Abstract
We report the complete sequence of an extreme halophile, Halobacterium sp. NRC-1, harboring a dynamic 2,571,010-bp genome containing 91 insertion sequences representing 12 families and organized into a large chromosome and 2 related minichromosomes. The Halobacterium NRC-1 genome codes for 2,630 predicted proteins, 36% of which are unrelated to any previously reported. Analysis of the genome sequence shows the presence of pathways for uptake and utilization of amino acids, active sodium-proton antiporter and potassium uptake systems, sophisticated photosensory and signal transduction pathways, and DNA replication, transcription, and translation systems resembling more complex eukaryotic organisms. Whole proteome comparisons show the definite archaeal nature of this halophile with additional similarities to the Gram-positive Bacillus subtilis and other bacteria. The ease of culturing Halobacterium and the availability of methods for its genetic manipulation in the laboratory, including construction of gene knockouts and replacements, indicate this halophile can serve as an excellent model system among the archaea.
Halobacterium specieso are obligately halophilic microorganisms that have adapted to optimal growth under conditions of extremely high salinity—10 times that of sea water. They contain a correspondingly high concentration of salts internally and exhibit a variety of unusual and unique molecular characteristics. Since their discovery, extreme halophiles have been studied extensively by chemists, biochemists, microbiologists, and molecular biologists to define both molecular diversity and universal features of life. A notable list of early research milestones on halophiles includes the discovery of a cell envelope composed of an S-layer glycoprotein, archaeol ether lipids and purple membrane, and metabolic and biosynthetic processes operating at saturating salinities (1). These early discoveries established the value of investigations directed at extremophiles and set the stage for pioneering phylogenetic studies leading to the three-domain view of life and classification of Halobacterium as a member of the archaeal domain (2, 3).
The Halobacterium genome was originally studied in the 1960s and found to be composed of two components, a GC-rich (68%) major fraction and a relatively AT-rich (58% GC) satellite (4, 5). Subsequent work showed that satellite DNA corresponded to the presence of large and variable covalently closed extrachromosomal circles and a large number of transposable insertion sequence (IS) elements, which explained the observed genetic plasticity of halophiles (6, 7). For Halobacterium NRC-1, 3 circular replicons were mapped, a ≈2-Mbp chromosome and 2 large replicons, pNRC200 and pNRC100, about 350 and 200 Kbp in size (8–11). We sequenced pNRC100 as a preliminary step in this genome project (12) and found a dynamic 191,346-bp replicon containing 176 putative genes, several of which are likely to be essential.
The complete genome sequence of Halobacterium NRC-1 is notable because of the excellent characteristics of halophiles as experimental organisms among the archaea (13). Culturing is facile, because they are both aerobic and mesophilic. DNA-mediated transformation may be accomplished at high efficiency, and cloning and expression vectors with selectable markers are readily available. Several gene replacement and knockout strategies have been used successfully, including a recently developed selectable and counterselectable method by using the yeast ura3 gene homolog, which should permit systematic knockout of all nonessential genes (14). Moreover, large-scale PCR amplification has been conducted successfully and DNA arrays constructed for interrogating patterns of gene expression. For biochemical analysis, Halobacterium proteins can be released by lysis in hypotonic medium and stabilized by addition of salts and other compatible solutes. Both membrane and soluble proteins have been useful for structural studies using electron and x-ray methods (15–17). These characteristics, coupled with the complete genome sequence, make Halobacterium NRC-1 an excellent experimental model among the archaea.
Genome Sequence, Annotation, and Organization
We sequenced the Halobacterium NRC-1 genome by using a whole genome shotgun strategy. Approximately 45,000 high-quality sequences were obtained by using automated Applied Biosystems sequencers, which provided ×7.5 coverage of the large chromosome. We used 505 oligonucleotides for directed sequencing of lower-quality regions or regions with single coverage. The remaining low-quality regions were covered by sequencing both ends of 124 PCR amplified genomic fragments. The shotgun Halobacterium NRC-1 sequences were assembled by using the phredphrap programs (19–21). Initially, all of the known and putative new IS elements were masked in the assembly. This resulted in 84 high-quality contigs, which were subsequently merged into groups of 2–10 adjacent contigs by a second round of assembly without repeat masking. Finally, the grouped contig consensus sequences were merged into three circular contigs by using a third round of assembly with the phredphrap programs. The sequences have been deposited in GenBank and assigned the following accession numbers: AE004437, AE004438, and AF016485.
Our results confirmed the expected size and structure of the Halobacterium NRC-1 genome. The genome was found to be 2,571,010 bp in size and composed of 3 circular replicons, a 2,014,239-bp-large chromosome and 2 smaller replicons, pNRC100 (191,346 bp) (12) and pNRC200 (365,425 bp). Interestingly, pNRC100 and pNRC200 contained a 145,428-bp region of identity, including 33- to 39-kb inverted repeats that mediate inversion isomerization (10). These two replicons were substantially less GC rich than the largest replicon (57.9% and 59.2% vs. 67.9%). The genome contained 91 IS elements representing 12 families, including 29 on pNRC100 (12), 40 on pNRC200, and 22 on the large chromosome. Two new elements, ISH5 and ISH10, were identified.
The program glimmer (22, 23) was used for gene prediction on the finished Halobacterium NRC-1 genome sequence. Predicted genes were translated and the resulting sequences used to search the nonredundant database of proteins (translation of GenBank CDS, Protein Data Bank, SwissProt, and Protein Identification Resource databases) available on the National Center for Biotechnology Information web site by using the netblast program (24) in the GCG software package (Genetics Computer Group, Madison, WI). To aid in the processing of large numbers of data files, we developed perl-based scripts to handle recursively the input of sequences and their analysis. Additional analysis was conducted by a consortium of 12 laboratories (http://zdna.micro.umass.edu/haloweb).
