host cells, Bl. floridanus uses the host cell machinery to sustain some essential cellular functions, e.g., the biosynthesis of most nonessential amino acids, vitamins, and cofactors (see Table 4). Blochmannia, similarly to W. glossinidia, has retained several reactions of the citrate cycle, which is almost entirely missing in Bu. aphidicola.In Bl. floridanus, only most of the energy-yielding steps are retained, being able to oxidize α-ketoglutarate to produce malate. Because it is unable to perform the acetyl-CoA-fixing steps, an intermediate compound of the cycle must be provided by the host cell, possibly glutamate or α-ketoglutarate itself, which may be imported by the bacteria via putative aspartate/glutamate carriers (such as GltP). Bl. floridanus then should return malate to the host cell to complete the deviation of the citrate cycle running in the mitochondrion.
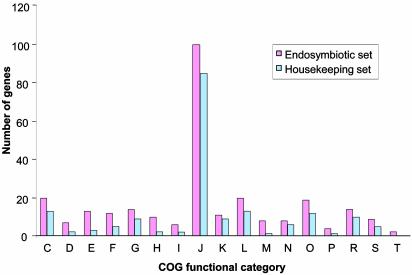
Comparative Genomics. Comparative analysis of all sequenced genomes of insect endosymbionts reveals they share only 277 (≈50%) protein-coding genes, and 36 RNA-specifying genes (90%), making a total of 313 shared genes (Fig. 3; see also Table 5, which is published as supporting information on the PNAS web site). However, the number increases up to ≈70% of protein-coding genes in pair-wise comparisons between genera. All these genomes are relatively similar in size (the smallest corresponding to Bu. aphidicola, with a more ancient endosymbiotic relationship with their hosts), and they encode a quite similar number of genes in each functional cluster of orthologous genes (COG) category (23). Of interest, in all these genomes ≈27% of the genes are devoted to information storage and processing (COG categories J, K, and L), and most genes in these categories are shared by all of them. Around 70% of the maintained genes involved in cell division processes are also shared by all five endosymbionts. In some other functional categories, such as molecular chaperones, ion transport, signal transduction, energy production, and carbohydrate metabolism, most of the functions represented in the five genomes are quite similar, although the individual genes shared among all of them do not represent >50%. Remarkable differences were found only among the genes that encode proteins involved in cell envelope and flagellar biosynthesis and in the metabolism of amino acids, nucleotides, and coenzymes. These findings suggest that the molecular mechanisms necessary for survival in an intracellular environment may be quite similar for any endosymbiotic association, whereas about one-third of the coding capacity of each endosymbiont seems to be dedicated to specific requirements of the respective symbiosis, mainly reflecting differences in host lifestyle, nutritional needs, and location within the host cells.
Fig. 3.
Distribution of the putative minimum gene set for insect endosymbiotic life in nonredundant functional categories and its comparison with M. genitalium. The endosymbiotic set corresponds to the 278 protein-coding genes shared by the five insect endosymbiont genomes. The housekeeping set corresponds to the 179 genes with a putative homologue in M. genitalium. Clusters of orthologous genes categories correspond to: C, energy production and conversion; D, cell division and chromosome partitioning; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme metabolism; I, lipid metabolism; J, translation, ribosomal structure and biogenesis; K, transcription; L, DNA replication, recombination and repair; M, cell envelope biogenesis, outer membrane; N, cell motility and secretion; O, posttranslational modification, protein turnover, chaperones; P, inorganic ion transport and metabolism; R, general function predicted only; S, function unknown; and T, signal transduction mechanisms.
When the complete set of protein-coding genes shared by all five sequenced endosymbiont genomes was compared with the genome of the obligate parasite M. genitalium (53), only 179 putative homologous genes were found (Fig. 3). These genes that are present in all resident genomes analyzed may represent the basic subset of genes required for bacterial cell life, whereas the rest of the genes shared by all five endosymbionts but absent in M. genitalium can be considered essential for endosymbiotic functions. However, these lists should be enlarged to include those genes whose essential function is performed by a nonhomologous gene in at least one of the genomes.
