Abstract
Triplex-forming oligonucleotides (TFOs) can bind to polypurine/polypyrimidine regions in DNA in a sequence-specific manner. The specificity of this binding raises the possibility of using triplex formation for directed genome modification, with the ultimate goal of repairing genetic defects in human cells. Several studies have demonstrated that treatment of mammalian cells with TFOs can provoke DNA repair and recombination, in a manner that can be exploited to introduce desired sequence changes. This review will summarize recent advances in this field while also highlighting major obstacles that remain to be overcome before the application of triplex technology to therapeutic gene repair can be achieved.
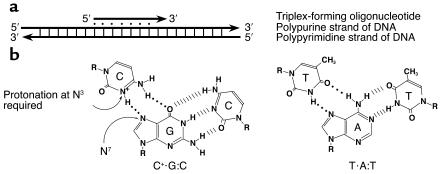
Triple-helix DNA
DNA triple helices form in a sequence-specific manner on polypurine:polypyrimidine tracts (1–3) which are widespread in mammalian genomes (4–6). The third strand lies in the major groove of an intact duplex (Figure 1) and is stabilized by two Hoogsteen hydrogen bonds between third strand bases and the purines in the duplex (3, 7). The third strand may consist of pyrimidines, or purines, depending on the nature of the target sequences (Figures 2 and 3). In the pyrimidine (or Y.R:Y) motif, a homopyrimidine oligonucleotide binds in a direction parallel to the purine strand in the duplex, with canonical base triplets of T.A:T and C.G:C. In the alternate purine motif (R.R:Y), a homopurine strand binds antiparallel to the purine strand, with base triplets of A.A:T and G.G:C (8, 9).
Figure 1.
Diagrammatic depiction of a DNA triple helix, with the third strand binding in the major groove.
Figure 2.
Diagram of the pyrimidine motif for triple helix formation. (a) Orientation of the third strand in the pyrimidine triple-helix motif. Note that the third strand is parallel in terms of 5′ to 3′ orientation with respect to the purine-rich strand of the duplex target. (b) Base triplets formed in the pyrimidine motif and illustration of the Hoogsteen hydrogen bonds that stabilize triple-helix formation.
Figure 3.
Diagram of the purine motif for triple-helix formation. (a) Orientation of the third strand in the purine triple-helix motif. Note that the third strand is oriented antiparallel in terms of 5′ to 3′ direction with respect to the purine-rich strand of the duplex target. (b) Base triplets formed in the purine motif and illustration of the reverse Hoogsteen hydrogen bonds that stabilize triple-helix formation.
The demonstration that synthetic oligonucleotides could form stable triplexes (10, 11) suggested that TFOs could be developed as sequence-specific gene targeting reagents in living cells (12–17). However, a number of obstacles have been, and still must be, overcome. Triplex chemistry and biochemistry impose fundamental limitations to TFO activity in the nuclear environment, and target options are limited to polypurine:polypyrimidine sequences. In addition, it has been shown that nucleosomes can inhibit triplex formation (18–20). Consequently, accessibility of genomic targets is an important issue.
Recent developments in nucleoside and oligonucleotide analogue chemistry show great promise for solving problems of TFO bioactivity and target options. We will discuss the challenges posed by the cellular environment and target restrictions, and some of the chemistry that may address these issues. We will also consider the activity of TFOs in biological assays. We will not attempt an in depth review of the chemistry of TFOs. Instead, we will stress some of the strategic themes and cite a few specific examples. The interested reader should see (21–23) for excellent and comprehensive discussions of TFO chemistry.
Obstacles to TFO activity under physiological conditions
Biological applications of TFOs are compromised by fundamental biophysical considerations, as well as limitations imposed by physiological conditions. Triplex formation involves the approach and binding of a negatively charged third strand to a double-negatively charged duplex. Neutralization of charge repulsion is typically provided experimentally by levels of Mg++ (5–10 mM) (24) that are much higher than what is thought to be available in cells (25). Furthermore, triplex formation involves conformational changes on the part of the third strand, and some distortion of the underlying duplex (26–28). Pyrimidine motif triplexes are unstable at physiological pH because of the requirement for cytosine protonation that occurs at relatively acidic pH (pKa = 4.5). This is necessary for the second Hoogsteen hydrogen bond, although the resultant positive charge apparently makes the more important contribution to triplex stability (29). Pyrimidine motif triplexes containing adjacent cytosines are often less stable than those with isolated cytosines. Traditionally this has been ascribed to charge-charge repulsion effects (30), although a recent study suggests incomplete protonation of adjacent cytosines may be the critical factor (31). In addition, purine motif third strands (which are G rich) may form G tetrads in physiological levels of K+, which inhibit triplex formation (32). All these factors impose kinetic barriers on triplex formation and reduce the stability of triplexes once formed (most triplexes, even under optimal conditions in vitro, are less stable than the underlying duplex).
Oligonucleotide modifications improve TFO activity under physiological conditions
The use of 5-methylcytosine partially alleviates the pH restriction of TFOs in the pyrimidine motif (33, 34). This is thought to be due to the contribution of the methyl group in the major groove to base stacking (35), and/or the exclusion of water molecules from the groove (36). Cytosine has also been replaced with analogues such as 8-oxoadenine (37), pseudoisocytidine (38), and a 6-keto derivative of 5-methylcytydine (39). These can form two hydrogen bonds at physiological pH, and may be useful for sequence targets with adjacent cytosines. Two groups have presented interesting data regarding 2-aminopyridine (2AP) (40, 41). This analogue is protonated at neutral pH and stable triplexes were formed by TFOs containing adjacent 2AP residues on targets with adjacent cytosines. These results are consistent with the argument that proton competition between adjacent cytosines is the basis for the difficulty of targets with adjacent cytosines (31). Although the biochemical data with these analogues are encouraging, only 5-methylcytosine has been tested in biological assays.
