Abstract
Replication through damaged sites in DNA requires in Escherichia coli the SOS stress-inducible DNA polymerase V (UmuC), which is specialized for lesion bypass. Homologs of the umuC gene were found on native conjugative plasmids, which often carry multiple antibiotic-resistant genes. MucB is a UmuC homolog present on plasmid R46, and its variant plasmid pKM101 has been introduced into Salmonella strains for use in the Ames test for mutagens. Using a translesion replication assay based on a gapped plasmid carrying a site-specific synthetic abasic site in the single-stranded DNA region, we show that MucB is a DNA polymerase, termed pol RI, which is specialized for lesion bypass. The activity of pol RI requires the plasmid-encoded MucA′ protein and the E. coli RecA and single-strand DNA binding proteins. Elimination of any of the proteins from the reaction abolished lesion bypass and polymerase activity. The unprocessed MucA could not substitute for MucA′ in the bypass reaction. The presence of a lesion bypass DNA polymerase on a native conjugative plasmid, which has a broad host range specificity and carries multiple antibiotic-resistant genes, raises the possibility that mutagenesis caused by pol RI plays a role in the spreading of antibiotic resistance among bacterial pathogens.
The SOS stress response in Escherichia coli is a global response, controlling the coordinated expression of 20–30 genes that function to increase cell survival under environmental stress conditions that cause DNA damage (1, 2). Most remarkably, mutagenesis caused by UV light and many chemical mutagens is regulated by the SOS response (3–5). The in vitro reconstitution of SOS-targeted mutagenesis (6–8) has provided strong biochemical proof that SOS mutagenesis targeted to DNA lesions occurs via translesion replication (lesion bypass) by a new DNA polymerase, pol V (the umuC gene product), with the obligatory participation of UmuD′, RecA, and single strand-binding protein (SSB) (9, 10). The same set of proteins, while replicating undamaged DNA, causes a high mutations frequency (11, 12) and is responsible for SOS mutagenesis on undamaged chromosomal regions (11). A second E. coli homolog of UmuC, DinB, is also a DNA polymerase (pol IV; ref. 13) and is involved in untargeted mutagenesis associated with undamaged phage-infecting SOS-induced cells (14, 15). Its role in chromosomal mutagenesis is not clear yet.
Two homologs of the E. coli umuC gene were recently found in the yeast Saccharomyces cerevisiae. The REV1 gene is required for UV mutagenesis and encodes a dCMP transferase (16). The RAD30 gene encodes DNA polymerase η, which effectively and accurately bypasses cyclobutyl pyrimidine dimers, the major UV lesions (17). In addition, yeast cells contain DNA polymerase ζ, which is required for UV mutagenesis but is not a homolog of umuC. It is encoded by the REV3 and REV7 genes (18). Human cells contain four proteins that belong to this superfamily: DNA polymerase η is encoded by the XP-V (hRAD30A) gene (19, 20). This protein is defective in the genetic disease xeroderma pigmentosum variant, which causes sunlight sensitivity and predisposition to skin cancer. The function of two other homologs, DNA polymerase ι, encoded by hRAD30B (21, 22), and DNA polymerase θ, encoded by hDINB1 (refs. 23 and 24; also termed DNA polymerase κ, ref. 25), is unknown. Human cells contain also the REV1 gene, which encodes a dCMP transferase (26, 27), and a homolog of the yeast REV3 gene, which was shown to be required for UV mutagenesis in human cells (28).
An interesting group of umuC and umuD homologs contains genes residing on native conjugative plasmids. These plasmids have a broad host range specificity, and they often carry multiple antibiotic-resistant genes (29). Their existence in human pathogenic bacteria may account, in part, for the growing problem of antibiotics resistance among bacterial pathogens (30–32). The most extensively studied of these is the mucAB operon, carried on plasmid pKM101 (33), which is a natural variant of plasmid R46. Plasmid pKM101 was introduced into the Salmonella strains used in the Ames test for mutagens, where it increased the sensitivity of the assay via mucAB-mediated mutagenesis (34). Other known plasmidic umuDC homologs include impCAB (35), Sam AB (36), and rum AB (37). We have previously overproduced MucA, MucA′, and MucB and showed that MucA′ forms a homodimer and that MucB is a single-stranded DNA (ssDNA)-binding protein (38). In addition, we found that MucB interacts with a SSB-coated ssDNA, causing a major conformational change, but without causing massive dissociation of SSB from the DNA (38). Here we show that MucB protein is a DNA polymerase, specialized for bypass of DNA lesions.