Our analysis identified 2,682 likely genes (including 52 RNA genes) in the Halobacterium NRC-1 genome, of which 1,658 coded proteins with significant matches to the databases. Of the matches, 591 were to conserved hypothetical proteins, and 1,067 were to proteins with known or predicted function. The large chromosome contained 2,111 putative genes, pNRC200 contained 374, and pNRC100 contained 197. A significantly larger fraction of the genes on the large chromosome (45%) matched to genes of known function in the databases than did genes on either pNRC200 (32%) or pNRC100 (26%). The complete genetic map and table of genes and genetic elements are available on the PNAS web site as supplementary material (www.pnas.org).
Interestingly, about 40 genes on pNRC100 and pNRC200 coded for proteins likely to be essential or important for cell viability such as a DNA polymerase, seven TBP and TFB transcription factors, and the arginyl-tRNA synthetase, indicating that these replicons are minichromosomes (12). A fraction of these genes have a G + C composition that is significantly higher than the minichromosome average, e.g., those coding for potassium and phosphate uptake, thioredoxin reductase, cytochrome oxidase, and Orc/Cdc6 cell division proteins. These results and the finding of many IS elements on pNRC100 and pNRC200 indicate that the minichromosomes contribute to Halobacterium genome evolution by facilitating the acquisition of new genes (12).
Energy Metabolism
Halobacterium NRC-1 is an aerobic chemoorganotroph, growing on the degradation products of less halophilic organisms as the salinity reaches near saturation. In the laboratory, cells are cultured best in a complex medium (13, 25). A minimal medium described for Halobacterium includes all but 5 of the 20 amino acids for growth (26). Several amino acids may be used as a source of energy, including arginine and aspartate, which are passed to the citric acid cycle via 2-oxoglutarate and oxaloacetate, respectively (Fig. 1). Under aerobic conditions, arginine is presumably converted to glutamate via the arginine deiminase pathway, and this amino acid then enters the cycle via glutamate dehydrogenase. The arginine deiminase pathway is coded by the arcRACB genes (27), which are found on pNRC200.
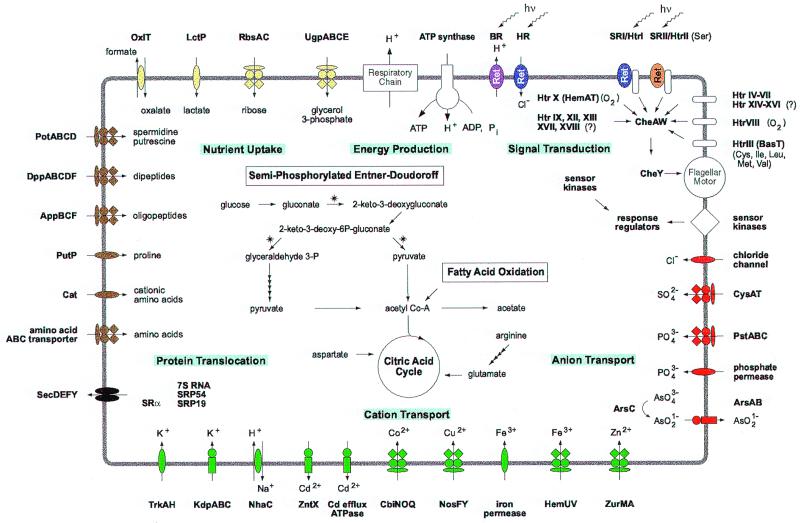
Figure 1.
An integrated view of the biology of Halobacterium NRC-1. Aspects of energy production, nutrient uptake, membrane assembly, cation and anion transport, and signal transduction are depicted. ATP synthesis by chemiosmotic coupling of proton transport by the respiratory chain and by light-driven proton pumping by bacteriorhodopsin (BR) (purple oval) or chloride transport by halorhodopsin (HR) (blue oval) is shown. Below, the semiphosphorylated Entner–Doudoroff pathway is shown, and the presence of fatty acid oxidation and the citric acid cycle is indicated. Enzymes not yet identified are marked with asterisks. A variety of nutrient uptake systems (represented by yellow or brown structures) coded by the genome including glycerol 3-phosphate (UgpABCE) and ribose (RbsAC) ABC transporters, a lactate (LctP) transporter, formate-oxalate antiporter (OxiT), spermidine and putrescine uptake ABC transporter (PotABCD), and amino acid (PutP, Cat) and dipeptide (DppABCDF) transporters are shown. Other amino acid uptake systems, represented by a generic ABC transporter, are also likely to exist. Components of the protein translocation machinery (SecDEFY, SRP19, 54, SRα) (in black) are shown. Cation transporters (in green) shown are for K+ (TrkAH, and KdpABC), Na+ (NhaC), Cd2+ (ZntX and Cd efflux ATPase), Co2+ (CbiNOQ), Cu2+ (NosFY), Fe3+ (iron permease and HemUV), and Zn2+ (ZurMA). Anion transporters shown (in red) are for SO42− (CysAT), PO43− (PstABC and phosphate permease), Cl− (chloride channel), and arsenate (ArsABC). A complex system of photoreceptors and signal transduction components are shown, including two sensory receptors (SRI shown in blue and SRII shown in orange), 17 transducers (Htr I, II, III, IV, V, VI, VII, VIII, IX, X, XII, XIII, XIV, XV, XVI, XVII, and XVIII) responding to light (hυ), O2, or amino acids, as indicated. Transmission of the motility signal to the flagellar motor via CheAW and CheY is shown by arrows. Single examples of sensor kinases, membrane bound (white rhombus) or cytoplasmic, and response regulators are identified.
