Abstract
The Ig and T cell receptor (TCR) loci have an exceptionally dynamic evolutionary history, but the mechanisms responsible remain a subject of speculation. Ig and TCR genes are unique in vertebrates in that they are assembled from V, D, and J segments by site-specific recombination in developing lymphocytes. Here we examine the extent to which the V(D)J recombination in germline cells may have been responsible for remodeling Ig and TCR loci in mammals by asking whether gene segments have evolved as a unit, or whether, instead, recombination signal sequences (RSSs) and coding sequences have different phylogenies. Four distinct types of RSS have been defined in the human Ig heavy-chain variable region (Vh) locus, namely H1, H2, H3, and H5, and no other RSS type has been detected in other mammalian species. There is a well-supported discrepancy between the evolutionary history of the RSSs as compared with the Vh coding sequences: the RSS type H2 of one Vh gene segment has clearly become replaced by a RSS type H3 during mammalian evolution, between 115 and 65 million years ago. Two general models might explain the RSS swap: the first involves an unequal crossing over, and the second implicates germline activation of V(D)J recombination. The Vh-H2/RSS-H3 recombination product has likely been selected during the evolution of mammals because it provides better V(D)J recombination efficiency.
The vertebrate immune system generates an extensive repertoire of Igs and T cell receptors (TCR) capable of engaging a wide variety of distinct antigens. Their diversity is produced in developing B and T lymphocytes by V(D)J recombination, which assembles the exons encoding the antigen-binding domains from separate V (variable), D (diversity), and J (joining) gene segments (1). Targets for this recombination reaction are the recombination signal sequences (RSSs) that lie immediately adjacent to the recombination site in the DNA. The RSSs are composed of conserved heptamer and nonamer motifs, separated by a spacer of either 12 or 23 base pairs in length.
V(D)J rearrangements are mediated by the recombination activating gene (RAG) proteins RAG-1 and RAG-2, which can carry out the recognition and cleavage events in the recombination reaction, generating specific double-strand DNA breaks at the borders between the RSSs and the coding segments (2).
V, D, and J coding segments are thought to evolve by the “birth-and-death model” (3, 4). According to this model, new gene segments are created by duplications. Although some are maintained in functional form in the genome, others are deleted or become pseudogenes because of deleterious mutations. In the case of V segments, the minimum DNA fragment that must be duplicated to give rise to a new functional gene includes the coding sequence, its adjacent RSS, as well as the 5′ regulatory region. These three different elements are presumed to have diverged together as a functional unit during evolution, implying they share the same phylogenetic history. But, because of well-established evidence of germline recombination in cartilaginous fish (5), we cannot exclude the possibility that RSS-mediated rearrangements in the Vh locus may have occurred during the evolution of mammals.
Here we have analyzed the evolution of the Ig heavy-chain variable region (Vh) gene segments to test coevolution between the Vh coding sequences and the adjacent RSSs. Two separate phylogenetic analyses have been performed, one based on the coding sequences and the other based on the RSSs. These analyses have defined four types of RSS in the human Vh locus (namely H1, H2, H3, and H5); no additional types have been seen for other species of mammals. Significantly, the phylogenetic tree constructed with the RSSs reveals a different evolutionary history than that of the Vh coding sequences. A recombination event appears to have resulted in the replacement of the H2 RSS by H3 during mammalian evolution. That the involved gene segments were divergent and that the apparent recombination site is located at the 3′ end of Vh, near or at the coding sequence/RSS border, suggest that site-specific rather than homology-based recombination created the change. Replacement of the H2 RSS by H3 would likely have been positively selected during mammalian evolution because it more closely resembles the canonical RSS sequence and thus should endow an associated Vh coding sequence with a higher probability of V(D)J rearrangement and expression.
Materials and Methods
Data.
Of the 123 Vh segments identified in the complete sequence of the human locus, 39 are functional (6) and were selected for phylogenetic analyses (Fig. 2).
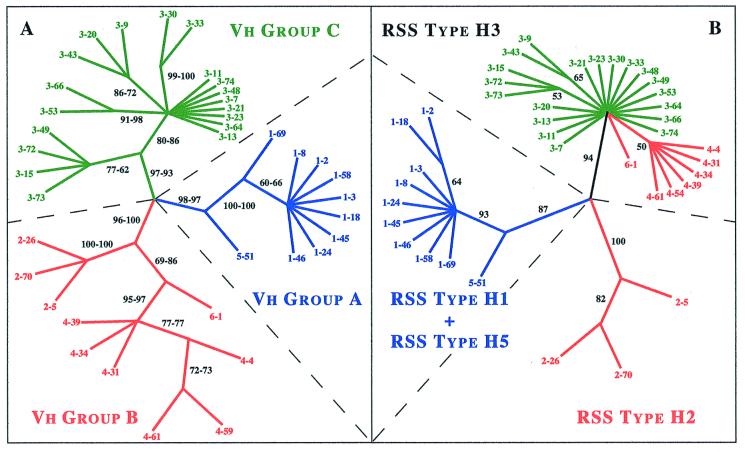
Figure 2.