Materials and Methods
Proteins.
MucB, MucA′, and MucA (38) as well as the fusion MBP-UmuC protein and UmuD′ were purified as previously described (7, 10). SSB and RecA were purified according to published procedures (refs. 39 and 40, respectively), except that a phosphocellulose purification step was added for RecA. Restriction nucleases, T4 DNA ligase and T4 polynucleotide kinase, were from New England Biolabs. T7 gp6 exonuclease was from Amersham Pharmacia, and S1 nuclease was from Promega.
DNA Substrates.
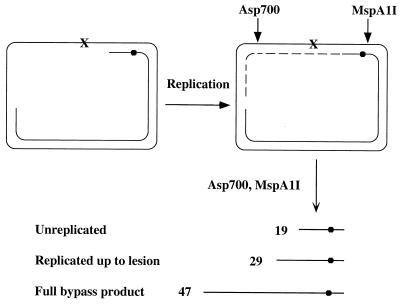
The preparation of the gapped plasmid carrying a site-specific lesion was recently described (41, 42). Gapped plasmid GP21 contained a site-specific synthetic (tetrahydrofuran) abasic site and an ssDNA region of approximately 350 nucleotides (Fig. 1).
Figure 1.
Outline of the translesion replication assay. X, site-specific synthetic abasic site; ●, the internal radiolabeled phosphate. See text for detail.
Translesion Replication Assay.
The translesion replication reaction was performed essentially as previously described (7, 10), except that MucA′ and MucB were used instead of UmuD′ and UmuC. The reaction mixture (25 μl) contained 20 mM Tris⋅HCl, pH 7.5/8 μg/ml BSA/5 mM DTT/0.1 mM EDTA/4% glycerol/1 mM ATP/10 mM MgCl2/0.1 mM each of dATP, dGTP, dTTP, and dCTP, 50 ng (1 nM) gapped plasmid, 600 nM SSB, 4 μM RecA, 2.5 μM MucA′, and 50–300 nM MucB. When used, MucA was at 1.5–5.0 μM. Reactions were carried out at 37°C for various periods of time. Analysis of the bypass products was done as described (10). Briefly, the reaction mixture was treated with proteinase K, followed by phenol/chloroform extraction and ethanol precipitation. The DNA was then treated with calf intestine alkaline phosphatase to hydrolyze remaining dNTPs, after which the DNA was digested with Asp-700 and MspA1I (Fig. 1). The DNA samples were fractionated by 15% PAGE-urea, followed by phosphorimager analysis (Fuji BAS 2500). The extent of bypass was calculated by dividing the amount of bypass products by the amount of the extended primers.
Results
We have previously overproduced MucB, MucA′, and MucA, purified them in denatured form, and refolded them (38). With the development of an effective lesion bypass in vitro assay system (7, 10) and the finding that UmuC is a lesion bypass DNA polymerase (9, 10), we explored the possibility that MucB is also a DNA polymerase. The experimental bypass assay system was previously described (7, 41). Briefly, the DNA substrate consists of a gapped plasmid carrying a site-specific synthetic abasic site in the ssDNA region and an internal radiolabeled phosphate in the primer strand (Fig. 1). Upon addition of a DNA polymerase, the 3′ primer terminus is extended up to the abasic site. Lesion bypass will yield extension past the lesion, with the formation of a longer nascent DNA strand. To facilitate analysis, after termination of the reaction the DNA products were extracted and restricted with MspA1I, which cleaves four nucleotides upstream the radiolabel, and with Asp-700, which cleaves downstream to the lesion. This yielded radiolabeled DNA fragments of 19, 29, and 47 nucleotides long for the uninitiated primer, the nascent strand blocked at the lesion, and the bypass product, respectively (Fig. 1). These products were fractionated by urea-PAGE and visualized and quantified by phosphorimaging.