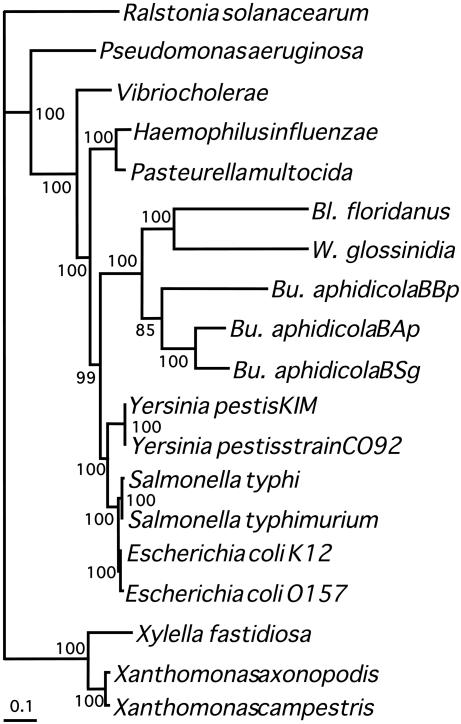
Phylogenetic Analysis. The phylogenetic relationship of Bl. floridanus with other γ-proteobacteria, including insect endosymbionts Bu. aphidicola and W. glossinidia, has been evaluated by analyzing a set of 61 conserved protein-coding genes. Bl. floridanus forms a monophyletic cluster with the other endosymbiotic bacteria closely related to Enterobacteriaceae (Fig. 4), thus supporting a common origin for these endosymbionts. Furthermore, Bl. floridanus seems to be more closely related to W. glossinidia than to Bu. aphidicola, although further analyses are needed to establish whether their common genome features (see above) result from a common ancestry or derive from adaptation to common environmental features such as their cytosolic location within the host cells.
Fig. 4.
Phylogenetic tree obtained by maximum likelihood with a trimmed alignment of 61 concatenated proteins. Numbers in nodes indicate proportion of quartets supporting the corresponding inner branch, as determined by the quartet-puzzling method. The same topology was obtained by using a Bayesian inference method in which all nodes had a posteriori probabilities equal to t.
Conclusion
The evolutionary forces that have led to the reduction in size of resident genomes are currently under discussion. Some authors have suggested these genomes evolve in a completely neutral way (54). However, there is evidence of an increase in the fixation rate of deleterious mutations by genetic drift, due to the existence of bottlenecks in the populations (5). It is reasonable to assume that, at the beginning of the symbiotic integration, the loss of the genes involved in DNA repair favored the bias toward A+T content, allowing an increase in the number of random deleterious mutations. Although purifying selection might purge many such mutations, these could accumulate in genes involved in the metabolism of compounds that can be obtained from the host. Furthermore, the faster replication of shorter genomes would probably favor the smallest molecules. According to the nearly neutral theory of molecular evolution, although most fixed mutations will be slightly deleterious, a small proportion of positive ones would also be present, which could compensate the detrimental effects of previously fixed mutations (55). This positive selection has been proven for GroEL (21, 28), a chaperone that helps to the correct folding of many damaged proteins and is overexpressed in analyzed endosymbiotic bacteria. It is likely that the initial acquisition of symbiotic bacteria provided the insects with important selective advantages, e.g., to exploit new nutritional sources. The loss of DNA repair mechanisms at the beginning of the symbiotic relationship started a process of continuous degeneration of resident genomes. Hence, the present contribution of endosymbionts to the host may not be as relevant as during the first stages of the symbiotic integration. What is evident, however, from their genome content is that they are still supplying essential metabolic capabilities to their hosts, because important functions related to each specific endosymbiotic relationship are retained.
Supplementary Material
Acknowledgments
We thank Profs. F. J. Ayala, P. Baumann, and R. E. Lenski for critical reading of the manuscript. This work was supported by Ministerio de Educación y Ciencia, Spain (BFM2000-1383); Comunidad Autónoma de Valenciana, Spain (GV01-177); Comunidad Autónoma de Madrid, Spain; Instituto Nacional de Técnica Aeroespacial, Spain; Fondo Social Europeo, European Union; Fondo Social Europeo para el Desarrollo de las Regiones, European Union; Universitat de València, Spain; Deutsche Forschungsgemeinschaft, Germany (SFB567/C2); Fonds der Chemischen Industrie, Germany; and Germany–Spain Joined Actions (HA2001-0048).