Phylogenetic trees constructed from all known functional human Vh coding sequences (A) and RSSs (B). Phylogenetic analyses for the 39 functional genes of the human Vh locus were performed by using the maximum parsimony method with paup 3.1.1 (8). The trees presented here correspond to the bootstrap 50% majority-rule consensus trees, i.e., only groups with bootstrap proportions >50% are retained. (A) Tree constructed from the 261-bp sequences of Vh exon 2; see Materials and Methods. The sequences were examined by unweighted or weighted maximum parsimony analyses. The bootstrap proportions for the two different analyses are indicated on the branches, respectively at the left and right. The three major groups of Vh genes thus defined have been previously reported (3) and are here distinguished by the colors, blue for group A, red for group B, and green for group C. (B) Tree constructed from the 39-bp sequences of the Vh RSSs. It can be seen here that groups B and C, which are found with Vh coding sequences, are not retrieved for the associated RSSs.
Other mammalian germ-line Vh sequences, with a complete ORF and an RSS at the 3′ end, were located in the GenBank database by searching all orders of Mammalia by using the key words “heavy chain,” “variable,” and “recombination” (or “heptamer” or “RSS”). Ten germ-line sequences for which RSS have not been determined were also included in the analyses (Fig. 3) to provide the greatest available diversity for group B (see below). Thus, a total of 83 mammal Vh sequences of which 73 included RSS elements were used for phylogenetic analyses. The tree was rooted with Vh 101 of Heterodontus francisci (horned shark) and Vh 102 of Raja erinacea (little skate), because these sequences branched off before the divergence of the mammalian Vh genes (3).
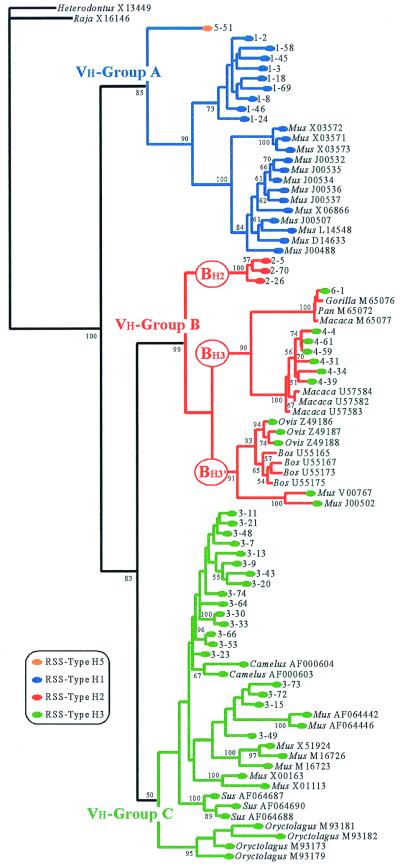
Figure 3.
Phylogenetic tree for mammalian Vh germ-line sequences. The phylogenetic analyses were performed for 85 Vh exon 2 sequences, each 261 bp, as described above (see Fig. 2 and Materials and Methods). The tree shown is the consensus of the 54 trees (length = 396,410 steps) obtained with the weighted maximum parsimony analysis. Bootstrap proportions are indicated on the branches in black. The colors of the branches indicate the group to which the human Vh coding sequences belong, as described in Fig. 2. At the branch extremity, a circle indicates that the RSS is known and is colored as shown in the key to indicate RSS type. Nonhuman sequences are designated with the genus of the animal followed by the GenBank accession number.
Phylogenetic Analyses of Vh Coding Sequences.
The nucleotide Vh coding sequences corresponding to the Vh exons 1 and 2 were first aligned by using clustalx ver. 1.8 (7) and were further refined by visual inspection to maximize the sequence homology and minimize the number of postulated insertion and deletion events. Vh exon 1 was not used for the phylogenetic analyses because of ambiguities in the alignments. For the same reason, three regions of Vh exon 2 were removed: the most variable positions within CDR1 (complementarity-determining region 1) (nucleotides 99–116), those in CDR2 (), and the extreme 3′ end of the Vh-encoded portion of CDR3 (nucleotides 315–325) (Supplementary Fig. 5; see www.pnas.org). Because estimates of homoplasy content (described below) required complete triplet codons, the two first nucleotides of Vh exon 2 were also eliminated. In total, the phylogenetic analyses of the Vh segments were conducted on a data matrix representing 261 nucleotides.