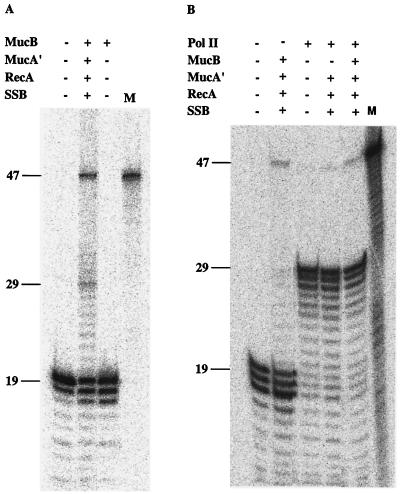
Incubation of the gap-lesion plasmid with MucB in the presence of dNTPs and Mg2+ did not reveal any polymerase activity, as indicated by the lack of extension of the DNA primer (Fig. 2A, lane 3). Therefore, MucB has very little or no polymerase activity on its own. Upon addition of MucA′, RecA, and SSB, there was a strong stimulation of DNA synthesis activity, indicating the activity of a DNA polymerase. This activity led to the extension of the radiolabeled primer up to the abasic site and past it, generating the full-length 47-nucleotide-long product (Fig. 2A, lane 2). For comparison, Fig. 2B shows the activity of DNA polymerase II (pol II) in the same assay system. Although primer utilization by pol II was high, polymerization was severely arrested at the abasic site, and very little lesion bypass was observed (Fig. 2B, lane 3), similar to previous results (42). In contrast, initiation of primer extension by MucB in the presence of MucA′, RecA, and SSB was low, but once DNA synthesis started, it showed little inhibition at the abasic site, leading to bypass of the abasic site (Fig. 2B, lane 2). When the two polymerases were mixed, there was generally little effect on lesion bypass, and in fact a slight inhibition was observed, probably due to competition between the polymerases for the primer terminus (Fig. 2B, lane 5). Omission of MucB from this mixture reduced bypass, suggesting that the stimulation of bypass caused by MucA′, RecA, and SSB is specific to MucB (Fig. 2B, lane 4).
Figure 2.
DNA polymerase activity of MucB. Gap-filling bypass replication was performed with MucB alone or in the presence of MucA′, RecA, and SSB using the gap-lesion plasmid GP21 as a substrate. Reactions were performed as described under Materials and Methods at 37°C for 10 min. When present, pol II was at a concentration of 100 nM. Reaction products were restricted, fractionated by urea-PAGE, and visualized by phosphorimaging. The DNA bands of 19, 29, and 47 nucleotides long represent the unextended primer, the replication product blocked at the abasic site, and the bypass product, respectively. M, size marker for the 47-mer bypass product.
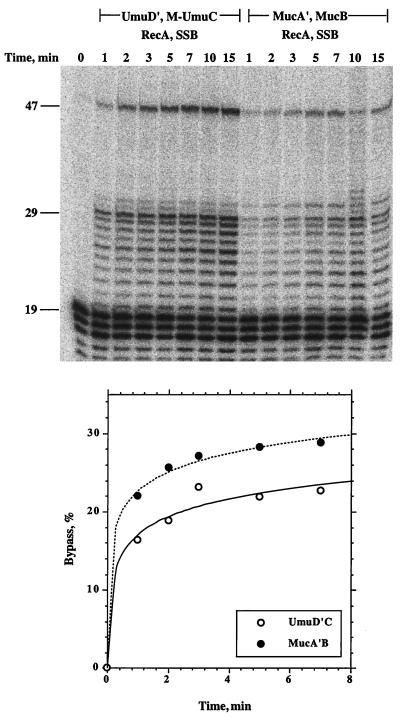
A time course of translesion replication by MucB, MucA′, RecA, and SSB revealed that 28% of the molecules on which DNA synthesis was initiated showed lesion bypass within 5 min (Fig. 3). For comparison, we performed the bypass reaction with UmuC, UmuD′, RecA, and SSB (Fig. 3). As can be seen, the two systems show generally similar results. The number of initiations in the pol V reaction was higher than with MucB, and therefore the bands of all of the extended primers are stronger than with MucB. However, when the extent of lesion bypass is calculated out of the initiated products, it is in fact slightly lower than with MucB. These results indicate that MucB is indeed a DNA polymerase. Notice that there was little inhibition of DNA synthesis at the synthetic abasic site, indicating a high propensity to bypass the synthetic abasic site.
