Abstract
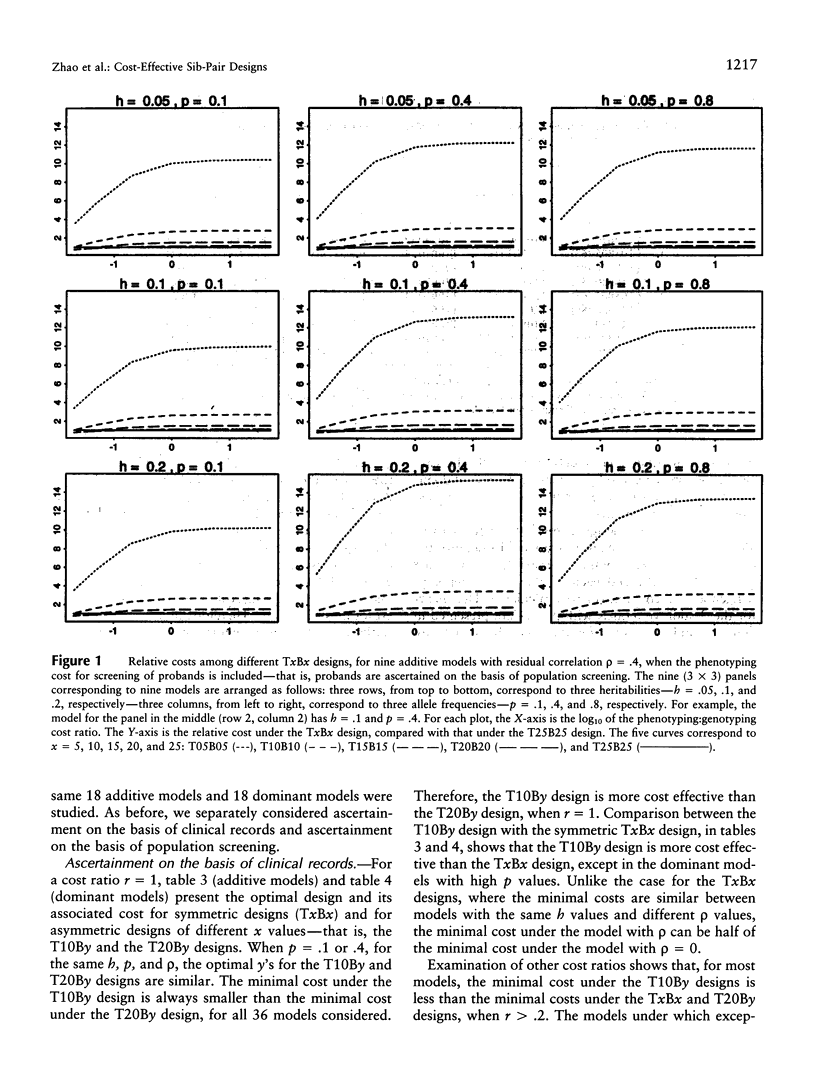
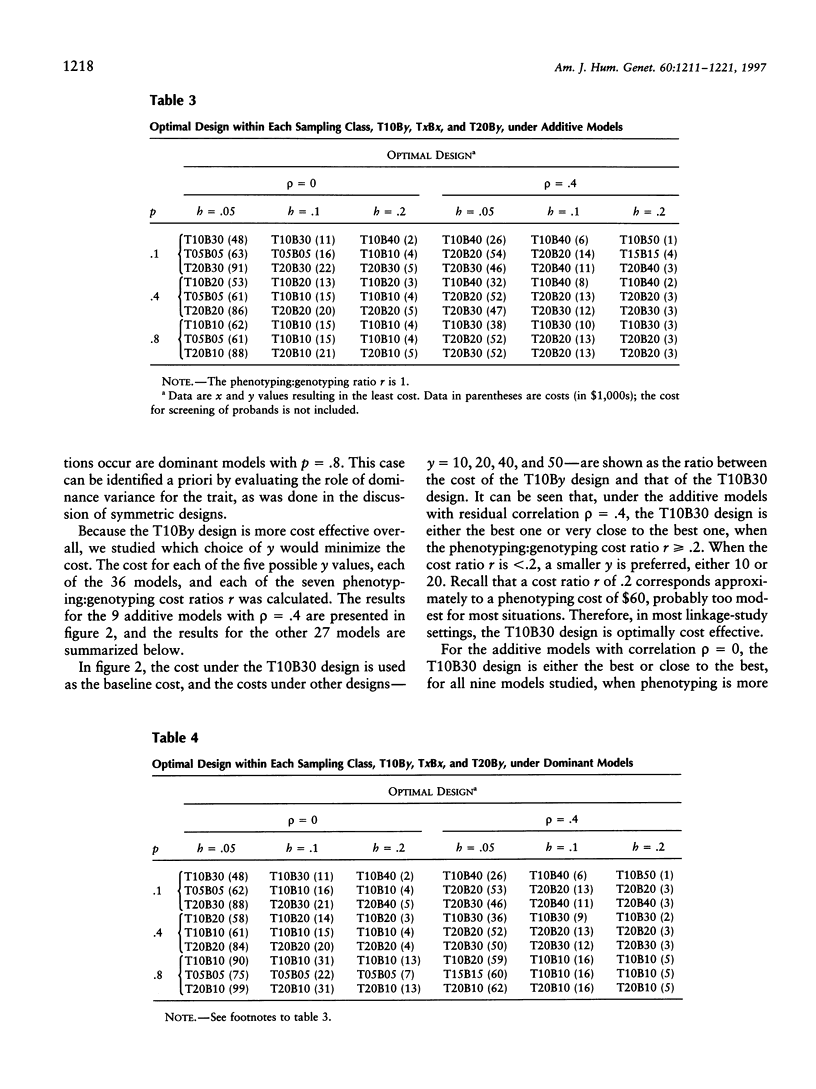
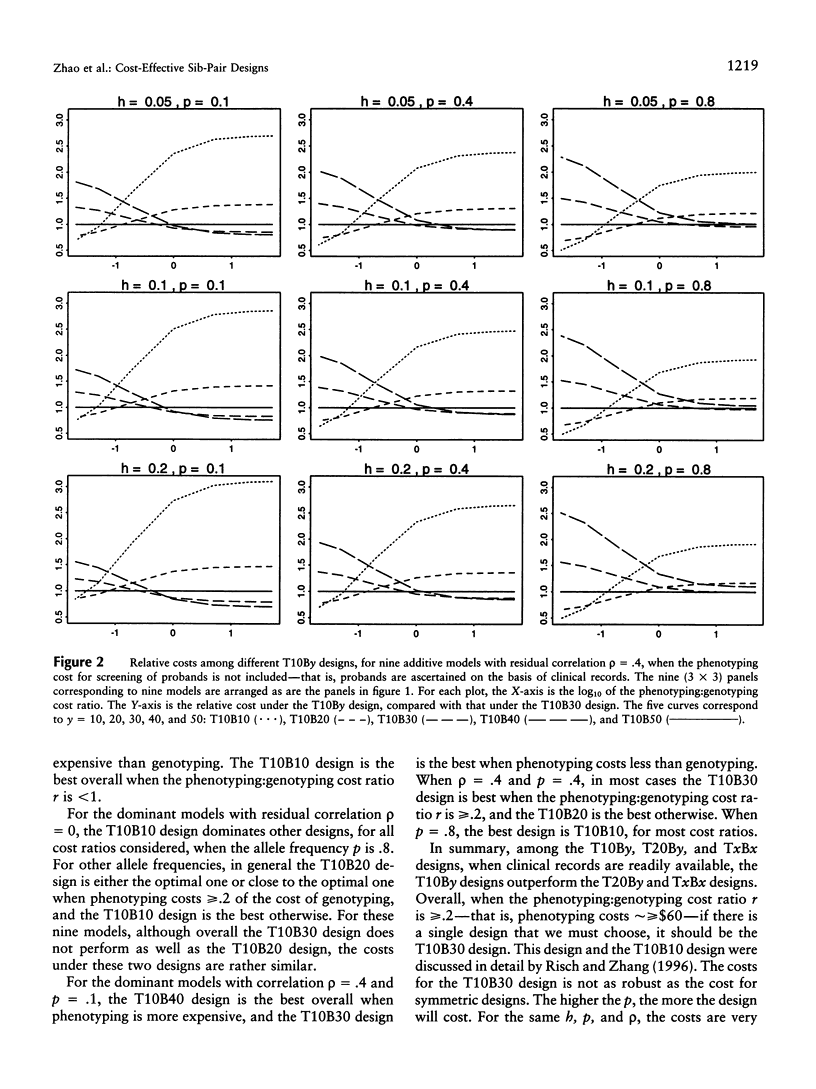
The extreme discordant-sib-pair design has been found to be the most powerful, across most genetic models. In this paper, we address two of the most frequently asked questions related to this design. First, under the extreme discordant-sib-pair design, a large number of people have to be screened for the phenotype of interest, before the desired number of discordant sibs can be collected for genotyping and linkage analysis. When the phenotyping cost is not negligible compared with the genotyping cost, such methods might not be cost effective. The second question is how sensitive the cost is to the genetic model and allele frequency. In this paper, we compare the cost under different sampling strategies, different genetic models, and different phenotyping:genotyping cost ratios. Because our knowledge of the underlying genetic model for a trait is limited, the discordant-sib-pair design proves to be the most robust. When the cost for screening probands is not included, the design that genotypes sibs with one sib in the top 10% and the other sib in the bottom 30% of the population with respect to the trait of interest