The 261 nucleotides of Vh exon 2 were examined by the maximum parsimony (MP) method by using the software paup 3.1.1 (8). The coding Vh sequences were examined with either unweighted or weighted MP analyses. For the unweighted MP analysis, the same weight was attributed to all of the six types of nucleotide substitution (i.e., C-T, A-G, A-C, A-T, C-G, and G-T). For the weighted MP analysis, the CI and S values (CI = consistency index, S = slope of saturation) of each of the six types of nucleotide substitution were calculated for the 261 nucleotides distinguishing the three codon positions (Supplementary appendix; see www.pnas.org). Then the products CI × S were used for phylogenetic reconstruction to weight each nucleotide substitution at the three codon positions (9, 10). The aim of the weighted MP analysis is to take into account the evolutionary effects of mutational constraints (e.g., transitions evolve faster than transversions) and selective constraints (i.e., nucleotide changes involving conservative amino acid replacements evolve faster).
Searches for the shortest tree(s) were performed with the default options of paup. The robustness of the nodes was assessed by the bootstrap method (11) with bootstrap proportions (BP) computed with paup after 100 replicates of the closest stepwise addition of taxa option. The bootstrap analysis is a computer-based technique introduced by Felsenstein (11) for the evaluation of phylogenetic trees. At each replicate of bootstrap, the nucleotide sites are sampled with replacement to build a new data set the same size as the original alignment. They are then subject to a phylogenetic search. The technique provides assessments of “confidence” for each node of an observed tree, based on the proportion of replicates showing that same node. In particular, the probability that a node is an erroneous one is <5%, if it is supported by 95% of the replicates. On the other hand, little confidence can be given to a node that is supported by <50% of the replicates.
Phylogenetic Analyses of RSSs.
The alignment of the 39 human RSSs was unambiguous except for the exact position of a 1-bp deletion in the spacer of 5–51 (Fig. 1). This deletion was not assumed by Matsuda et al. (6) but is introduced here to maximize the sequence homology on the basis of the high similarity between 5–51 and all members of the Vh family 1, especially for the 3′ and 5′ ends of the spacer and nonamer.
Figure 1.
Sequence alignment of the 39 functional genes of the human Vh locus. The alignment includes the 3′ end of Vh exon 2, the three RSS regions (heptamer, 23-bp spacer, and nonamer), and the 3′ flanking sequence. All sequences were taken from the complete sequence of the human Vh locus (6). The human Vh gene segments are classified into seven families (of which family 7 is not included in this alignment because no functional Vh gene segment has been described for this family); the first number of each gene name refers to the family, and the second number designates the ordinal position on chromosome 14 (6).
The 39 nucleotides of RSSs were examined by unweighted MP analysis. Searches for the shortest tree(s) and BP analyses were carried out as previously specified for the Vh exon 2.
Characterization of RSS Types.
One can assume that the evolution of RSSs is strongly affected by reversions and convergences (homoplasy) because of constraints that maintain V(D)J rearrangement capability. High levels of homoplasy can mask the phylogenetic signal contained in the data resulting either in lack of resolution or in incorrect groupings of sequences (12–14). Homoplasy can particularly confound the analysis when numerous sequences are compared by using only a small number of nucleotide sites, as is the case for the RSSs (39 bp). Therefore, the sequence flanking the 3′ end of the RSS was used to define the different types of RSS. No known function is associated with this region, and a free accumulation of mutations is expected to ensure lower levels of homoplasy. According to this criterion, a group of sequences is a new RSS type when all members exhibit a similar 3′ flanking region that cannot be aligned with the 3′ flanking region of any other RSSs.
Results and Discussion
Vh Coding Sequence Tree vs. RSS Tree.
The phylogenetic tree based on the 39 human functional Vh coding sequences (Fig. 2A) is in agreement with previous proposals that have shown the existence of seven Vh families distributed in three major clusters (3, 6, 15–17) corresponding to clans I, II, and III for mammals (15) or groups A, B, and C for vertebrates (3): (i) group A includes families 1 and 5 only; (ii) group B allies families 2, 4, and 6; and (iii) group C is composed of a single family, 3. Bootstrap analyses reveal that these three groups of Vh genes are statistically well supported (BP between 93 and 100%; Fig. 2A).