In accordance with the ability of Halobacterium NRC-1 to grow on amino acids, which ultimately are catabolized by the citric acid cycle, the genes coding all of the enzymes for an aerobic cycle are present (Fig. 1). In common with all archaea, the conversion of pyruvate to acetyl-CoA (before the citric acid cycle) and of 2-oxoglutarate to succinyl-CoA are catalyzed by the respective 2-oxoacid ferredoxin oxidoreductases (28, 29). Interestingly, genes encoding malate ferredoxin oxidoreductase and fumarate reductase are also present, so that when combined with the 2-oxoglutarate oxidoreductase, they could form a partial reverse citric acid cycle from oxaloacetate to 2-oxoglutarate under anaerobic conditions, as has been found in a number of methanogenic archaea (30, 31). In connection with the citric acid cycle, the key enzymes of the glyoxylate cycle, isocitrate lyase, and malate synthase, could not be identified in the genome sequence. This is in accord with an inability of Halobacterium to grow on acetate (32, 33).
Growth on amino acids requires a gluconeogenic pathway for carbohydrate synthesis, and the genes for a reverse Embden–Meyerhof glycolytic pathway have been identified except for fructose-1,6-bisphosphate aldolase. The inability to find this gene was unexpected, particularly as those for triose-phosphate isomerase and fructose-1,6-bisphosphatase are present. However, an unusual class I aldolase found in eukaryotic organisms has been detected in some related Haloarcula species (34), and it may be that a similar enzyme is present in Halobacterium NRC-1 but is too divergent in sequence to permit assignment.
Although Halobacterium is reported to be unable to metabolize sugars, genes coding for glucose dehydrogenase and 2-keto-3-deoxygluconate kinase appear to be present in NRC-1. These are enzymes of the semiphosphorylated Entner–Doudoroff pathway shown to be present in several halophilic archaea (25, 35), although the gene for 2-keto-3-deoxy-6-phosphogluconate aldolase remains to be assigned in NRC-1. With respect to glucose catabolism via an Embden–Meyerhof glycolytic pathway, a 6-phosphofructokinase gene could not be found by using both ATP- and ADP-dependent homologs as queries. The genes for the catabolism of glyceraldeyde 3-phosphate (the product of glucose catabolism via Entner–Doudoroff and/or Embden–Meyerhof pathways) to pyruvate are all present, and it is these same enzymes that function to effect gluconeogenesis.
Halobacterium NRC-1 also possesses genes encoding enzymes of the bacterial-like fatty acid β-oxidation pathway. Both medium-chain and long-chain acyl-CoA ligases, 3 acyl-CoA dehydrogenases, enoyl-CoA hydratase, 2 3-hydroxyacyl-CoA dehydrogenases, and 2 3-ketoacyl-CoA thiolases are present. However, despite the presence of these genes, there are no reports of the oxidation of fatty acids by NRC-1. Finally, a gene cluster coding for proteins similar to a 2-oxoacid dehydrogenase complex in Bacillus species was identified in NRC-1, including pyruvate decarboxylase (a and b chains), lipoyl acyltransferase, and dihydrolipoamide dehydrogenase, as has also been reported in Haloferax volcanii (36, 37).
Cell Envelope Components and Transport
The cell envelope of Halobacterium NRC-1 consists of a single lipid bilayer membrane surrounded by an S-layer assembled from the cell-surface glycoprotein (38). Although the cytoplasm is in osmotic equilibrium with the hypersaline environment, the cell maintains a high (≈4 M) intracellular K+ concentration that is equivalent to the external Na+ concentration (39). The passive permeability of the membrane to K+ and Na+ ions is low (40), so active transport is required to maintain the ionic distribution. Accordingly, NRC-1 has multiple active K+ transporters, including KdpABC, an ATP-driven K+ transport system, and TrkAH, a low-affinity K+ transporter driven by the membrane potential (Fig. 1). Active Na+ efflux is probably mediated by NhaC proteins, which likely correspond to the unidirectional Na+/H+ antiporter activity described previously (41). Interestingly, KdpABC, TrkA (three of five copies), and NhaC (one of three copies) are coded by pNRC200.
At least 27 members of the ABC transporter superfamily are present in Halobacterium NRC-1. Among active transporters for nutrient uptake identified were those for cationic amino acids (Cat) and proline (PutP), dipeptides (DppABCDF), oligopeptides (AppACF), and a sugar transporter (Rbs) (Fig. 1). Among small-ion transporters, most were closely related to bacterial proteins. Genes for exporting heavy metals (arsenite and cadmium) and other toxic compounds (multidrug-resistance homologs) are present (Fig. 1). Four ars genes (arsRDAC) are clustered on pNRC100 (12), whereas a fifth gene (arsB) resides on the large chromosome. Phosphate transport is mediated by at least two systems, including PstABC (two copies) and phosphate permease (three copies); all but one copy of pstABC are coded by pNRC200.