RNA third strands formed more stable pyrimidine-motif triplexes than the corresponding DNA strands (42), which prompted the use in third strands of 2′-O-Methoxy (OMe) sugar residues (27, 43), and recently, 2′, 4′ bridged ribose substitutions (44). These modifications preorganize the third strand in a conformation that is compatible with triplex formation and imposes minimal distortion on the underlying duplex (28, 44). Other modifications also improve stability. Intercalators linked to TFOs improve binding (45), propynyl-deoxyuridine reduces the Mg++ dependence (46), as does replacement of the ribose with a morpholino analogue (47).
Backbone modifications that replace the phosphate linkage (48), or a bridging oxygen atom with a nitrogen (49) improve TFO binding in vitro. Replacement of a nonbridging oxygen atom in the backbone with a charged amine reduces the likelihood of self-structure formation of purine TFOs in physiological K+ (50, 51). A positive charge on a thymidine analogue (52), or linkage of positively charged moieties to TFOs, also enhances triplex stability (53, 54).
A positive charge and an RNA-like sugar conformation have been joined in the 2′-O-(2-aminoethyl) (AE) ribose derivatives developed by Cuenoud and colleagues (55–57). TFOs carrying these substitutions show enhanced kinetics of triplex formation and greater stability of the resultant complex at physiological pH and low Mg++ concentration. NMR analysis indicates a specific interaction between the positively charged amines (at physiological pH) and phosphate groups in the purine strand of the duplex (58). A related approach has been described in a thymidine analogue containing both 5-aminopropylargyl and 2′-aminoethoxy moieties (59).
Biological activity of TFOs
In early biological studies, TFOs were envisioned as tools to inhibit gene expression by blocking transcription initiation or elongation (the “antigene” strategy) (60). Triplex formation within promoter sites has been shown to block transcription factor access and inhibit gene activation in vitro, and several studies have demonstrated that TFOs can decrease gene expression in mammalian cells in a directed way (see ref. 61 for a recent description of this approach). As an alternative strategy for genetic manipulation, we, and others, have investigated the use of TFOs to mediate genome modification, resulting in a change in target sequence (13, 17, 62–70). This has the advantage of introducing permanent changes in the target sequence, which simplifies interpretation of the experiments. However, more importantly, it also has potential as a gene knockout tool and as a means for gene correction.
In our initial work, the third strand was conjugated to a mutagen, such as psoralen, so that the sequence specificity of the third strand binding could be conferred on the action of the mutagen (71, 72). Psoralen-conjugated TFOs transfected into monkey COS cells could induce base pair–specific mutations within a supF mutation reporter gene in a simian virus 40-based plasmid episome in the cells, at frequencies in the range of 1–5% (16). The key finding in this work was that the binding affinity of the TFO to its target site, as measured in vitro, was highly correlated with its intracellular activity. TFOs with equilibrium dissociation constants (Kd’s) of approximately 10–9 M were active; those with Kd’s of 10–6 M were not.
This work was subsequently extended to psoralen-TFO–mediated knockout of chromosomal genes. In one study, the supFG1 reporter integrated into the chromosome of mouse fibroblasts was used as a target (73). Again, only high-affinity TFOs were active, achieving targeted mutagenesis frequencies of 0.1%. In the supFG1 experiments, essentially unmodified G-rich oligonucleotides (except for 3′ end capping) designed to bind in the antiparallel purine motif were used, but it should be noted that in those experiments the 30-bp polypurine site in the supFG1 gene afforded the possibility of high affinity third-strand binding by G-rich TFOs synthesized with standard DNA chemistry (16).
We have determined the activity of variously modified pyrimidine motif psoralen-TFOs, designed to target a sequence in the endogenous hamster hypoxanthine phosphoribosyltransferase (HPRT) gene, in HPRT knockout assays (17). TFOs with uniform 2′-O-methyl substitutions were inactive, but addition of a pyrene intercalator conferred knockout activity with HPRT –4 clones recovered at frequencies in the range of 10–4 to 10–3. Sequence analysis of the mutant clones confirmed the localization of mutations to the target region (17). More recently, we have prepared psoralen-TFOs containing several AE residues as well as 2′-O-methyl substitutions. These mixed substitution TFOs form triplexes at lower levels of Mg++ than required for the TFOs with only the 2′-O-methyl residues. Thermal melting experiments showed that the AE triplexes were more stable than the 2′-O-methyl only triplexes, and, notably, were more stable than the underlying duplex at physiological pH. They were also more stable in the cellular compartment in which replication and mutagenesis occur (62, 74). Most importantly they are quite active in the HPRT knockout assay (62). These results indicate that the introduction of positive charge can confer biological activity on pyrimidine motif TFOs.
Other oligonucleotide modifications also support biological activity. For example, a psoralen-linked pyrimidine TFO with a phosphoramidate backbone (in this case, substitution of a bridging oxygen with nitrogen) was shown to have statistically significant activity in a yeast reversion assay (75). Psoralen-linked G-rich TFOs in the purine motif, with a nonbridging phosphoramidate substitution (N,N-diethyl-ethylenediamine), were active in targeted mutagenesis experiments at a short (10 bp) homopurine/homopyrimidine site in mammalian cells that otherwise is not susceptible to targeting by unmodified phosphodiester oligonucleotides (67). Improved binding activity in reduced Mg++ is probably the basis for the bioactivity of the TFOs carrying these modifications.
In addition to the use of psoralen-conjugated TFOs in site-directed mutagenesis studies in mammalian cells and in yeast, a few groups have used these reagents as tools for direct physical demonstrations of triplex formation in permeabilized mammalian cells. Such studies have used PCR and restriction enzyme protection assays to argue for the formation of TFO-targeted crosslinks in genomic DNA in chromatin (76–78). These results, and the mutagenesis experiments, show that the packaging of the DNA into chromatin is not an absolute barrier to gene targeting with TFOs.
Triplex formation by itself can promote DNA metabolism
In the course of our work with psoralen-TFOs, we observed that unconjugated TFOs were also capable of inducing mutations in the target gene in an SV40 vector, at least when the binding affinity was sufficiently high (79). This effect was shown to be a consequence of the stimulation of DNA repair by the formation of the triple helix, which seems to be recognized by the nucleotide excision repair complex as a lesion.