Data deposition: The sequences reported in this paper have been deposited in the GenBank/EMBL/DDBJ database (accession no. BX248583).
References
- 1.Margulis, L. & Fester, R., eds. (1991) Symbiosis as a Source of Evolutionary Innovation (MIT Press, Cambridge, MA). [PubMed]
- 2.Buchner, P. (1965) Endosymbiosis of Animals with Plant Microorganisms (Interscience, New York).
- 3.Moran, N. A. & Telang, A. (1998) BioScience 48, 295–304. [Google Scholar]
- 4.Baumann, P., Moran, N. A. & Baumann, L. (2000) in The Prokaryotes, ed. Dworkin, M. (Springer, New York).
- 5.Moran, N. A. (1996) Proc. Natl. Acad. Sci. USA 93, 2873–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva, F. J., Latorre, A. & Moya, A. (2001) Trends Genet. 17, 615–618. [DOI] [PubMed] [Google Scholar]
- 7.Gil, R., Sabater-Muñoz, B., Latorre, A., Silva, F. J. & Moya, A. (2002) Proc. Natl. Acad. Sci. USA 99, 4454–4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blochmann, F. (1892) Zentralbl. Bakteriol. Mikrobiol. Hyg. Ser. A 11, 234–240. [Google Scholar]
- 9.Sauer, C., Stackebrandt, E., Gadau, J., Hölldobler, B. & Gross, R. (2000) Int. J. Syst. Evol. Microbiol. 50, 1877–1886. [DOI] [PubMed] [Google Scholar]
- 10.Pfeiffer, M. & Linsenmair, K. E. (2000) Insectes Soc. 47, 123–132. [Google Scholar]
- 11.Sauer, C., Dudaczek, D., Hölldobler, B. & Gross, R. (2002) Appl. Environ. Microbiol. 68, 4187–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison, C. P., Douglas, A. E. & Dixon, A. F. G. J. (1989) Invertebr. Pathol. 53, 427–428. [Google Scholar]
- 13.Sambrook, J., Fritsch, E. F. & Maniatis, T., eds. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 14.van Ham, R. C. H. J., Kamerbeek, J, Palacios, C., Rausell, C., Abascal, F., Bastolla, U., Fernández, J. M., Jiménez, L., Postigo, M., Silva, F. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staden, R., Beal, K. F. & Bonfield, J. K. (1998) in Bioinformatics: Methods and Protocols, eds. Misener, S. & Krawetz, S. A. (Humana, Totowa, NJ), Vol. 132, pp. 115–130. [Google Scholar]
- 16.Delcher, A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashin, A. & Borodovsky, M. (1998) Nucleic Acids Res. 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson, J. D., Higgins, D. G. & Gibson., T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fares, M. A., Barrio, E., Sabater Muñoz, B. & Moya, A. (2002) Mol. Biol. Evol. 19, 1162–1170. [DOI] [PubMed] [Google Scholar]
- 22.Abril, J. F. & Guigó, R. (2000) Bioinformatics 16, 743–744. [DOI] [PubMed] [Google Scholar]
- 23.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley, M. & Labedan, B. (1996) in Escherichia coli and Salmonella, ed. Neitdhardt, F. C. (Am. Soc. Microbiol., Washington, DC), pp. 2118–2202.