For polypeptide translocation across the membrane, the general secretory (Sec) machinery for Halobacterium NRC-1 appears to be a hybrid of the eukaryotic and bacterial systems (44). The core components, Sec61α/SecY and Sec61γ/SecE, as well as those of the signal recognition particle, SRP54/Ffh and its 7S RNA scaffold, are related to the corresponding eukaryotic factors (Fig. 1). The SRP complex also includes SRP19, a subunit found in eukaryotes but not in bacteria. On the other hand, like bacteria, NRC-1 contains the universally conserved SRP-receptor subunit SRPα/FtsY and lacks the eukaryotic β-subunit homolog. The bacterial translocase protein homologs SecD and SecF are also present, but the essential bacterial ATPase SecA is absent. In addition, a gene closely related to tatC of A. fulgidus (45) was found, suggesting the presence of the twin-arginine protein export pathway.
The polar lipids of Halobacterium include phospholipids and glycolipids based on archaeol, a glycerol diether lipid containing phytanyl chains derived from C20 isoprenoids (46). All of the key enzymes of isoprenoid synthesis were identified, including HMG-CoA reductase (MvaA), the target of the growth inhibitor mevinolin (47). Interestingly, two genes for this pathway, coding for mevalonate pyrophosphate decarboxylase and isopentenyl pyrophosphate isomerase, have not been found in the genomes of other archaea. Enzymes catalyzing formation of polar lipids, which have been outlined by metabolic labeling from mevalonate and dihydroxyacetone (48), are coded in NRC-1. For synthesis of phospholipids, proteins related to bacterial and archaeal phosphatidyl transferases (PgsA and PssA) are present, although CDP-archaeol synthase has not been identified.
Because the apolar lipids of Halobacterium are isoprenoids, their synthesis likely requires some of the same machinery needed to synthesize the phytanyl chains of archaeol. Additional enzymes are required to synthesize the C30 isoprenoid squalene, the C40 retinal-precursor β-carotene, and C50 bacterioruberins, which are thought to act as photoprotectants (49). We identified two phytoene synthase and three phytoene dehydrogenase homologs in NRC-1.
Signal Transduction and Photobiology
Halobacterium inhabits a harsh environment with extreme solar radiation and dynamic nutritional conditions. Accordingly, Halobacterium cells have developed sophisticated sensory pathways for color-sensitive phototaxis, chemotaxis to a large variety of substances, aerotaxis, osmotaxis, and thermotaxis (Fig. 1). Compared with the 5 methyl-accepting taxis transducers in Escherichia coli, the NRC-1 genome reveals at least 17 homologous methyl-accepting proteins, 13 of which had been previously identified (50–52). One transducer reported in other Halobacterium strains (htrXI or car) is not present in NRC-1. Unlike in bacteria, transducer and flagellin genes are not clustered in one or two operons, although a single large cluster of genes in Halobacterium NRC-1 includes nine che genes and two fla genes. The cluster includes a complete set of Bacillus subtilis che gene homologs, consisting of cheA, B, C (named cheJ in Halobacterium), D, R, Y, W, as well as a second cheC (cheC1 and cheC2) and a second cheW (cheW1 and cheW2). There is no cheZ, which encodes a phospho-CheY phosphatase important in Escherichia coli taxis adaptation that is also present in several other bacteria, but not in B. subtilis. There are six flagellin genes, but the numerous flagellar apparatus and motility genes of bacteria are not evident in the sequence.
Halobacterium has been studied heavily with regard to its photoactive visual pigment-like seven-transmembrane-helix retinal proteins, the archaeal rhodopsins, which have been demonstrated in several archaeal halophiles as well as in the eukaryote Neurospora crassa and other fungi (53). Only the four members of this family previously identified in Halobacterium are present [the light-driven ion transporters bacteriorhodopsin and halorhodopsin, and the phototaxis receptors, sensory rhodopsins I and II (Fig. 1)]. Other possible photoreceptor genes identified include those homologous to genes encoding the flavoprotein cryptochromes, which serve as circadian photoregulators in Arabidopsis and mammals (54). There are two homologous genes in NRC-1; however, it should be noted that they are also homologous to E. coli photolyase, and photolyase and cryptochromes cannot be unequivocally distinguished on the basis of primary sequence alone. A homolog of KaiC that generates circadian oscillation in cyanobacteria is present in NRC-1 (55). At least 6 response regulator genes and 14 histidine kinase-encoding genes were found in the NRC-1 genome.
DNA Replication, Repair, and Recombination
The Halobacterium NRC-1 genome revealed three DNA polymerase types (56), two family B polymerases (one coded by pNRC200), a bacteriophage-like family A polymerase, as well as the heterodimeric family D polymerase. The large subunit of the latter contains an intein similar to the hyperthermophilic archaeon Pyrococcus horikoshii. Additional proteins that may be active at the replication fork include a putative DNA ligase, primase, type I topoisomerase (TopA), and two type II topoisomerases (GyrA and B, and Top6A and B). We also observed the presence of the following: Pcna, sliding clamp, Rfc, clamp loader, and Rpa, replication protein A involved in single-strand DNA binding, Mcm minichromosome maintenance protein, and Orc/Cdc6, origin recognition complex proteins. Nine copies of orc are present including three scattered on the large chromosome, suggesting the possibility of multiple replication origins.
For DNA repair, Halobacterium NRC-1 possesses two of the three genes involved in the guanine oxidization pathway, mutT and mutY. In addition, both the nucleotide and base excision pathways appear to be complete as copies of the uvrABC nuclease and uvrD helicase, and endonucleases and glycosylase genes are present. Two of the three genes of methyl-directed mismatch repair were found, mutL and mutS (three copies), but the nuclease gene mutH was missing. The E. coli-type dam methylase (recognizing GATC) is absent in NRC-1. However, a putative CTAG-specific methylase gene is present, which has also been found in Methanobacterium thermoformicicum (57).