Significantly, the phylogenetic tree based on the RSSs (Fig. 2B) does not possess the same topology as that constructed with the Vh coding sequences, and unexpectedly the monophylies of the groups B and C are not retrieved. Instead, three major associations, which are highly supported in terms of bootstrap values, are observed in the RSS tree: (i) the first encompasses all members of Vh families 1 and 5, i.e., the totality of group A (BP = 87) (ii); the second encompasses all elements of Vh family 2 (BP = 100); and (iii) the third encompasses all representatives of Vh family 3 as well as Vh families 4 and 6 (BP = 94).
Human RSS Types.
Four types of RSS have been identified in the human Vh locus, namely H1, H2, H3, and H5 (H refers to the human, and the number designates the most representative Vh family). RSS type H3 is the largest, with 26 members corresponding to Vh families 3, 4, and 6 (belonging to Vh groups C and B). RSS type H2 associates with Vh family 2 (Vh group B). RSS type H1 includes all representatives of Vh family 1 (Vh group A). RSS type H5 is composed of the single Vh 5–51 (Vh group A). Although types H1 and H5 appear to be related on the sole basis of the RSS, that their 3′-flanking sequences cannot be aligned (Fig. 1) supports their separation into two distinct types.
The four RSS types, defined according to the 3′ flanking sequences, have some notable type-specific differences in their heptamer and nonamer elements.
With respect to the heptamer (canonical sequence: CACAGTG), the CAC sequence bordering the recombination site is perfectly conserved for all of the 39 functional Vh segments of the human locus (Fig. 1). This conservation of CAC is not exceptional and mirrors the situation for all RSSs, regardless of spacer length, associated coding sequence, or specific locus (18, 19). The notion that the CAC sequence is essential for efficient assembly of Ig and TCR genes is consistent with functional tests; alteration of any of the CAC nucleotides severely depressed RSS proficiency in recombination assays (18, 20). The four nucleotides completing the heptamer motif are also well conserved among the 39 human Vh RSSs (Fig. 1). The canonical sequence CACAGTG appears in 9 of 9 H1 RSSs, in the single H5 RSS, and in 24 of 26 H3 RSSs. This somewhat lower conservation for the last four heptamer nucleotides is also generally observed for RSSs at most Ig and TCR loci (18, 19). Again, mutational tests have shown that alteration of any of the last four heptamer nucleotides caused diminished recombination frequencies (18, 20). Significantly, although almost all of the 39 human Vh RSSs have canonical heptamers, none of the type H2 RSSs follow suit: the heptamer sequence for this type is either CACAAAG or CACAGAG.
With respect to the nonamer (canonical sequence: ACAAAAACC), variability among the 39 RSSs is much more marked than for the heptamers. This is particularly true for the four spacer-proximal nucleotides, which show type-specific differences: TCAG for H1, TCTA for H5, ACAA for H2, and ACAC for H3 (in 20/26 sequences). By contrast, the last five nucleotides of the nonamer are well conserved; AAACC is found in 35 of the 39 human sequences. The functional asymmetry of the nonamer suggested by the evolutionary conservation has been demonstrated previously; mutation experiments have highlighted the particular importance of the AAA nucleotides that fall within the last spacer-distal five positions of the nonamer (18, 20). Here, the AAA nucleotides in question are present in all type H1, H5, and H3 RSSs, and again the exceptions are found within type H2. Instead of ACAAAAACC, two of the three members (2–26 and 2–70) have the sequence ACAAGAACC. This exact nonamer sequence when previously tested in a V(D)J recombination assay was seen to lower RSS function to 3–4% that of a canonical RSS (18). In contrast, although none of the type-specific nonamer sequences seen for H1, H3, and H5 RSSs has been tested directly, RSSs mutated to other sequences at the corresponding sites are not greatly affected (having 27 to 100% of wild-type function) (18, 20).
Consequently, a consistent result that emerges from RSS comparisons is that the heptamer and nonamer sequences of the type H2 RSSs are exceptional. None of the heptamer sequences is canonical, and the variant nonamers are predicted to have particularly poor recombination function. In addition, two of the three type H2 RSSs are noncanonical for both the heptamer and nonamer. It has been shown that heptamer and nonamer variations with a relatively minor effect on recombination function can synergize to depress greatly recombination efficiency when each change is present within a single RSS (18).
Evolution of Vh Genes in Mammals.