Recently, Vasquez et al. demonstrated that systemically administered TFOs (without psoralen or any other DNA reactive conjugate) could induce mutations at specific genomic sites in the somatic cells of adult mice (68). In this work, a 30-mer purine TFO, with a 3′ propanolamine group to prevent exonuclease-mediated degradation, was injected intraperitoneally for 5 consecutive days. The TFO was targeted to chromosomal copies of an integrated supF reporter gene. After an additional 10 days, the mice were sacrificed, and tissues were taken for mutation analysis. In general, mice treated with the sequence-specific TFO had a fivefold elevated mutation rate in the targeted supF gene, but not in the nontargeted cII gene. All tissues tested showed TFO-induced mutagenesis except the brain, which had no mutagenesis over background, consistent with TFOs inability to cross the blood-brain barrier. This work established that site-directed DNA binding molecules, upon systemic administration, can mediate gene targeting and gene modification in vivo in whole animals.
Triplex-induced recombination: gene correction via recombinational repair
Based on the concept that third-strand binding, with or without psoralen coupling, can trigger DNA repair, we hypothesized that such binding might also be recombinogenic due to the production of repair-dependent DNA strand breaks. Using an SV40 vector containing two mutant copies of the supF gene, we found that both psoralen-TFOs (64) and non-psoralen TFOs (63) could trigger recombination within an SV40 virus genome. This induced recombination, in the case of the TFOs, without psoralen, was found to be dependent on the presence of functional Xeroderma pigmentosum group A protein (XPA) protein (63), the key recognition factor in nucleotide excision repair, a result consistent with our hypothesis.
These results were extended to a chromosomal target, in which two mutant thymidine kinase (TK) genes were integrated into a single chromosomal site in mouse fibroblasts (69). Transfection (via cationic liposomes) of the cells with high-affinity TFOs targeting a region between the two TK genes yielded recombination at a frequency of approximately 30 × 10–6, about sevenfold above background. When the TFOs were microinjected into the nuclei of the cells (about 70,000 copies/cell), the yield of recombinants increased to 1–2%, more than 1000-fold over background. Analysis of the recombinant clones revealed all the recombination events involved gene conversion rather than cross-over recombination. Similar work to use TFOs to stimulate recombination has been described by Wilson and colleagues using a locus in CHO cells containing duplicated APRT genes as a target (80). Besides providing proof-of-principle, the results obtained in the TK system highlight another challenge in using triplex technology for gene repair: achieving efficient delivery into the cell nucleus.
Bifunctional oligonucleotides for gene correction
The observation of the ability of third strand binding to provoke DNA repair and stimulate recombination led us to develop a strategy to mediate targeted gene conversion using a TFO linked to a short DNA fragment homologous to the target site (except for the base pair to be corrected) (70). In this bifunctional molecule, the TFO domain mediates site-specific binding to target the molecule to the desired gene. This binding also triggers repair to sensitize the target site to recombination. The tethered homologous donor fragment can participate in recombination and/or gene conversion with the target gene to correct or alter the nucleotide sequence.
Using a bifunctional oligomer with a 40-mer donor domain and a 30-mer TFO domain, correction of a single base-pair mutation in the supF reporter gene within an SV40 vector in COS cells was achieved (70). Correction frequencies were in the range of 0.1–0.5% with the full bifunctional molecule. Oligomers consisting of either domain above or of either domain substituted with heterologous sequences reduced activity by tenfold or more. The donor domain alone consistently did mediate some gene correction, as would be expected, based on the known ability of short DNA fragments to mediate some level of recombination (81–83). However, there was a clear synergism due to combination with the TFO domain.
In vitro studies in human cell-free extracts further demonstrated that triplex formation could induce recombination of the target site with a short donor oligonucleotide, and they also revealed a requirement for XPA and Rad51 function in the pathway (84). Importantly, the in vitro work also revealed that the donor DNA does not need to be covalently linked to the TFO, suggesting that the ability of the third strand to stimulate repair and recombination is a key property, presumably because of the production of strand breaks as recombinogenic intermediates.
Several other in vitro studies have demonstrated synergistic binding of bifunctional oligonucleotides containing domains designed to form triplexes as well as duplexes with a target DNA (66, 85, 86). Interestingly, in one strategy, the triplex-forming domain was linked to the duplex-forming domain by annealing via a short stretch of complementary nucleotides (86), rather than via covalent linkage (the latter either by postsynthetic coupling or by cosynthesis as a single, long oligonucleotide).
Prospects for the expansion of the triplex-binding code
Although we have just begun to explore the applications of TFOs for genome modification, we would argue that two fundamental issues have been resolved in recent years: that it is possible to prepare TFOs with gene-targeting activity in vivo, and that bioactivity implies at least some degree of target accessibility. Thus, although there is a great deal to be done to extend these conclusions, expansion of target options to mixed purine/pyrimidine sequences would appear to be the most important challenge facing the field. We will discuss some of the efforts to expand the triplex-binding code, although it should be pointed out that only a few compounds have been tested in bioassays. For thorough accounts of this aspect of the field the reader should consult recent reviews (21, 23).
Purine bases engaged in Watson Crick pairing can form two additional hydrogen bonds, while duplex pyrimidines can only form one additional hydrogen bond. These hydrogen bonding patterns are the basis for much of the stability of conventional triplexes, and one of the reasons for the instability of triplexes on mixed sequences. An additional problem presented by T:A inversions is the projection into the major groove by the 5-methyl group of T resulting in steric hindrance to groove occupancy by third strands. Candidate compounds for binding at inversion sites fall into different categories: natural bases, intercalators, analogues that make a single hydrogen bond with the inverted pyrimidine base, analogues that make hydrogen bonds with the purine base, and, perhaps, also with the pyrimidine base (base pair binders).