Figure 3.
Kinetics of translesion replication by MucB in the presence of MucA′, RecA, and SSB. The translesion replication assay was performed as described under Materials and Methods, with 2.5 μM MucA′ and 250 nM MucB for the indicated periods of time. The phosphorimage of the replication products is shown in the upper panel, and their quantification is shown in the lower panel. The percent of lesion bypass is calculated as the total amount of DNA extended beyond the abasic site, divided by the total amount of extended primers.
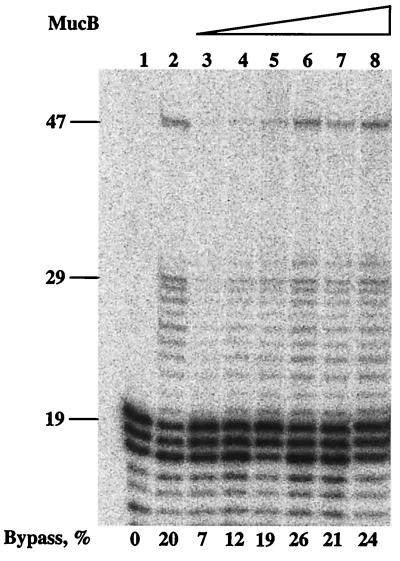
The experiment described in Fig. 3 was performed using 250 nM MucB. Titration of MucB to lower concentration showed bypass at concentrations as low as 50 nM (Fig. 4). In this context, it is interesting to note that the intracellular concentrations of MucA and MucB in constitutively SOS-induced cells were reported to be very high, approximately 60 μM and 20 μM, respectively (43). Similarly to what was seen in Figs. 2 and 3, there seem to be replication pauses up to the lesion; however, once the lesion is bypassed, the pauses in synthesis are largely reduced. At this point, we do not know the reason for this behavior except that it was also observed during bypass by pol III holoenzyme alone (41, 42) and by pol V (10). It is possible that these polymerases can sense the downstream lesion as they are approaching it, leading to synthesis pauses.
Figure 4.
Titration of MucB in translesion replication in the presence of MucA′, RecA, and SSB. The translesion replication assay was performed as described under Materials and Methods, with 2.5 μM MucA′, for 5 min. The concentrations of MucB were as follows: lane 3, 50 nM; lane 4, 100 nM; lane 5, 150 nM; lane 6, 200 nM; lane 7, 250 nM; lane 8, 300 nM. Lane 1 is a control without proteins, whereas lane 2 contains products of a reaction promoted by pol V (250 nM) and UmuD′ (2.5 μM) in the presence of RecA and SSB.
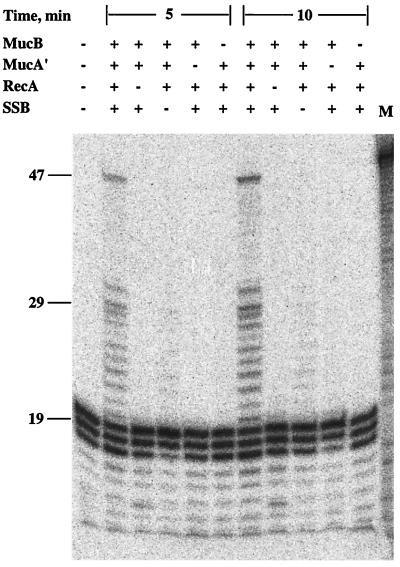
To examine the requirement for each of the components, the lesion bypass experiments were performed under conditions in which single components were omitted, one at a time. As can be seen in Fig. 5, elimination of each of the components led to the abolition of lesion bypass, indicating that each of the four proteins was absolutely required for lesion bypass. In fact, DNA synthesis up to the lesion was also greatly reduced when any of the proteins was omitted. Thus, the MucB DNA polymerase is highly activated by MucA′, RecA, and SSB, which potentiate it as an effective lesion bypass DNA polymerase. We term it DNA polymerase RI because it is a polymerase encoded by a native R plasmid.