To extend this analysis, a phylogenetic tree was constructed for available mammalian germ-line Vh segments (Fig. 3). The tree was similar to the one generated with human sequences alone. Three major clusters emerge, designated groups A (BP = 85), B (BP = 99), and C (BP = 50). In group A, the human Vh family 1 is associated with a large cluster of mouse sequences, and the human Vh 5–51 is found in basal position. Group B assembles three major subgroups: (i) BH3 contains sequences of Primates (chimpanzee, gorilla, and macaque) related to the human Vh families 4 and 6; (ii) BH3′ unites the Vh genes of the bovids (cow and sheep) with two mouse sequences; and (iii) BH2 is composed of the single human Vh family 2. Group C comprises all Vh sequences of the camel, pig, and rabbit, human Vh family 3, and several Vh genes of mouse. The presence of Vh sequences belonging to both human and mouse in all three groups, A, B, and C, has led to the conclusion that these groups arose before the mammalian radiation 65 million years (My) ago (15). Extension of the sampling to all vertebrate taxa has shown that groups A and B contain not only mammals but also amphibians, whereas group C includes also birds, “reptiles,” amphibians, and “bony fishes” (3, 4). It has been concluded therefore that the three groups, A, B, and C, had already diverged before the radiation of tetrapods, around 350 My ago (3).
Conservation of RSS Within the Mammals.
All RSSs identified in mammals other than humans can be linked to the types H1 and H3 defined within the human Vh locus (Fig. 3). Type H1 was also found in the mouse (Mus), whereas type H3 was encountered in mammals belonging to four different orders: cow (Bos), sheep (Ovis), camel (Camelus), and pig (Sus) for Artiodactyla; rabbit (Oryctolagus) for Lagomorpha; chimpanzee (Pan), gorilla (Gorilla), and macaques (Macaca) for Primates, and mouse for Rodentia. It must be noted that none of the four RSS types found in the mammals can be related to any RSS identified in birds, “reptiles,” amphibians, and “fishes.” This result likely reflects a limitation inherent in the analysis, because we must rely on the functionally nonconserved 3′ flanks to define RSS types. The long time since the divergence of mammals from the other vertebrates leads to the accumulation of a lot of mutations in the 3′ flanking region, including nucleotide transformations and insertions and deletions that have completely obscured the ancient phylogenetic signal.
The distribution of the four RSS types in the mammalian Vh gene tree (Fig. 3) can be summarized as follows: (i) coding sequences of group A are flanked by the related RSS types H1 and H5; (ii) coding sequences of group C are flanked by RSS type H3; and (iii) coding sequences of group B are flanked by two unrelated types of RSS: H2 and H3.
RSS Replacement in the Evolution of Mammals.
According to the “birth-and-death model of evolution” (3, 4), new V gene segments are produced by tandem or block duplications. To be functional, a V gene segment must have an intact exon–intron structure with a complete ORF, an intact 5′ regulatory region including two cis-acting transcriptional elements—the octamer motif and the TATA box—and a functional RSS. All elements must be duplicated together to give rise to a new functional gene segment. The birth-and-death model in its simplest form predicts that distinct groups of RSS should be coincident with the three groupings of A, B, and C based on the Vh coding sequences. Our analyses in contrast established that two unrelated types of RSS were found within group B (H2 and H3). In this group, three nodes are strongly supported by the BP analyses, namely subgroup BH2 (BP = 100), which has a RSS type H2, and subgroups BH3 (BP = 90) and BH3′ (BP = 91), which have the RSS type H3. Two alternative topologies are in conflict for the relationships between these three subgroups: (BH2(BH3,BH3′) in the weighted MP analysis (Fig. 3) and (BH3(BH2,BH3′) in the unweighted MP analysis (data not shown). These two different topologies indicate two distinct hypotheses for RSS evolution. The first, as seen in Fig. 3, implies that the ancestral Vh gene of group B was flanked by a RSS type H2, which was later replaced by an H3 RSS through recombination with a Vh gene of group C. By contrast, (BH3(BH2,BH3′) implies that the common Vh ancestor of groups B and C had an H3 RSS, which was later replaced by H2 RSS in the ancestor of subgroup BH2.
Four major points argue in favor of the (BH2(BH3′,BH3) hypothesis. (i) The alternative (BH3(BH2,BH3′) topology involves a highly unlikely and indeed exceptional conservation of the RSS spacer and nonfunctional 3′ flanking sequence during vertebrate evolution, because groups B and C had already diverged before the tetrapod radiation, around 350 My ago (3). If this were the case, we would expect to find the RSS type H3 not only in all or most Vh genes of groups B and C in mammals but also in Vh genes of other tetrapod groups. However, none of the germ-line Vh genes of groups B and C reported for birds, crocodilians, and amphibians are characterized by RSS and 3′ flanking region of type H3. (ii) In hypothesizing a replacement of the H3 RSS by the H2 RSS (rather than vice versa), one must suppose the type H2 sequence was inserted from a donor of unknown origin. In the case of the alternative hypothesis (BH2(BH3,BH3′), the origin of H2 RSS is known: it corresponds to the ancestral RSS type of group B. In this context, the absence of RSS type H2 in tetrapod species other than mammals is consistent with the long time divergence that separates mammals from other groups (around 350 My). (iii) In terms of exclusive synapomorphies (i.e., nucleotide states characteristic of a cluster of sequences), the grouping of BH3 with BH3′ is better supported than the grouping of BH2 with BH3′: two A→T nucleotide substitutions in positions 54 and 66, corresponding to T→S amino acid replacements, for the first, and only one A→C nucleotide substitution in position 228, corresponding to V→L amino acid replacement, for the second. (iv) Group C and subgroups BH3 and BH3′ share many sequence similarities for the RSS and 3′ flanking region but are clearly distinct for the regions located 5′ of the RSS (exon 2, intron, exon 1, and the 5′ regulatory region; data not shown). Such differences are not explained by a simple process of sequence divergence. We suggest that the common ancestor of subgroups BH3 and BH3′ was originally flanked by an H2 RSS, which was replaced by an H3 RSS originally associated with a group C via recombination.