Natural bases in parallel triplexes have been shown to form triplets at inverted base pairs, specifically G.T:A, and T.C:G (87, 88). Although these have been incorporated in TFOs employed in biological experiments, triplexes containing them are not as stable as perfectly matched triplexes (89, 90). Thus we found that a TFO containing G, for triplet formation at a T:A inversion site, was not effective in our hprt gene knockout assay (17). The T.C:G triplet can form in both parallel and antiparallel triplexes. Use in parallel triplexes is compromised by the lack of sequence stringency (since T also forms triplets with A:T pairs). However this limitation does not apply to antiparallel triplexes and we have used T in a biologically active purine motif TFO (AG30, described above) for triplet formation at T:A sites in an otherwise perfect polypurine target (16).
Intercalators, incorporated into the TFOs and located adjacent to mismatch sites, have been used to stabilize natural base triplets with inverted base pairs. An acridine derivative stabilized either inversion depending on its position relative to the site (45). A naphthalene-based intercalator linked to 5-methylcytosine, designed to form one hydrogen bond and also intercalate, showed selectivity for C:G inversions (91). We introduced a pyrene derivative at a T:A inversion site in our active TFO in our initial demonstration of HPRT gene knockout (17). It is clear that intercalators can enhance triplex stability, however it is likely that this will be at the cost of some degree of sequence specificity. Precisely this observation was made with one of the earliest efforts to construct an analogue for expansion of target options. A benzamidophenyl derivative of imidazole formed stable triplets at both T:A and C:G base pairs by intercalation next to the target base pairs (92, 93).
There have been a number of attempts to synthesize compounds with hydrogen bonding potential that would form triplets with inverted pairs with good affinity and specificity. There have been some promising developments with the C:G inversion, particularly in the pyrimidine motif. A thymidine analogue designed to form a hydrogen bond with the C showed selectivity relative to the other base pairs (94). Similar results with a related compound have been reported in one of the few instances in which the new sugar chemistry has been combined with a base analogue (95). A base pair recognition strategy was pursued by Miller and colleagues who prepared several N4-substituted cytosine derivatives with side-chain extensions designed to make hydrogen bonds with both the cytosine and the guanine in the inverted base pair (96, 97). This approach is conceptually attractive because formation of hydrogen bonds with both bases should enhance the specificity of the interaction with the intended base pair, as indeed was observed with some of the derivatives (97). While these and other analogues show promise for solving the C:G inversion they do not form triplets as strong as the canonical C+.G:C. However, this is, in part, because they do not carry a positive charge. Combination with sugar and backbone modifications that introduce positive charge may overcome this limitation.
Analogues that bind C:G interruptions in antiparallel purine–motif triplexes have also been described. An example of an analogue that can reach across the major groove and form hydrogen bonds with the guanine base in the inverted C:G pair is 2′-deoxyformycin A (98). Another interesting approach involved the replacement of natural bases with azole derivatives. The use of the smaller aromatic ring was intended to overcome some of the steric problems associated with base pair inversions (99). They were able to demonstrate enhanced binding by the substituted TFOs to targets containing inversions, but base pair discrimination was poor. However, this may be a useful point of departure for side-chain derivatives that would improve selectivity.
Current efforts to overcome the T:A inversion have been less successful. A recent effort employed a post synthetic modification strategy, at an internal acyclic linker, to prepare a number of candidates (100). Although this approach has a great deal of potential, the most effective binders recovered in the study were intercalators, which failed to distinguish between T:A and C:G base pairs in the target sequence. This result is similar to those reported previously and suggests that in further work analogues with intercalative potential should be avoided. Finally, a pyridazine derivative on an acyclic linker has been shown to form triplets with T:A pairs (101). This analogue showed reasonable base pair discrimination, but was analyzed in the context of a peptide nucleic acid, and has not been examined in an oligonucleotide.
While this is a cursory overview, it is striking how much thoughtful chemistry has been described, some with considerable promise, and how rare has been the analysis of these developments in biological assays.
Summary and future directions
Overall, the work to date suggests that TFOs can be used to mediate site-specific genome modification. This capacity derives not only from the ability of TFOs to bind as third strands with sequence specificity but also from the ability of the resulting triple helices, or TFO-mutagen complexes, to provoke repair and recombination, leading to directed mutagenesis, recombination, and, potentially, gene correction. It seems likely that recent advances in oligonucleotide chemistry have considerable potential for the development of TFOs with robust gene targeting activity. This will require coordinated effort between the chemists and biologists, but recent data suggest that this effort will be rewarded.
Note added in proof.
B.J. Gold and colleagues have recently described a novel approach towards the development of TFOs that can bind general sequences (102).
Acknowledgments
This work was supported by the NIH (grants GM54731 and CA64186 to P.M. Glazer).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: triplex-forming oligonucleotide (TFO); 2-aminopyridine (2AP); 2′-O-methoxy (OMe); 2-aminoethyl (AE); equilibrium dissociation constant (Kd); hypoxanthine phosphoribosyltransferase (HPRT); Xeroderma pigmentosum group A protein (XPA); thymidine kinase (TK).