Repair genes similar to those in yeast are present in Halobacterium NRC-1, including rad2, rad3, rad24, and rad25. Several of these proteins appear to be active in the excision repair pathway. Products of rad3 and rad25 have been identified as repair helicases and Rad2 is a single-stranded DNA endonuclease. This suggests that Halobacterium NRC-1 has developed multiple pathways to repair UV-induced damage as a means for survival. Cell-cycle genes in Halobacterium NRC-1 include five copies of cdc48, one of which is on pNRC200.
The search for genes encoding proteins involved in recombination yielded two RadA genes, with homology to both the yeast protein Rad51 and the E. coli protein RecA (58) and a homolog of the putative Holliday junction resolvase from Pyrococcus furiosus (59).
Transcription
Halobacterium NRC-1, like other archaea, drives regulated transcription by using a single version of a eukaryotic RNA polymerase II-like transcription system. The information for the multisubunit RNA polymerase II is coded by 12 genes located at 6 loci. Genes encoding Rpo subunits A, C, B′, B′′, and H are present in a gene cluster (60), as are the genes for subunits E′ and E′′, and subunits K and N. Subunit M, which has also been annotated as TFIIS (61), is also present.
An interesting finding is the presence of multiple copies of TBP and TFB transcription factor genes. Five complete tbp genes and one partial gene that has one-half of the two stirrups were identified. Four of the six tbp genes were reported previously on pNRC100 (12); additional single genes were found on both the large chromosome and pNRC200. In contrast, five of the seven tfb genes are present on the large chromosome, and the other two are on pNRC200. The possibility of a novel regulatory system involving up to 42 different TBP-TFB combinations has been discussed recently (62). The finding of alternate TATA box and possibly BRE sequences on the basis of saturation mutagenic analysis of the bacterio-opsin gene (bop) promoter supports this hypothesis (63, 64). At least 27 transcriptional regulators were also identified. Transcription factors known to be required for polymerase II transcription in other systems (TFIIF, TFIIH, and TFIIEβ) were not evident. A TFIIEα homolog was identified by using the pfam search tool (65). Additional factors present include termination/antitermination factor homologs NusA and NusG (66).
Translation
Translational components of Halobacterium NRC-1, like other archaea, have both bacterial and eukaryotic homologs. We identified 47 tRNA genes for all 20 amino acids and all 61 possible codons, by using the tRNA scan-se program (67), including tRNAs with 44 unique anticodons, 1 methionine initiator tRNA, 1 redundant tRNA (Ala-CGC), and 1 tRNA (anticodon CAU), which is predicted to be converted from methionine to isoleucine specificity posttranscriptionally as in E. coli (68). Three tRNA genes contain introns, Trp-tRNA-CCA, elongator Met-tRNA-CAU, and Ile-tRNA-CAU. Aminoacyl tRNA synthetases are present for all amino acids except asparagine and glutamine, which likely require amidotransferases. Homologs of the gatA, gatB, and gatC genes, similar to other archaea that lack AsnRS and GlnRS genes, are present (69). Interestingly, one aminoacyl tRNA synthetase, ArgRS, closely related to the E. coli and other Gram-negative bacterial and yeast mitochondrial enzymes, is coded by pNRC200.
The single-copy rRNA operon is bacterial-like in its organization and gene content: 5′ 16S, tRNA (Ala-UGC), 23S, 5S, tRNA (Cys-GCA) (70). The RNA component but not protein components of RNaseP was detected. Genes coding homologs of the eukaryotic nucleolar proteins fibrillarin and Nop56/58 were also identified in NRC-1. The occurrence of these proteins in other archaea and the recent identification of C/D box snoRNAs in thermophilic archaea (71) suggest that the snoRNA-mediated 2-O-methylribose modification system is generally present, although none could be identified in NRC-1.
Generally, the protein components of the translation apparatus of archaea resemble more closely those of eukaryotes than those of bacteria (72). In our annotation of ribosomal (r-) proteins, we used the nomenclature for Haloarcula marismortui (71), a related halophile where 25 30S subunit and 28 50S subunit r-proteins have been enumerated by purification, partial or complete amino acid sequence analysis, and gene sequence analysis (73), and where the crystal structure of the 50S subunit has been determined (17). Despite their generally higher sequence similarity to eukaryotes, the r-protein genes of Halobacterium NRC-1 are organized into multigene clusters that resemble operons of E. coli. In one of these clusters, the L1P, L10P, and L12P genes are cotranscribed, and the 5′ leader of the mRNA contains a bacterial-like L1 translational operator, a structural mimic of the site in 23S rRNA that is used to autogenously regulate translation of the mRNA (74). Genes coding homologs of eukaryotic eIF1A, eIF2 α, β, and γ subunits, eIF4, eIF5, and eIF2B α and δ are also present.
Evolutionary Comparisons
The Halobacterium NRC-1-predicted proteome was compared with 11 other complete microbial genomes by using the darwin suite of programs (75, 76). The results shown in Table 1 confirm the archaeal nature of Halobacterium NRC-1, showing closest similarities to Archaeoglobus fulgidus and Methanococcus jannaschii. We also found homologs to many of the archaeal “signature” proteins recently reported (77). The NRC-1-predicted proteins were also similar to the Gram-positive bacterium, B. subtilis, more than to any other bacteria, and displayed a large number of unique homologs with the radiation-resistant bacterium Deinococcus radiodurans, suggesting that NRC-1 may have acquired a substantial number of genes from certain bacteria, possibly by lateral gene transfer. Additional findings were that the NRC-1 proteome is highly acidic (average pI of 5.1), consistent with protein stabilization and adaptation to a high-salt environment (41), and that there is a high degree of redundancy among many protein classes. A more detailed comparative genomics investigation should provide further insights into evolutionary and adaptative forces operating in this extremophile.