Although it is difficult to give a precise date for the recombination event that replaced an H2 RSS with an H3 RSS, three clues based on the fossil record (21, 22) indicate that it probably occurred between 115 and 65 My ago. (i) The absence of RSS type H3 in amphibians, “reptiles,” and birds suggests the event happened after the emergence of mammals, around 200 My ago. (ii) That groups BH3 and BH3′ consist of species belonging to three different orders of mammals (Artiodactyla, Primates, and Rodentia) indicates the event took place before the adaptive radiation of eutherian mammals, 65 My ago. (iii) Because none of the available marsupial Vh coding sequences is related to group B, it can be assumed that the event arose after the emergence of the placental mammals, around 115 My ago.
The Evolutionary Success of RSS Type H3.
RSS type H3 was found in a great variety of mammal species, including in group B, the human, mouse, and sheep, and in group C, camel, the human, mouse, pig, and rabbit. In addition, phylogenetic analyses that also included cDNA sequences (data not shown) revealed candidate gene segments in other animals that may contain RSS type H3. Diverse species express Vh sequences closely related to the human Vh family 3: the Australian brush-tailed possum for Diprotodontia, the South American short-tailed gray opossum for Didelphimorphia, the dog and American mink for Carnivora, and the llama for Artiodactyla. Certain Vh genes of the cow (Artiodactyla) and horse (Perissodactyla) may also be endowed with type H3 RSSs, because they are more closely related to human Vh families 4 and 6 than human Vh family 2. The rat (Rodentia), like the human and mouse, is also characterized by a large spectrum of Vh diversity, and several different cDNA sequences have been reported for each of the three groups A, B, and C.
To summarize the situation in the mammals: (i) RSS types H2 and H5 have to date been reported only for humans, but the analyses of cDNA lead us to suspect their existence in rodents (mouse and rat). (ii) RSS type H1 is present in the Vh locus of the human and mouse, and presumably in the rat. (iii) RSS type H3 is the most dispersed in the class Mammalia, because it is found without doubt in the orders Artiodactyla, Lagomorpha, Primates, and Rodentia, and possibly in the Carnivora and Perissodactyla, as well as possibly also in the marsupial orders Didelphimorphia and Diprotodontia. With respect to mammal history, the evolutionary success of the RSS type H3 is consequently undeniable.
Is RSS Type H3 the Most Efficient for V(D)J Recombination?
The success of RSS type H3 contrasts with the infrequent occurrence of type H2 among mammalian Vh gene segments. Indeed, type H2 is seen only in the three gene segments of the human Vh family 2, and in other species examples of the H2 RSS have not been documented. (Vh coding sequences found in cDNA indicate the possibility that one might find perhaps some type H2 RSSs in rats and mice.) The limited representation of RSS type H2 in contemporary mammals may be traced perhaps to the atypical sequence features found in the heptamer and nonamer. Two of the changes are known to reduce V(D)J rearrangement as compared with a canonical RSS: (i) the substitution of A for G in the fifth position of the nonamer (ACAAAAACC to ACAAGAACC), and (ii) the substitution of T for A at the sixth position of the heptamer (CACAGTG to CACAGAG) (18, 20). RSSs with the particular combination of CACAGAG for the heptamer and ACAAGAACC for the nonamer have not been tested in functional assays directly but, consistent with all in vivo and in vitro studies to date, are predicted to be even more compromised for joining than those tested RSSs where each change was present alone. Thus the 2–26 and 2–70 genes are not associated with a competent RSS, and one would predict these genes are not frequently used in Vh to Dh recombination. For the remaining 2–5 gene, we might also expect a low efficiency for recombination because of the two substitutions within the RSS of G for A and T for A respectively at the fifth and sixth heptamer positions. In contrast, RSS type H3 is characterized by a canonical heptamer CACAGTG in almost all examples (25/26 sequences). Further, although all of the H3 nonamers are different from the canonical motif (ACACAAACC vs. ACAAAAACC), this nonamer position, to the extent it has been studied in vivo, has a far less measurable impact on recombination than the change that interrupts the critical succession of three A nucleotides in positions 5 to 7 (18, 20).