References
- 1.Felsenfeld G, Davies DR, Rich A. Formation of a three stranded polynucleotide molecule. J. Am. Chem. Soc. 1957;79:2023–2024. [Google Scholar]
- 2.Thuong NT, Helene C. Sequence specific recognition and modification of double helical DNA by oligonucleotides. Angewandte Chemie. Intl. Ed. Eng. 1993;32:666–690. [Google Scholar]
- 3.Frank-Kamenetskii MD, Mirkin SM. Triplex DNA structures. Annu. Rev. Biochem. 1995;64:65–95. doi: 10.1146/annurev.bi.64.070195.000433. [DOI] [PubMed] [Google Scholar]
- 4.Manor H, Rao BS, Martin RG. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J. Mol. Evol. 1988;27:96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- 5.Schroth GP, Ho PS. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behe MJ. An overabundance of long oligopurine tracts occurs in the genome of simple and complex eukaryotes. Nucleic Acids Res. 1995;23:689–695. doi: 10.1093/nar/23.4.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radhakrishnan I, Patel DJ. DNA triplexes: solution structures, hydration sites, energetics, interactions, and function. Biochemistry. 1994;33:11405–11416. doi: 10.1021/bi00204a001. [DOI] [PubMed] [Google Scholar]
- 8.Fossella JA, Kim YJ, Shih H, Richards EG, Fresco JR. Relative specificities in binding of Watson-Crick base pairs by third strand residues in a DNA pyrimidine triplex motif. Nucleic Acids Res. 1993;21:4511–4515. doi: 10.1093/nar/21.19.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Letai AG, Palladino MA, Fromm E, Rizzo V, Fresco JR. Specificity in formation of triple-stranded nucleic acid helical complexes: studies with agarose-linked polyribonucleotide affinity columns. Biochemistry. 1988;27:9108–9112. doi: 10.1021/bi00426a007. [DOI] [PubMed] [Google Scholar]
- 10.Moser HE, Dervan PB. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- 11.Le Doan T, et al. Sequence-specific recognition, photocrosslinking and cleavage of the DNA double helix by an oligo-[alpha]-thymidylate covalently linked to an azidoproflavine derivative. Nucleic Acids Res. 1987;15:7749–7760. doi: 10.1093/nar/15.19.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helene C, Thuong NT, Harel-Bellan A. Control of gene expression by triple helix–forming oligonucleotides. The antigene strategy. Ann. N. Y. Acad. Sci. 1992;660:27–36. doi: 10.1111/j.1749-6632.1992.tb21054.x. [DOI] [PubMed] [Google Scholar]
- 13.Vasquez KM, Wilson JH. Triplex-directed modification of genes and gene activity. Trends Biochem. Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 14.Maher LJ., III Prospects for the therapeutic use of antigene oligonucleotides. Cancer Invest. 1996;14:66–82. doi: 10.3109/07357909609018437. [DOI] [PubMed] [Google Scholar]
- 15.Neidle S. Recent developments in triple-helix regulation of gene expression. Anticancer Drug Des. 1997;12:433–442. [PubMed] [Google Scholar]
- 16.Wang G, Levy DD, Seidman MM, Glazer PM. Targeted mutagenesis in mammalian cells mediated by intracellular triple helix formation. Mol. Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majumdar A, et al. Targeted gene knockout mediated by triple helix forming oligonucleotides. Nat. Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 18.Brown PM, Fox KR. Nucleosome core particles inhibit DNA triple helix formation. Biochem. J. 1996;319:607–611. doi: 10.1042/bj3190607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espinas ML, Jimenez-Garcia E, Martinez-Balbas A, Azorin F. Formation of triple-stranded DNA at d(GA.TC)n sequences prevents nucleosome assembly and is hindered by nucleosomes. J. Biol. Chem. 1996;271:31807–31812. doi: 10.1074/jbc.271.50.31807. [DOI] [PubMed] [Google Scholar]
- 20.Bailey CP, Dagle JM, Weeks DL. Cationic oligonucleotides can mediate specific inhibition of gene expression in Xenopus oocytes. Nucleic Acids Res. 1998;26:4860–4867. doi: 10.1093/nar/26.21.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gowers DM, Fox KR. Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res. 1999;27:1569–1577. doi: 10.1093/nar/27.7.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox KR. Targeting DNA with triplexes. Curr. Med. Chem. 2000;7:17–37. doi: 10.2174/0929867003375506. [DOI] [PubMed] [Google Scholar]
- 23.Luyten IA, Herdewijn P. Hybridization properties of base-modified oligonucleotides within the double and triple helix motif. Eur. J. Med. Chem. 2002;33:515–576. [Google Scholar]
- 24.Blume SW, et al. The integral divalent cation within the intermolecular purine*purine.pyrimidine structure: a variable determinant of the potential for and characteristics of the triple helical association. Nucleic Acids Res. 1999;27:695–702. doi: 10.1093/nar/27.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pesco J, Salmon JM, Vigo J, Viallet P. Mag-indo1 affinity for Ca(2+), compartmentalization and binding to proteins: the challenge of measuring Mg(2+) concentrations in living cells. Anal. Biochem. 2001;290:221–231. doi: 10.1006/abio.2000.4983. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert DE, Feigon J. Multistranded DNA structures. Curr. Opin. Struct. Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 27.Shimizu M, Konishi A, Shimada Y, Inoue H, Ohtsuka E. Oligo(2′-O-methyl)ribonucleotides. Effective probes for duplex DNA. FEBS Lett. 1992;302:155–158. doi: 10.1016/0014-5793(92)80428-j. [DOI] [PubMed] [Google Scholar]
- 28.Asensio JL, Carr R, Brown T, Lane AN. Conformational and thermodynamic properties of parallel intramolecular triple helixes containing a DNA, RNA, or 2′-OMeDNA third strand. J. Am. Chem. Soc. 1999;121:11063–11070. [Google Scholar]