is, across most models studied, the optimum among the designs considered in this paper. The cost under this design, across different genetic models, appears to be relatively robust to allele frequency and model type, whether additive or dominant. If probands initially must be screened as well, then 25% appears to be the optimal portions of the upper and lower distributions to be studied.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carey G., Williamson J. Linkage analysis of quantitative traits: increased power by using selected samples. Am J Hum Genet. 1991 Oct;49(4):786–796. [PMC free article] [PubMed] [Google Scholar]
- Eaves L., Meyer J. Locating human quantitative trait loci: guidelines for the selection of sibling pairs for genotyping. Behav Genet. 1994 Sep;24(5):443–455. doi: 10.1007/BF01076180. [DOI] [PubMed] [Google Scholar]
- Fulker D. W., Cardon L. R. A sib-pair approach to interval mapping of quantitative trait loci. Am J Hum Genet. 1994 Jun;54(6):1092–1103. [PMC free article] [PubMed] [Google Scholar]
- Haseman J. K., Elston R. C. The investigation of linkage between a quantitative trait and a marker locus. Behav Genet. 1972 Mar;2(1):3–19. doi: 10.1007/BF01066731. [DOI] [PubMed] [Google Scholar]
- Lander E., Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995 Nov;11(3):241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Laskarzewski P. M., Khoury P., Morrison J. A., Kelly K., Mellies M. J., Glueck C. J. Prevalence of familial hyper- and hypolipoproteinemias: the Princeton School District Family Study. Metabolism. 1982 Jun;31(6):558–577. doi: 10.1016/0026-0495(82)90095-6. [DOI] [PubMed] [Google Scholar]
- Namboodiri K. K., Green P. P., Kaplan E. B., Tyroler H. A., Morrison J. A., Chase G. A., Elston R. C., Rifkind B. M., Glueck C. J. Family aggregation of high density lipoprotein cholesterol. Collaborative lipid research clinics program family study. Arteriosclerosis. 1983 Nov-Dec;3(6):616–626. doi: 10.1161/01.atv.3.6.616. [DOI] [PubMed] [Google Scholar]
- Risch N., Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995 Jun 16;268(5217):1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- Whittemore A. S. Genome scanning for linkage: an overview. Am J Hum Genet. 1996 Sep;59(3):704–716. [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Risch N. Mapping quantitative-trait loci in humans by use of extreme concordant sib pairs: selected sampling by parental phenotypes. Am J Hum Genet. 1996 Oct;59(4):951–957. [PMC free article] [PubMed] [Google Scholar]