In support of the supposition that H2 RSSs are less proficient and that this would represent a selective disadvantage, it has been seen that Vh family 2 is underrepresented among the Vh genes used by IgM-expressing peripheral blood B cells of normal human adults (23). We suggest that the replacement of the H2 RSS by H3 was selected during the evolution of mammals, because H3 provides for a higher recombination efficiency. In favor of this hypothesis, we note that for all mammalian species that express only a small number of Vh genes (camel, pig, rabbit, and sheep), RSS type H3 is found exclusively (see Fig. 3). Several groups have emphasized that the level at which certain B cell receptor specificities occur in vivo might be dictated to some extent by family-specific RSS characteristics (24–26). RSS replacement during evolution means that under some circumstances, V genes with advantageous binding properties may evolve to have an appropriate level of representation in the immune repertoire through shuffling of RSS elements. Our study here, as discussed below, suggests that the V(D)J recombination mechanism itself may have played an active role in shuffling RSS sequences.
Models of RSS Replacement.
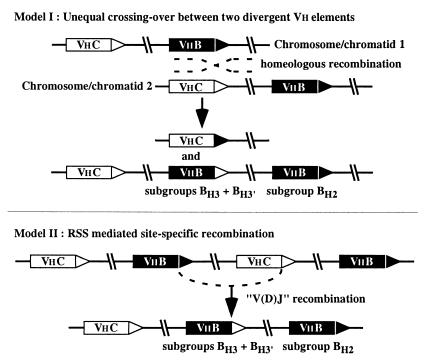
Two general models are proposed to explain the presence of two different gene structures in group B (i.e., Vh genes flanked by H2 RSSs and by H3 RSSs) (Fig. 4). The first model postulates an unequal crossing over in germ cells (or precursors) involving two divergent Vh genes of groups B and C. According to this model, unequal crossing over resulted in a tandem gene duplication on the chromosome that was selected during evolution and in Vh gene deletion on the other chromosome, presumably lost. RSS-targeted recombination is not implicated in this mechanism. The model, although it can account formally for the apparent reshuffling of RSS elements, must presume that “homeologous” recombination took place between distantly related Vh coding sequences.
Figure 4.
Models for how type H3 RSSs came to be associated with Vh genes of group B. Two models are proposed to explain the presence of the RSS type H3 within group B: the first involves an unequal crossing over (model I), and the second postulates a germline site-specific V(D)J recombination event (model II). Type H2 and H3 RSSs are indicated by solid and open triangles, respectively.
The second general model supposes that RSS shuffling came through the activity of the V(D)J recombination machinery in the germline (Fig. 4, II). In developing lymphoid cells, V(D)J recombination involves introduction of a double-strand break between the RSS heptamer and the adjacent coding segment. There is evidence that for some species, V(D)J recombination has occurred in the germline. This is demonstrated by the presence of joined and unjoined Ig and TCR genes in the genomes of cartilaginous fish (27, 28). The event shown in Fig. 4, model II, supposes that a germline V(D)J recombination event occurred before mammalian radiation. However, in place of the usual joined gene produced by V(D)J recombination, the recombination resulted in an alternative “hybrid joint” product. Germline joining with a hybrid joint outcome has been observed in sharks (5). Additionally, the particular variation proposed here, that is, hybrid joint formation between two 23-bp spacer RSSs, has been demonstrated to occur, albeit at a low frequency, in an in vivo plasmid recombination system (29). Thus, although the second model presupposes a V(D)J recombination event that is nonstandard in two respects, each possibility is supported by experimental evidence. That the involved gene segments were divergent and that the apparent recombination site is located at the 3′ end of Vh, near or at the coding sequence/RSS border, suggest that site-specific rather than homology-based recombination created the change of RSS type.
Regardless of the mechanism by which it occurred, because, as shown here, RSS elements have a different evolutionary history from coding sequences, we propose that RSS swapping has been an important adjunct to “birth and death” in the evolution of mammalian Ig loci.
Supplementary Material
Acknowledgments
This study was supported by funds from the Medical Research Council of Canada (MRC), from the National Cancer Society of Canada to S.L. and M.R.C., from the Terry Fox Marathon of Hope, and from the Cancer Research Society of Canada (G.W.). R.G. is a recipient of an MRC fellowship. We thank Dr. Ana Cumano and Pr. Jacques Charlemagne for their helpful comments on the manuscript.