- 29.Asensio JL, Lane AN, Dhesi J, Bergqvist S, Brown T. The contribution of cytosine protonation to the stability of parallel DNA triple helices. J. Mol. Biol. 1998;275:811–822. doi: 10.1006/jmbi.1997.1520. [DOI] [PubMed] [Google Scholar]
- 30.Volker J, Klump HH. Electrostatic effects in DNA triple helices. Biochemistry. 1994;33:13502–13508. doi: 10.1021/bi00249a039. [DOI] [PubMed] [Google Scholar]
- 31.Sugimoto N, Wu P, Hara H, Kawamoto Y. pH and cation effects on the properties of parallel pyrimidine motif DNA triplexes. Biochemistry. 2001;40:9396–9405. doi: 10.1021/bi010666l. [DOI] [PubMed] [Google Scholar]
- 32.Arimondo PB, Garestier T, Helene C, Sun JS. Detection of competing DNA structures by thermal gradient gel electrophoresis: from self-association to triple helix formation by (G,A)-containing oligonucleotides. Nucleic Acids Res. 2001;29:E15. doi: 10.1093/nar/29.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JS, Woodsworth ML, Latimer LJ, Morgan AR. Poly(pyrimidine).poly(purine) synthetic DNAs containing 5 - methylcytosine form stable triplexes at neutral pH. Nucleic Acids Res. 1984;12:6603–6614. doi: 10.1093/nar/12.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Povsic TJ, Dervan PB. Triple helix formation by oligonucleotides on DNA extended to the physiological range. J. Am. Chem. Soc. 1989;111:3059–3061. [Google Scholar]
- 35.Singleton SF, Dervan PB. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992;31:10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- 36.Xodo LE, Manzini G, Quadrifoglio F, van der Marel GA, van Boom JH. Effect of 5-methylcytosine on the stability of triple-stranded DNA—a thermodynamic study. Nucleic Acids Res. 1991;19:5625–5631. doi: 10.1093/nar/19.20.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller PS, Bi G, Kipp SA, Fok V, DeLong RK. Triplex formation by a psoralen-conjugated oligodeoxyribonucleotide containing the base analog 8-oxo-adenine. Nucleic Acids Res. 1996;24:730–736. doi: 10.1093/nar/24.4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono A, Ts’o PO, Kan L. Triple helix formation of oligonucleotides containing 2′-O-methylpseudoisocytidine in substitition for 2′-deoxycytidine. J. Am. Chem. Soc. 1991;113:4032–4033. [Google Scholar]
- 39.Xiang GB, McLaughlin LW. A cytosine analogue containing a conformationally flexible acyclic linker for triplex formation at sites with contiguous G-C base pairs. Tetrahedron. 1998;54:375–392. [Google Scholar]
- 40.Cassidy SA, et al. Recognition of GC base pairs by triplex forming oligonucleotides containing nucleosides derived from 2-aminopyridine. Nucleic Acids Res. 1997;25:4891–4898. doi: 10.1093/nar/25.24.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hildebrandt S, Blaser A, Parel SP, Leumann CJ. 5-substituted 2-aminopyridine C-nucleosides as protonated cytidine equivalents: increasing efficiency and selectivity in DNA triple helix formation. J. Am. Chem. Soc. 1997;119:5499–5511. [Google Scholar]
- 42.Roberts RW, Crothers DM. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 43.Escude C, Sun JS, Rougee M, Garestier T, Helene C. Stable triple helices are formed upon binding of RNA oligonucleotides and their 2′-O-methyl derivatives to double-helical DNA. C. R. Acad. Sci. III. 1992;315:521–525. [PubMed] [Google Scholar]
- 44.Torigoe H, Hari Y, Sekiguchi M, Obika S, Imanishi T. 2′-O,4’-C-methylene bridged nucleic acid modification promotes pyrimidine motif triplex DNA formation at physiological pH: thermodynamic and kinetic studies. J. Biol. Chem. 2001;276:2354–2360. doi: 10.1074/jbc.M007783200. [DOI] [PubMed] [Google Scholar]
- 45.Kukreti S, Sun JS, Garestier T, Helene C. Extension of the range of DNA sequences available for triple helix formation: stabilization of mismatched triplexes by acridine-containing oligonucleotides. Nucleic Acids Res. 1997;25:4264–4270. doi: 10.1093/nar/25.21.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lacroix L, et al. Triplex formation by oligonucleotides containing 5-(1-propynyl)-2’- deoxyuridine: decreased magnesium dependence and improved intracellular gene targeting. Biochemistry. 1999;38:1893–1901. doi: 10.1021/bi982290q. [DOI] [PubMed] [Google Scholar]
- 47.Lacroix L, Arimondo PB, Takasugi M, Helene C, Mergny JL. Pyrimidine morpholino oligonucleotides form a stable triple helix in the absence of magnesium ions. Biochem. Biophys. Res. Commun. 2000;270:363–369. doi: 10.1006/bbrc.2000.2438. [DOI] [PubMed] [Google Scholar]
- 48.Arya DP, Bruice TC. Triple-helix formation of DNA oligomers with methylthiourea-linked nucleosides (DNmt): a kinetic and thermodynamic analysis. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4384–4389. doi: 10.1073/pnas.96.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escude C, et al. Stable triple helices formed by oligonucleotide N3′ → P5’ phosphoramidates inhibit transcription elongation. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4365–4369. doi: 10.1073/pnas.93.9.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dagle JM, Weeks DL. Positively charged oligonucleotides overcome potassium-mediated inhibition of triplex DNA formation. Nucleic Acids Res. 1996;24:2143–2149. doi: 10.1093/nar/24.11.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrenmann F, Vasseur JJ, Debart F. Alpha-oligonucleotides with anionic phosphodiester and cationic phosphoramidate linkages enhanced stability of DNA triple helix. Nucleosides Nucleotides Nucleic Acids. 2001;20:797–799. doi: 10.1081/NCN-100002432. [DOI] [PubMed] [Google Scholar]