Figure 5.
Protein requirement of MucB-promoted translesion replication. The translesion replication assay was performed as described under Materials and Methods, with 2.5 μM MucA′ and 250 nM MucB, for the indicated periods of time. Parallel reactions were run, in which individual components were omitted, one at a time. M, size marker for the 47-mer bypass product.
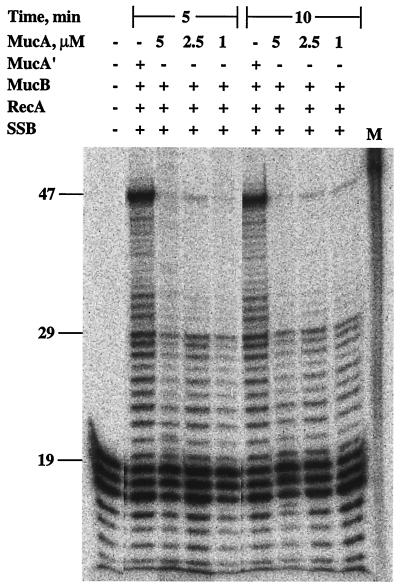
MucA′ is obtained from MucA by posttranslational processing promoted by RecA (44, 45). We examined whether lesion bypass could be promoted with MucA instead of MucA′. As can be seen in Fig. 6, translesion replication was strongly reduced under these conditions. The residual bypass activity may be attributed to residual MucA′ present in the MucA preparation due to autocleavage of MucA (38, 45) and to cleavage of MucA promoted by RecA under our assay conditions (38).
Figure 6.
MucA cannot substitute for MucA′ in MucB-promoted lesion bypass. The translesion replication assay was performed as described under Materials and Methods, for the indicated periods of time, with 250 nM MucB and 4 μM MucA′ or the indicated concentrations of MucA. Reaction products were restricted and analyzed by urea-PAGE followed by phosphorimaging. M, size marker for the 47-mer bypass product.
Discussion
Homologs of UmuC and UmuD′ were found to be present on native conjugative plasmids, which have a broad host range specificity (29). The interest in these plasmids stems from the fact that they often carry multiple antibiotic-resistant genes and that they are frequently found among bacterial pathogens. We have previously overexpressed and purified the MucA, MucA′, and MucB proteins and showed that MucA′ forms a homodimer and that MucB interacts with SSB-coated ssDNA and alters its conformation without inducing gross dissociation of SSB from DNA (38). Here, we used the translesion replication system that we have developed (7) to examine whether MucB is a DNA polymerase. Based on the data presented, MucB is indeed a lesion bypass DNA polymerase. We term it DNA polymerase RI, as it is a DNA polymerase encoded by the native conjugative R46 plasmid. It is the second known prokaryotic lesion bypass DNA polymerase; however, it is likely that the other bacterial and plasmidic homologs of UmuC are also DNA polymerases.
MucB is a dormant DNA polymerase. Its activation requires MucA′, RecA, and SSB. However, once activated it shows a high propensity to replicate a synthetic abasic site, which is known to severely block DNA polymerase I (46–48), DNA polymerase II (42, 49), and DNA polymerase III (42). In this sense, the MucA′B system is a functional homolog of the UmuD′C system. The activity of MucA′, RecA, and SSB in lesion bypass by pol RI remains to be elucidated. However, it is well established that RecA forms a helical nucleoprotein filament along single-stranded DNA and that the assembly of this filament is stimulated by SSB (reviewed in refs. 50 and 51). Therefore, it is possible that pol RI acts on RecA-coated DNA. MucA′ is known to interact with pol RI (38) and with RecA (52). Therefore, its role may be to mediate the interaction between pol RI and the RecA nucleoprotein filament. These rather complex requirements for lesion bypass by pol RI (and by pol V) may be required to achieve tight control over the activity of these polymerases.