Abbreviations
- RSS
recombination signal sequence
- BP
bootstrap proportions
- MP
maximum parsimony
- My
million years
- TCR
T cell receptor
- Vh
variable region of Ig heavy chains
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lewis S M. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. Adv Immunol. 1997;64:39–64. doi: 10.1016/s0065-2776(08)60886-x. [DOI] [PubMed] [Google Scholar]
- 3.Ota T, Nei M. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- 4.Nei M, Gu X, Sitnikova T. Proc Natl Acad Sci USA. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kokubu F, Litman R, Shamblott M J, Hinds K, Litman G W. EMBO J. 1988;11:3413–3422. doi: 10.1002/j.1460-2075.1988.tb03215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda F, Ishii K, Bourvagnet P, Kuma K-I, Hayashida H, Miyata T, Honjo T. J Exp Med. 1998;188:2151–2162. doi: 10.1084/jem.188.11.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champaign, IL: Illinois Natural History Survey; 1993. , Ver. 3.1.1. [Google Scholar]
- 9.Hassanin A, Lecointre G, Tillier S. C R Acad Sci Ser III. 1998;321:611–620. doi: 10.1016/s0764-4469(98)80464-2. [DOI] [PubMed] [Google Scholar]
- 10.Hassanin A, Pasquet E, Vigne J D. J Mamm Evol. 1998;5:217–236. [Google Scholar]
- 11.Felsenstein J. Evolution (Lawrence, KS) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. Syst Zool. 1978;27:401–410. [Google Scholar]
- 13.Naylor G J P, Brown W M. Syst Biol. 1998;47:61–76. doi: 10.1080/106351598261030. [DOI] [PubMed] [Google Scholar]
- 14.Philippe H, Lecointre G, L, Le Guyader H. Mol Biol Evol. 1996;13:1174–1186. [Google Scholar]
- 15.Schroeder H W J, Hillson J L, Perlmutter R M. Int Immunol. 1990;2:41–50. doi: 10.1093/intimm/2.1.41. [DOI] [PubMed] [Google Scholar]
- 16.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller C. Sequences of Proteins of Immunological Interest. 5th Ed. Bethesda: Natl. Inst. of Health; 1991. [Google Scholar]
- 17.Haino M, Hayashida H, Miyata T, Shin E K, Matsuda F, Nagaoka H, Matsumura R, Taka-ishi S, Fukita Y, Fujikura J, Honjo T. J Biol Chem. 1994;269:2619–2626. [PubMed] [Google Scholar]
- 18.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 19.Ramsden D A, Baetz K, Wu G E. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akamatsu Y, Tsurushita N, Nagawa F, Matsuoka M, Okazaki K, Imai M, Sakano H. J Immunol. 1994;153:4520–4529. [PubMed] [Google Scholar]
- 21.Benton M J. Vertebrate Palaeontology. 2nd Ed. London: Chapman & Hall; 1997. [Google Scholar]
- 22.Rich T H, Vickers-Rich P, Constantine A, Flannery T F, Kool L, van Klaveren N. Science. 1997;278:1438–1442. doi: 10.1126/science.278.5342.1438. [DOI] [PubMed] [Google Scholar]
- 23.Rassenti L Z, Kohsaka H, Kipps T J. Ann N Y Acad Sci. 1995;764:463–473. doi: 10.1111/j.1749-6632.1995.tb55866.x. [DOI] [PubMed] [Google Scholar]
- 24.Schroeder H W, Jr, Hillson J L, Perlmutter R M. Int Immunol. 1989;2:41–50. doi: 10.1093/intimm/2.1.41. [DOI] [PubMed] [Google Scholar]
- 25.Ramsden D A, Wu G E. Res Immunol. 1992;143:811–817. doi: 10.1016/0923-2494(92)80096-4. [DOI] [PubMed] [Google Scholar]
- 26.Larijani M, Yu C C, Golub R, Lam Q L, Wu G E. Nucleic Acids Res. 1999;27:2304–2309. doi: 10.1093/nar/27.11.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S S, Fitch D, Flajnik M F, Hsu E. J Exp Med. 2000;191:1637–1648. doi: 10.1084/jem.191.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litman G W, Anderson M K, Rast J P. Annu Rev Immunol. 1999;17:109–147. doi: 10.1146/annurev.immunol.17.1.109. [DOI] [PubMed] [Google Scholar]
- 29.Lewis S M, Hesse J E, Mizuuchi K, Gellert M. Cell. 1988;55:1099–1107. doi: 10.1016/0092-8674(88)90254-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.