- 52.Bijapur J, Keppler MD, Bergqvist S, Brown T, Fox KR. 5-(1-propargylamino)-2′-deoxyuridine (UP): a novel thymidine analogue for generating DNA triplexes with increased stability. Nucleic Acids Res. 1999;27:1802–1809. doi: 10.1093/nar/27.8.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rajeev KG, Jadhav VR, Ganesh KN. Triplex formation at physiological pH: comparative studies on DNA triplexes containing 5-Me-dC tethered at N4 with spermine and tetraethyleneoxyamine. Nucleic Acids Res. 1997;25:4187–4193. doi: 10.1093/nar/25.21.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sund C, Puri N, Chattopadhyaya J. Synthesis of C-branched spermine tethered oligo-DNA and the thermal stability of the duplexes and triplexes. Tetrahedron. 1996;52:12275–12290. [Google Scholar]
- 55.Cuenoud B, et al. Dual recognition of double stranded DNA by 2’-aminoethoxy-modified oligonucleotides. Angew. Chem. Int. Ed. Engl. 1998;37:1288–1291. doi: 10.1002/(SICI)1521-3773(19980518)37:9<1288::AID-ANIE1288>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 56.Blommers MJ, Natt F, Jahnke W, Cuenoud B. Dual recognition of double-stranded DNA by 2′-aminoethoxy-modified oligonucleotides: the solution structure of an intramolecular triplex obtained by NMR spectroscopy. Biochemistry. 1998;37:17714–17725. doi: 10.1021/bi9816352. [DOI] [PubMed] [Google Scholar]
- 57.Stutz AM, Hoeck J, Natt F, Cuenoud B, Woisetschlager M. Inhibition of interleukin-4- and CD40-induced IgE germline gene promoter activity by 2′-aminoethoxy-modified triplex-forming oligonucleotides. J. Biol. Chem. 2001;276:11759–11765. doi: 10.1074/jbc.M010260200. [DOI] [PubMed] [Google Scholar]
- 58.Carlomagno T, Blommers MJ, Meiler J, Cuenoud B, Griesinger C. Determination of aliphatic side-chain conformation using cross- correlated relaxation: application to an extraordinarily stable 2′- aminoethoxy-modified oligonucleotide triplex. J. Am. Chem. Soc. 2001;123:7364–7370. doi: 10.1021/ja002592r. [DOI] [PubMed] [Google Scholar]
- 59.Sollogoub M, Dominguez B, Fox KR, Brown T. Synthesis of a novel bis-amino-modified thymidine monomer for use in DNA triplex stabilization. Chemical Communications. 2000;23:2315–2316. [Google Scholar]
- 60.Helene C. The anti-gene strategy: control of gene expression by triplex-forming- oligonucleotides. Anticancer Drug Des. 1991;6:569–584. [PubMed] [Google Scholar]
- 61.Rapozzi V, et al. Antigene effect in K562 cells of a PEG-conjugated triplex-forming oligonucleotide targeted to the bcr/abl oncogene. Biochemistry. 2002;41:502–510. doi: 10.1021/bi011314h. [DOI] [PubMed] [Google Scholar]
- 62.Puri N, et al. Targeted gene knockout by 2′-O-aminoethyl modified triplex forming oligonucleotides. J. Biol. Chem. 2001;276:28991–28998. doi: 10.1074/jbc.M103409200. [DOI] [PubMed] [Google Scholar]
- 63.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol. Cell Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Faruqi AF, Seidman MM, Segal DJ, Carroll D, Glazer PM. Recombination induced by triple-helix-targeted DNA damage in mammalian cells. Mol. Cell Biol. 1996;16:6820–6828. doi: 10.1128/mcb.16.12.6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandor Z, Bredberg A. Repair of triple helix directed psoralen adducts in human cells. Nucleic Acids Res. 1994;22:2051–2056. doi: 10.1093/nar/22.11.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Broitman S, Amosova O, Dolinnaya NG, Fresco JR. Repairing the sickle cell mutation. I. Specific covalent binding of a photoreactive third strand to the mutated base pair. J. Biol. Chem. 1999;274:21763–21768. doi: 10.1074/jbc.274.31.21763. [DOI] [PubMed] [Google Scholar]
- 67.Vasquez KM, Dagle JM, Weeks DL, Glazer PM. Chromosome targeting at short polypurine sites by cationic triplex- forming oligonucleotides. J. Biol. Chem. 2001;276:38536–38541. doi: 10.1074/jbc.M101797200. [DOI] [PubMed] [Google Scholar]
- 68.Vasquez KM, Narayanan L, Glazer PM. Specific mutations induced by triplex-forming oligonucleotides in mice. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 69.Luo Z, Macris MA, Faruqi AF, Glazer PM. High-frequency intrachromosomal gene conversion induced by triplex- forming oligonucleotides microinjected into mouse cells. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9003–9008. doi: 10.1073/pnas.160004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan PP, et al. Targeted correction of an episomal gene in mammalian cells by a short DNA fragment tethered to a triplex-forming oligonucleotide. J. Biol. Chem. 1999;274:11541–11548. doi: 10.1074/jbc.274.17.11541. [DOI] [PubMed] [Google Scholar]
- 71.Havre PA, Glazer PM. Targeted mutagenesis of simian virus 40 DNA mediated by a triple helix- forming oligonucleotide. J. Virol. 1993;67:7324–7331. doi: 10.1128/jvi.67.12.7324-7331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Havre PA, Gunther EJ, Gasparro FP, Glazer PM. Targeted mutagenesis of DNA using triple helix-forming oligonucleotides linked to psoralen. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vasquez KM, Wang G, Havre PA, Glazer PM. Chromosomal mutations induced by triplex-forming oligonucleotides in mammalian cells. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lin FL, et al. Stability of DNA triplexes on shuttle vector plasmids in the replication pool in mammalian cells. J. Biol. Chem. 2000;275:39117–39124. doi: 10.1074/jbc.M005404200. [DOI] [PubMed] [Google Scholar]