The functional similarity of pol RI and pol V is manifested by the fact that the mucA′B operon complements a ΔumuDC mutant (33). In fact, mucA′B was reported to be more effective in promoting UV mutagenesis as compared with umuD′C (53). This higher efficiency to promote mutagenesis was used in the Ames test for mutagens, where the tester strains carry plasmid pKM101, a natural variant of plasmid R46, which harbors mucAB (34). The reason for the higher effectiveness of MucB to promote mutagenesis is not clear yet, but it was attributed to a faster processing of MucA to MucA′, as compared with UmuD processing to UmuD′ (45). We observed comparative bypass efficiencies by MucA′B and UmuD′C. However, it is difficult to draw conclusions from this comparison, as UmuC was purified in soluble form as a fusion to maltose binding protein (7), whereas MucB was used without a tag but was obtained by refolding of the denatured protein (38). The greater effect of MucA′B may be attributed also to their higher level of expression in SOS-induced cells (43).
Based on the results presented above, pol RI is a functional homolog of pol V, and like pol V, it requires the host SSB and RecA proteins. Goodman and coworkers have reported that lesion bypass by a UmuD′C complex required, in addition to SSB and RecA, six additional proteins, which are subunits of DNA polymerase III holoenzyme: the β subunit processivity clamp and the 5-subunit γ complex clamp loader (9). We have clearly obtained lesion bypass in the absence of these proteins, both with pol V (10) and pol RI. Recently, Goodman and coworkers have reported that they could obtain bypass with pol V in the absence of β subunit and the γ complex when they used ATPγS instead of ATP (12). ATPγS is known to stabilize RecA–ssDNA interactions (50). This suggests that the requirement for the β subunit and the γ complex was due to the inability to form a stable and functional RecA–ssDNA complex on the particular DNA substrate used in Goodman's studies. In that substrate, the lesion is located only 50 nucleotides from the 5′ end of the DNA (8, 9). Because RecA assembly occurs in the 5′ → 3′ direction (50), it may not fully cover the DNA 5′ to the lesion, and this causes a difficulty for the stable assembly of RecA near the lesion. In the substrate used in our studies, there is no problem of loading of RecA because the DNA is circular, and the ssDNA region extends over 300 nucleotides 5′ to the lesion. Taken together, it is clear that the basic lesion bypass reaction requires in vitro UmuD′C or MucA′B, as well as SSB and RecA, and no other proteins. However, it is possible that the processivity proteins increase the efficiency of the lesion bypass reaction, e.g., by increasing the efficiency of initiation of at the primer terminus. Alternatively, the processivity proteins may be required under special conditions or in a different Umu-promoted reaction, e.g., a DNA damage checkpoint activity (54, 55).
The presence of a lesion bypass polymerase on a native conjugative plasmid is intriguing. Having a limited size, such plasmids are expected to carry only genes with an unusual importance for the propagation of the plasmids in host cells. Why would lesion bypass proteins be selected to reside on plasmids? At least two answers come to mind. 1) Lesion bypass may represent a generic and simple, even “primitive” mode of DNA repair; it enables to preserve the continuity of the plasmid, even when it is damaged, by using replication read through, without actually removing the lesion. 2) Lesion bypass is usually associated with mutagenesis. The mutagenesis function may be beneficial for a plasmid, which is transmitted among a broad range of bacterial hosts, by allowing faster adaptation to foreign intracellular environments. A similar inducible mutator function for cellular adaptation was suggested for pol V (56–58). Because these plasmids often carry multiple antibiotic-resistant genes, the mucAB genes and their homologs may play an role in the spreading of antibiotic resistance among bacterial pathogens, a phenomenon that is becoming a growing threat to human health (30–32, 59).
Acknowledgments
We thank M. Goodman (University of Southern California, Los Angeles) for his generous gift of pol II. This research was supported by US–Israel Binational Science Foundation Grant 96-00448. Z.L. is the incumbent of The Maxwell Ellis Professorial Chair in Biomedical Research.