Table 1.
Pairwise comparison of Halobacterium NRC-1 proteome and 11 other microbial proteomes
| Proteome* | Genome size, MB | Number of homologs† | Number of unique homologs‡ | Average accepted point mutation (PAM)§ |
|---|---|---|---|---|
| Mge | 0.58 | 252 | 0 | 148.9 |
| Ctr | 1.05 | 429 | 4 | 153.6 |
| Tpa | 1.14 | 524 | 12 | 157.3 |
| Mja | 1.66 | 876 | 0 | 128.8 |
| Hin | 1.83 | 710 | 2 | 149.4 |
| Afu | 2.18 | 1,243 | 44 | 129.8 |
| Dra | 3.28 | 1,503 | 35 | 152.0 |
| Syn | 3.57 | 1,013 | 34 | 146.5 |
| Bsu | 4.20 | 1,027 | 29 | 141.0 |
| Eco | 4.60 | 1,190 | 25 | 149.5 |
| Sce | 13.12 | 1,183 | 79 | 153.0 |
Peptide sequences of 11 microorganisms [Mycoplasma genitalium (Mge), Chlamydia trachomatis (Ctr), Treponema pallidum (Tpa), M. jannaschii (Mja), Haemophilus influenzae (Hin), Archaeoglobus fulgidus (Afu), D. radiodurans (Dra), Synechocystis sp. PCC6803 (Syn), B. subtilis (Bsu), E. coli K12 (Eco), Saccharomyces cerevisiae (Sce)] were retrieved either from the National Center for Biotechnology Information Entrez Genome web site or from web sites for the specific genome projects.
The number of Halobacterium proteins that have at least one qualified homolog from the specified proteome.
The number of Halobacterium proteins that are uniquely homologous to the other organism.
To determine PAM values, only matches satisfying the minimum alignment length of 100 residues and maximum PAM value at 200 were collected. For each Halobacterium protein, the best match with the lowest PAM value from each compared proteome was used for generating the data in this table. The average PAM value between the Halobacterium proteome and each of the other 11 proteomes was obtained by dividing the sum of the best PAM values for all Halobacterium proteins by the number of proteins that have at least one qualified homolog in the compared proteome (the numbers in column 3). To determine whether the average PAMs for each proteome are significantly different from each other, ANOVA post-hoc tests were performed by using StartView from SAS Institute, Cary, NC. Afu or Mja vs. any other proteomes: P value <0.0001; Bsu vs. all other bacteria and Sce: P value less than or equal to 0.0024.
Future Prospects
The sequence of Halobacterium NRC-1 has revealed 3 large replicons, a large chromosome and 2 novel minichromosomes, and 2,682 putative genes, including 972 novel genes, with no homologs in the databases. Because this halophile is amenable to experimental analysis by using a battery of approaches such as gene knockouts, DNA arrays, and proteomics, future studies should yield significant insights into the functions of conserved unknown and hypothetical genes among the archaea. Moreover, because the halophilic proteins are highly negatively charged with enhanced solubility, they lend themselves readily to the determination of high-throughput three-dimensional structure by experimental and theoretical approaches (structural genomics). Also, this system should serve as an excellent model of aspects of eukaryotic biology, e.g., DNA replication, transcription, and translation. Comparison of a halophile genome to other prokaryotic genomes should lead to a better understanding of microbial adaptation to extreme conditions, such as hypersalinity, damaging radiation, and an oxidizing atmosphere. Indeed, the availability of the complete genome sequence for this easily cultured and tractable microbe should facilitate a wide range of studies and establish this halophile as a model organism among the archaea.
Supplementary Material
Acknowledgments
This work was supported by collaborative research grants from the National Science Foundation to S.D. (MCB-97022066 and MCB-9812330) and L.H. (MCB-9900497).
Abbreviation
- IS
insertion sequence
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AE004437, AE004438, and AF016485).