- 75.Barre FX, et al. Unambiguous demonstration of triple-helix-directed gene modification. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3084–3088. doi: 10.1073/pnas.050368997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giovannangeli C, et al. Accessibility of nuclear DNA to triplex-forming oligonucleotides: the integrated HIV-1 provirus as a target. Proc. Natl. Acad. Sci. U. S. A. 1997;94:79–84. doi: 10.1073/pnas.94.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Belousov ES, et al. Triplex targeting of a native gene in permeabilized intact cells: covalent modification of the gene for the chemokine receptor CCR5. Nucleic Acids Res. 1998;26:1324–1328. doi: 10.1093/nar/26.5.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oh DH, Hanawalt PC. Triple helix-forming oligonucleotides target psoralen adducts to specific chromosomal sequences in human cells. Nucleic Acids Res. 1999;27:4734–4742. doi: 10.1093/nar/27.24.4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang G, Seidman MM, Glazer PM. Mutagenesis in mammalian cells induced by triple helix formation and transcription-coupled repair. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 80.Vasquez KM, Marburger K, Intody Z, Wilson JH. Manipulating the mammalian genome by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 2001;98:8403–8410. doi: 10.1073/pnas.111009698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin FL, Sperle K, Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol. Cell Biol. 1990;10:113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Campbell CR, Keown W, Lowe L, Kirschling D, Kucherlapati R. Homologous recombination involving small single-stranded oligonucleotides in human cells. New Biol. 1989;1:223–227. [PubMed] [Google Scholar]
- 83.Goncz KK, Kunzelmann K, Xu Z, Gruenert DC. Targeted replacement of normal and mutant CFTR sequences in human airway epithelial cells using DNA fragments. Hum. Mol. Genet. 1998;7:1913–1919. doi: 10.1093/hmg/7.12.1913. [DOI] [PubMed] [Google Scholar]
- 84.Datta HJ, Chan PP, Vasquez KM, Gupta RC, Glazer PM. Triplex-induced recombination in human cell-free extracts: Dependence on XPA and HsRad51. J. Biol. Chem. 2001;276:18018–18023. doi: 10.1074/jbc.M011646200. [DOI] [PubMed] [Google Scholar]
- 85.Gamper H, Hou Y-M, Stamm MR, Podyminogin M, Meyer RB. Strand invasion of supercoiled DNA by oligonucleotide with a triplex guide sequence. J. Am. Chem. Soc. 1998;120:2182–2183. [Google Scholar]
- 86.Biet E, Maurisse R, Dutreix M, Sun J. Stimulation of RecA-mediated D-loop formation by oligonucleotide- directed triple-helix formation: guided homologous recombination (GOREC) Biochemistry. 2001;40:1779–1786. doi: 10.1021/bi001605a. [DOI] [PubMed] [Google Scholar]
- 87.Griffin LC, Dervan PB. Recognition of thymine adenine.base pairs by guanine in a pyrimidine triple helix motif. Science. 1989;245:967–971. doi: 10.1126/science.2549639. [DOI] [PubMed] [Google Scholar]
- 88.Greenberg W, Dervan PB. Energetics of formation of sixteen triple helical complexes which vary at a single posiiton within a purine motif. J. Am. Chem. Soc. 1995;117:5016–5022. [Google Scholar]
- 89.Wang E, Malek S, Feigon J. Structure of a G.T.A triplet in an intramolecular DNA triplex. Biochemistry. 1992;31:4838–4846. doi: 10.1021/bi00135a015. [DOI] [PubMed] [Google Scholar]
- 90.Jiang L, Russu IM. Proton exchange and local stability in a DNA triple helix containing a G.TA triad. Nucleic Acids Res. 2001;29:4231–4237. doi: 10.1093/nar/29.20.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gianolio DA, McLaughlin LW. Selective recognition of a dC-dG base pair by oligonucleotide directed triplex formation using a dC residue tethering an intercalator. J. Am. Chem. Soc. 1999;121:6334–6335. [Google Scholar]
- 92.Griffin LC, Kiessling LL, Beal PA, Gillespie P, Dervan PB. Recognition of all four base pairs of double helical DNA by triple helix formation: design of nonnaturaldeoxyribonucleosides for pyrimidine-purine base pair binding. J. Am. Chem. Soc. 1992;114:7976–7982. [Google Scholar]
- 93.Koshlap KM, Gillespie P, Dervan PB, Feigon J. Nonnatural deoxyribonucleoside D3 incorporated in an intramolecular DNA triplex binds sequence-specifically by intercalation. J. Am. Chem. Soc. 1993;115:7908–7909. [Google Scholar]
- 94.Prevot-Halter I, Leumann CJ. Selective recognition of a C-G base-pair in the parallel DNA triple- helical binding motif. Bioorg. Med. Chem. Lett. 1999;9:2657–2660. doi: 10.1016/s0960-894x(99)00451-5. [DOI] [PubMed] [Google Scholar]
- 95.Okiba S, Hari Y, Sekiguchi M, Imanishi T. A 2′,4′-bridged nucleic acid containing 2-pyridone as a nucleobase: efficient recognition of a C-C interruption by triplex formation with a pyrimidine motif. Angew. Chem. Int. Ed. Engl. 2002;40:2079–2081. doi: 10.1002/1521-3773(20010601)40:11<2079::AID-ANIE2079>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 96.Huang CY, Bi G, Miller PS. Triplex formation by oligonucleotides containing novel deoxycytidine derivatives. Nucleic Acids Res. 1996;24:2606–2613. doi: 10.1093/nar/24.13.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang CY, Cushman CD, Miller PS. Triplex formation by an oligonucleotide containing N4-(3-acetamidopropyl)cytosine. J. Org. Chem. 1993;58:5048–5049. [Google Scholar]
- 98.Rao TS, Hogan ME, Revankar GR. Synthesis of triple helix forming oligonucleotides containing 2′-deoxyformycin. Nucleosides Nucleotides. 1994;13:95–107. [Google Scholar]
- 99.Durland RH, et al. Azole substituted oligonucleotides promote antiparallel triplex formation at non-homopurine duplex targets. Nucleic Acids Res. 1995;23:647–653. doi: 10.1093/nar/23.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mokhir AA, Connors WH, Richert C. Synthesis and monitored selection of nucleotide surrogates for binding T:A base pairs in homopurine-homopyrimidine DNA triple helices. Nucleic Acids Res. 2001;29:3674–3684. doi: 10.1093/nar/29.17.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eldrup AB, Dahl O, Nielsen PB. A novel peptide nucleic acid monomar for recognition of thymine in triple helix structures. J. Am. Chem. Soc. 2002;119:11116–11117. [Google Scholar]
- 102.Li J-S, Fan Y-H, Zhang Y, Marky LA, Gold B. J. Am. Chem. Soc. 2003;125:2084–2093. doi: 10.1021/ja028033x. [DOI] [PubMed] [Google Scholar]