Abbreviations
- ssDNA
single-stranded DNA
- SSB
ssDNA-binding protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.200361997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.200361997
References
- 1.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. [Google Scholar]
- 2.Fernandez De Henestrosa A R, Ogi T, Aoyagi S, Chafin D, Hayes J J, Ohmori H, Woodgate R. Mol Microbiol. 2000;35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 3.Witkin E M. Bacteriol Rev. 1976;40:869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Livneh Z, Cohen-Fix O, Skaliter R, Elizur T. CRC Crit Rev Biochem Mol Biol. 1993;28:465–513. doi: 10.3109/10409239309085136. [DOI] [PubMed] [Google Scholar]
- 5.Walker G C. Trends Biochem Sci. 1995;20:416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan M, Lu C, Woodgate R, O'Donnell M, Goodman M, Echols M. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reuven N B, Tomer G, Livneh Z. Mol Cell. 1998;2:191–199. doi: 10.1016/s1097-2765(00)80129-x. [DOI] [PubMed] [Google Scholar]
- 8.Tang M, Bruck I, Eritja R, Turner J, Frank E G, Woodgate R, O'Donnell M, Goodman M F. Proc Natl Acad Sci USA. 1998;95:9755–9760. doi: 10.1073/pnas.95.17.9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. Proc Natl Acad Sci USA. 1999;96:8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. J Biol Chem. 1999;274:31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- 11.Maor-Shoshani A, Reuven N B, Tomer G, Livneh Z. Proc Natl Acad Sci USA. 2000;97:565–570. doi: 10.1073/pnas.97.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang M, Pham P, Shen X, Taylor J S, O'Donnell M, Woodgate R, Goodman M F. Nature (London) 2000;404:1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- 13.Wagner J, Gruz P, Kim S R, Yamada M, Matsui K, Fuchs R P P, Nohmi T. Mol Cell. 1999;4:281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 14.Brotcorne-Lannoye A, Maenhaut-Michel G. Proc Natl Acad Sci USA. 1986;83:3904–3908. doi: 10.1073/pnas.83.11.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S R, Maenhaut-Michel G, Yamada M, Yamamoto Y, Matsui K, Sofuni T, Nohmi T, Ohmori H. Proc Natl Acad Sci USA. 1997;94:13792–13797. doi: 10.1073/pnas.94.25.13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson J R, Lawrence C W, Hinkle D C. Nature (London) 1996;382:729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- 17.Johnson R E, Prakash S, Prakash L. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 18.Nelson J R, Lawrence C W, Hinkle D C. Science. 1996;272:1646–1649. doi: 10.1126/science.272.5268.1646. [DOI] [PubMed] [Google Scholar]
- 19.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. Nature (London) 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 20.Johnson R E, Kondratick C M, Prakash S, Prakash L. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- 21.McDonald J P, Rapic-Otrin V, Epstein J A, Broughton B C, Wang X, Lehmann A R, Wolgemuth D J, Woodgate R. Genomics. 1999;60:20–30. doi: 10.1006/geno.1999.5906. [DOI] [PubMed] [Google Scholar]
- 22.Tissier A, McDonald J P, Frank E G, Woodgate R. Genes Dev. 2000;14:1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlach V L, Aravind L, Gotway G, Schultz R A, Koonin E V, Friedberg E C. Proc Natl Acad Sci USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson R E, Prakash S, Prakash L. Proc Natl Acad Sci USA. 2000;97:3838–3843. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 26.Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z. Nucleic Acids Res. 1999;27:4468–4475. doi: 10.1093/nar/27.22.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibbs P E, Wang X D, Li Z, McManus T P, McGregor W G, Lawrence C W, Maher V M. Proc Natl Acad Sci USA. 2000;97:4186–4191. doi: 10.1073/pnas.97.8.4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbs P E, McGregor W G, Maher V M, Nisson P, Lawrence C W. Proc Natl Acad Sci USA. 1998;95:6876–6880. doi: 10.1073/pnas.95.12.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woodgate R, Sedgwick S G. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 30.Davies J. Science. 1994;264:375–382. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 31.Dennesen P J, Bonten M J, Weinstein R A. Ann Med. 1998;30:176–185. doi: 10.3109/07853899808999401. [DOI] [PubMed] [Google Scholar]