Halobacterium species are referred to in the literature by a variety of designations, including H. halobium, H. cutirubrum, H. salinarium, and H. salinarum. The precise relationships among these organisms and Halobacterium sp. strain NRC-1 are not entirely clear (18). Strain NRC-1 was a gift from W. F. Doolittle, Dalhousie University, Halifax, Canada. The strain has been deposited with the American Type Culture Collection, Manassas, VA (reference no. ATCC 700922).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.190337797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.190337797
References
- 1.Bayley S T, Morton R A. CRC Crit Rev Microbiol. 1978;6:151–205. doi: 10.3109/10408417809090622. [DOI] [PubMed] [Google Scholar]
- 2.Woese C R. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doolittle W F. Science. 1999;25:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 4.Joshi J G, Guild W R, Handler P. J Mol Biol. 1963;6:34–38. doi: 10.1016/s0022-2836(63)80079-0. [DOI] [PubMed] [Google Scholar]
- 5.Moore R L, McCarthy B J. J Bacteriol. 1969;99:248–254. doi: 10.1128/jb.99.1.248-254.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charlebois R L, Doolittle W F. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 297–307. [Google Scholar]
- 7.DasSarma S. Experientia. 1993;49:482–486. doi: 10.1007/BF01955149. [DOI] [PubMed] [Google Scholar]
- 8.Bobovnikova Y, Ng W-L, DasSarma S, Hackett N R. Sys Appl Microbiol. 1994;16:597–604. [Google Scholar]
- 9.Hackett N R, Bobovnikova Y, Heyrovska N. J Bacteriol. 1994;176:7711–7718. doi: 10.1128/jb.176.24.7711-7718.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng W-L, Kothakota S, DasSarma S. J Bacteriol. 1991;173:1958–1964. doi: 10.1128/jb.173.6.1958-1964.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng W-L, Arora P, DasSarma S. Syst Appl Microbiol. 1994;16:560–568. [Google Scholar]
- 12.Ng W V, Ciufo S A, Smith T M, Bumgarner R E, Baskin D, Faust J, Hall B, Loretz C, Seto J, Slagel J, Hood L, DasSarma S. Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- 13.DasSarma S, Robb F T, Place A R, Sowers K R, Schreier H J, Fleischmann E M. Archaea: A Laboratory Manual—Halophiles. Plainview, NY: Cold Spring Harbor Lab. Press; 1995. [Google Scholar]
- 14.Peck R F, DasSarma S, Krebs M P. Mol Microbiol. 2000;35:667–676. doi: 10.1046/j.1365-2958.2000.01739.x. [DOI] [PubMed] [Google Scholar]
- 15.Subramaniam S, Henderson R J. J Struct Biol. 1999;128:19–25. doi: 10.1006/jsbi.1999.4178. [DOI] [PubMed] [Google Scholar]
- 16.Luecke H, Schobert B, Richter H T, Cartailler J P, Lanyi J K. Science. 1999;286:255–261. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- 17.Ban N, Sissen P, Hansen J, Capel M, Moore P B, Steitz T A. Nature (London) 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 18.Tindall B J. In: The Prokaryotes, A Handbook on the Biology of Bacteria. Balows A, Truper H J, Dworkin M, Harder K-H, Schleifer K-H, editors. New York: Springer; 1992. pp. 768–808. [Google Scholar]
- 19.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 20.Ewing B, Hillier L, Wendl M C, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 21.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 22.Salzberg S L, Delcher A L, Kasif S, White O. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delcher A L, Harmon D, Kasif S, White O, Salzberg S L. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang A, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawal N, Kelkar S M, Altekar W. Ind J Biochem Biophys. 1988;25:674–686. [PubMed] [Google Scholar]
- 26.Grey V L, Fitt P S. Can J Microbiol. 1976;22:440–442. doi: 10.1139/m76-068. [DOI] [PubMed] [Google Scholar]
- 27.Ruepp A, Soppa J. J Bacteriol. 1996;178:4942–4947. doi: 10.1128/jb.178.16.4942-4947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plaga W, Lottspeich F, Oesterhelt D. Eur J Biochem. 1992;205:391–397. doi: 10.1111/j.1432-1033.1992.tb16792.x. [DOI] [PubMed] [Google Scholar]
- 29.Adams M W W, Kletzin A. Adv Prot Chem. 1996;48:101–180. doi: 10.1016/s0065-3233(08)60362-9. [DOI] [PubMed] [Google Scholar]
- 30.Sprott G D, Ekiel I, Patel G B. Appl Environ Microbiol. 1993;59:1092–1098. doi: 10.1128/aem.59.4.1092-1098.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blaut M. Antonie Leeuwenhoek. 1994;66:187–208. doi: 10.1007/BF00871639. [DOI] [PubMed] [Google Scholar]
- 32.Oren A, Gurevich P. FEMS Microbiol Lett. 1995;130:91–95. [Google Scholar]
- 33.Serrano J A, Camacho M, Bonete M J. FEBS Lett. 1998;434:13–16. doi: 10.1016/s0014-5793(98)00911-9. [DOI] [PubMed] [Google Scholar]
- 34.Krishnan G, Altekar W. Eur J Biochem. 1991;195:343–350. doi: 10.1111/j.1432-1033.1991.tb15712.x. [DOI] [PubMed] [Google Scholar]
- 35.Tomlinson G A, Koch T K, Hochstein L I. Can J Microbiol. 1974;20:1085–1091. doi: 10.1139/m74-170. [DOI] [PubMed] [Google Scholar]
- 36.Danson M J, Jolley K A, Maddocks D G, Dyall-Smith M L, Hough D W. In: Microbiology and Biogeochemistry of Hypersaline Environments. Oren A, editor. Boca Raton, FL: CRC; 1999. pp. 239–248. [Google Scholar]