- 25.Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H. & Kanehisa, M. (1999) Nucleic Acids Res. 27, 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frank, C. A. & Lobry, J. R. (2000) Bioinformatics 16, 560–561. [DOI] [PubMed] [Google Scholar]
- 27.Tillier, E. R. & Collins, R. A. (2000) Nat. Genet. 26, 195–197. [DOI] [PubMed] [Google Scholar]
- 28.Fares, M. A., Ruiz González, M. X., Moya, A., Elena, S. F. & Barrio, E. (2002) Nature 417, 398. [DOI] [PubMed] [Google Scholar]
- 29.Castresana, J. (2000) Mol. Biol. Evol. 17, 540–552. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. (2002) Bioinformatics 18, 502–504. [DOI] [PubMed] [Google Scholar]
- 31.Muller, T., Spang, R. & Vingron, M. (2002) Mol. Biol. Evol. 19, 8–13. [DOI] [PubMed] [Google Scholar]
- 32.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754–755. [DOI] [PubMed] [Google Scholar]
- 33.Jones, D. T., Taylor, W. R. & Thornton, J. M. (1992) Comput. Appl. Biosci. 8, 275–282. [DOI] [PubMed] [Google Scholar]
- 34.Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. (2000) Nature 407, 81–86. [DOI] [PubMed] [Google Scholar]
- 35.Tamas, I., Klasson, L., Canbäck, B., Näslung, A. K., Eriksson, A. S., Wernegreen, J. J., Sandström, J. P., Moran, N. A. & Andersson, S. G. E. (2002) Science 296, 2376–2379. [DOI] [PubMed] [Google Scholar]
- 36.Akman, L., Yamashita, A. Watanabe, H., Oshima, K., Shiba, T., Hattori, M. & Aksoy, S. (2002) Nat. Genet. 32, 402–407. [DOI] [PubMed] [Google Scholar]
- 37.Blattner, F. R., Plunkett, G., III, Bloch, C. A., Perna, N. T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J. D., Rode, C. K., Mayhew, G. F., et al. (1997) Science 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- 38.Kogoma, T. (1997) Microbiol. Mol. Biol. Rev. 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schröder, D., Deppisch, H., Obermayer, M., Krohne, G., Stackebrant, E., Hölldobler, B., Goebel, W. & Gross, R. (1996) Mol. Microbiol. 21, 479–489. [DOI] [PubMed] [Google Scholar]
- 40.Reinhardt, C., Steiger, R. & Hecker, H. (1972) Acta Trop. 29, 280–288. [PubMed] [Google Scholar]
- 41.Patzer, S. I. & Hantke, K. (2000) J. Biol. Chem. 275, 24321–24332. [DOI] [PubMed] [Google Scholar]
- 42.Schell, M. A. (1993) Annu. Rev. Microbiol. 47, 597–626. [DOI] [PubMed] [Google Scholar]
- 43.Sulavik, M. C., Gambino, L. F. & Miller, P. F. (1995) Mol. Med. 1, 436–446. [PMC free article] [PubMed] [Google Scholar]
- 44.Spory, A., Bosserhoff, A., von Rhein, C., Goebel, W. & Ludwing, A. (2002) J. Bacteriol. 184, 3549–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yakushi, T., Masuda, K., Narita, S., Matsuyama, S. & Tokuda, H. (2000) Nat. Cell Biol. 2, 212–218. [DOI] [PubMed] [Google Scholar]
- 46.Lazzaroni, J. C., Germon, P., Ray, M. C. & Vianney, A. (1999) FEMS Microbiol. Lett. 177, 191–197. [DOI] [PubMed] [Google Scholar]
- 47.Sturgis, J. N. (2001) J. Mol. Biotechnol. 3, 113–122. [PubMed] [Google Scholar]
- 48.Goetz M., Bubert, A., Wang, G., Chico-Valero, I., Vazquez-Boland, J. A., Beck, M., Slaghuis, J., Szalay, A. A. & Goebel, W. (2001) Proc. Natl. Acad. Sci. USA. 98, 12221–12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young, G. M., Schmiel, D. H. & Miller, V. L. (1999) Proc. Natl. Acad. Sci. USA 96, 6456–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wigglesworth, W. B. (1934) The Principles of Insect Physiology (Methuen, London).
- 51.Collins, C. M. & D'Orazio, S. E. (1993) Mol. Microbiol. 9, 907–913. [DOI] [PubMed] [Google Scholar]
- 52.Sirko, A., Zatyka, M., Sadowy, E. & Hulanicka, D. (1995) J. Bacteriol. 177, 4134–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fraser, C. M., Gocayne, J. D., White, O., Adams, M. D., Clayton, R. A., Fleischmann, R. D., Bult, C. J., Kerlavage, A. R., Sutton, G., Kelley, J. M., et al. (1995) Science 270, 397–403. [DOI] [PubMed] [Google Scholar]
- 54.Itoh, T., Martin, W. & Nei, M. (2002) Proc. Natl. Acad. Sci. USA 99, 12944–12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hartl, D. L. & Taubes, C. H. (1996) J. Theor. Biol. 182, 303–309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.