- 32.Swartz M N. Proc Natl Acad Sci USA. 1994;91:2420–2427. doi: 10.1073/pnas.91.7.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perry K L, Walker G C. Nature (London) 1982;300:278–281. doi: 10.1038/300278a0. [DOI] [PubMed] [Google Scholar]
- 34.McCann J, Spingarn N E, Kobori J, Ames B N. Proc Natl Acad Sci USA. 1975;72:979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lodwick D, Owen D, Strike P. Nucleic Acids Res. 1990;18:5045–5050. doi: 10.1093/nar/18.17.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nohmi T, Hakura A, Nakai Y, Watanabe M, Murayama S Y, Sofuni T. J Bacteriol. 1991;173:1051–1063. doi: 10.1128/jb.173.3.1051-1063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarov-Blat L, Livneh Z. J Biol Chem. 1998;273:5520–5527. doi: 10.1074/jbc.273.10.5520. [DOI] [PubMed] [Google Scholar]
- 39.Lohman T M, Overman L B. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 40.Cox M M, McEntee K, Lehman I R. J Biol Chem. 1981;256:4676–4678. [PubMed] [Google Scholar]
- 41.Tomer G, Reuven N B, Livneh Z. Proc Natl Acad Sci USA. 1998;95:14106–14111. doi: 10.1073/pnas.95.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomer G, Livneh Z. Biochemistry. 1999;38:5948–5958. doi: 10.1021/bi982599+. [DOI] [PubMed] [Google Scholar]
- 43.Venderbure C, Chastanet A, Boudsocq F, Sommer S, Bailone A. J Bacteriol. 1999;181:1249–1255. doi: 10.1128/jb.181.4.1249-1255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hauser J, Levine A S, Ennis D G, Chumakov K M, Woodgate R. J Bacteriol. 1992;174:6844–6851. doi: 10.1128/jb.174.21.6844-6851.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kunkel T A, Shearman C W, Loeb L A. Nature (London) 1981;291:349–351. doi: 10.1038/291349a0. [DOI] [PubMed] [Google Scholar]
- 47.Sagher D, Strauss B. Biochemistry. 1983;22:4518–4526. doi: 10.1021/bi00288a026. [DOI] [PubMed] [Google Scholar]
- 48.Paz-Elizur T, Takeshita M, Livneh Z. Biochemistry. 1997;36:1766–1773. doi: 10.1021/bi9621324. [DOI] [PubMed] [Google Scholar]
- 49.Paz-Elizur T, Takeshita M, Goodman M, O'Donnell M, Livneh Z. J Biol Chem. 1996;271:24662–24669. doi: 10.1074/jbc.271.40.24662. [DOI] [PubMed] [Google Scholar]
- 50.Roca A I, Cox M M. CRC Crit Rev Biochem Mol Biol. 1990;25:415–456. doi: 10.3109/10409239009090617. [DOI] [PubMed] [Google Scholar]
- 51.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank E G, Hauser J, Levine A S, Woodgate R. Proc Natl Acad Sci USA. 1993;90:8169–8173. doi: 10.1073/pnas.90.17.8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanco M, Herrera G, Aleixandre V. Mol Gen Genet. 1986;205:234–239. doi: 10.1007/BF00430433. [DOI] [PubMed] [Google Scholar]
- 54.Opperman T, Murli S, Smith B T, Walker G C. Proc Natl Acad Sci USA. 1999;96:9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sutton M D, Opperman T, Walker G C. Proc Natl Acad Sci USA. 1999;96:12373–12378. doi: 10.1073/pnas.96.22.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radman M. In: Molecular Mechanisms for Repair of DNA. Hanawalt P, Setlow R B, editors. New York: Plenum; 1975. pp. 355–367. [Google Scholar]
- 57.Witkin E M, Wermundsen I E. Cold Spring Harbor Symp Quant Biol. 1979;43:881–886. doi: 10.1101/sqb.1979.043.01.095. [DOI] [PubMed] [Google Scholar]
- 58.Echols H. Cell. 1981;25:1–2. doi: 10.1016/0092-8674(81)90223-3. [DOI] [PubMed] [Google Scholar]
- 59.O'Brien T F. Clin Infect Dis. 1997;24, Suppl. 1:S2–S8. doi: 10.1093/clinids/24.supplement_1.s2. [DOI] [PubMed] [Google Scholar]