- 37.Jolley K A, Maddocks D G, Gyles S L, Mullan Z, Tang S L, Dyall-Smith M L, Hough D W, Danson M J. Microbiology. 2000;146:1061–1069. doi: 10.1099/00221287-146-5-1061. [DOI] [PubMed] [Google Scholar]
- 38.Kushner D J. In: The Archaebacteria. Woese C R, Wolfe R S, editors. Vol. 8. Orlando, FL: Academic; 1985. pp. pp.171–215. [Google Scholar]
- 39.Christian J H B, Waltho J A. Biochim Biophys Acta. 1962;65:506–508. doi: 10.1016/0006-3002(62)90453-5. [DOI] [PubMed] [Google Scholar]
- 40.Stoeckenius W, Lozier R H, Bogomolni R A. Biochim Biophys Acta. 1979;505:215–278. doi: 10.1016/0304-4173(79)90006-5. [DOI] [PubMed] [Google Scholar]
- 41.Lanyi J K. Microbiol Rev. 1978;42:682–706. doi: 10.1128/mr.42.4.682-706.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami N, Konishi T. Arch Biochem Biophys. 1990;281:13–20. doi: 10.1016/0003-9861(90)90406-o. [DOI] [PubMed] [Google Scholar]
- 43.MacDonald R E, Greene R V, Lanyi J K. Biochemistry. 1977;16:3227–3235. doi: 10.1021/bi00633a029. [DOI] [PubMed] [Google Scholar]
- 44.Pohlschroder M, Prinz W A, Hartmann E, Beckwith J. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 45.Berks B C, Sargent F, Palmer T. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 46.Kates M. Experientia. 1993;49:1027–1036. doi: 10.1007/BF01929909. [DOI] [PubMed] [Google Scholar]
- 47.Cabrera J A, Bolds J, Shields P E, Havel C M, Watson J A. J Biol Chem. 1986;261:3578–3583. [PubMed] [Google Scholar]
- 48.Kamekura M, Kates M. In: Halophilic Bacteria. Rodriguez-Valera F, editor. II. Boca Raton, FL: CRC; 1988. pp. 25–54. [Google Scholar]
- 49.Shahmohammadi H R, Asgarani E, Terato H, Saito T, Ohyama Y, Gekko K, Yamamoto O, Ide H. J Radiat Res. 1998;39:251–262. doi: 10.1269/jrr.39.251. [DOI] [PubMed] [Google Scholar]
- 50.Yao V J, Spudich J L. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rudolph J, Nordmann B, Storch K F, Gruenberg H, Rodewald K, Oesterhelt D. FEMS Microbiol Lett. 1996;139:161–168. doi: 10.1111/j.1574-6968.1996.tb08197.x. [DOI] [PubMed] [Google Scholar]
- 53.Spudich E N, Yang C S, Jung K H, Spudich J L. Annu Rev Cell Dev Biol. 2000;16:365. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 54.Cashmore A R, Jarillo J A, Wu Y J, Liu D. Science. 1999;284:760–765. doi: 10.1126/science.284.5415.760. [DOI] [PubMed] [Google Scholar]
- 55.Johnson C H, Golden S S. Annu Rev Microbiol. 1999;53:389–409. doi: 10.1146/annurev.micro.53.1.389. [DOI] [PubMed] [Google Scholar]
- 56.Cann I K O, Ishino Y. Genetics. 1999;152:1249–1267. doi: 10.1093/genetics/152.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nolling J, de Vos W M. Nucleic Acids Res. 1992;20:5047–5052. doi: 10.1093/nar/20.19.5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandler S J, Satin L H, Clark A J. Nucleic Acids Res. 1996;24:2125–2132. doi: 10.1093/nar/24.11.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komori K, Sakae S, Shinagawa H, Morikawa K, Ishino Y. Proc Natl Acad Sci USA. 1999;96:8873–8878. doi: 10.1073/pnas.96.16.8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leffers H, Gropp F, Lottspeich F, Zillig W, Garrett R A. J Mol Biol. 1989;206:1–17. doi: 10.1016/0022-2836(89)90519-6. [DOI] [PubMed] [Google Scholar]
- 61.Hausner W, Lange U, Musfeldt M. J Biol Chem. 2000;275:12393–12399. doi: 10.1074/jbc.275.17.12393. [DOI] [PubMed] [Google Scholar]
- 62.Baliga N S, Goo Y A, Ng W V, Hood L, Daniels C J, DasSarma S. Mol Microbiol. 2000;36:1184–1185. doi: 10.1046/j.1365-2958.2000.01916.x. [DOI] [PubMed] [Google Scholar]
- 63.Baliga N S, DasSarma S. J Bacteriol. 1999;181:2513–2518. doi: 10.1128/jb.181.8.2513-2518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baliga N S, DasSarma S. Mol Microbiol. 2000;36:1175–1183. doi: 10.1046/j.1365-2958.2000.01915.x. [DOI] [PubMed] [Google Scholar]
- 65.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bermudez-Cruz R M, Chamberlin M J, Montanez C. Biochimie. 1999;81:757–764. doi: 10.1016/s0300-9084(99)80134-5. [DOI] [PubMed] [Google Scholar]
- 67.Lowe T M, Eddy S R. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muramatsu T, Nishikava K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyoma S. Nature (London) 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 69.Tumbula D, Vothknecht U C, Kim H S, Ibba M, Min B, Li T, Pelaschier J, Stathopoulos C, Becker H, Soll D. Genetics. 1999;152:1269–1276. doi: 10.1093/genetics/152.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hui I, Dennis P P. J Biol Chem. 1985;260:899–906. [PubMed] [Google Scholar]
- 71.Omer A D, Lowe T M, Russell A G, Ebhardt H, Eddy S R, Dennis P P. Science. 2000;288:517–522. doi: 10.1126/science.288.5465.517. [DOI] [PubMed] [Google Scholar]
- 72.Dennis P P. Cell. 1997;89:1007–1010. doi: 10.1016/s0092-8674(00)80288-3. [DOI] [PubMed] [Google Scholar]
- 73.Engemann S, Noelle R, Herfurth E, Briesemeister U, Grelle G, Wittmann-Liebold B. Eur J Biochem. 1995;234:24–31. doi: 10.1111/j.1432-1033.1995.024_c.x. [DOI] [PubMed] [Google Scholar]
- 74.Shimmin L C, Dennis P P. EMBO J. 1989;8:1225–1235. doi: 10.1002/j.1460-2075.1989.tb03496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonnet G H, Cohen M A, Benner S A. Science. 1992;256:1443–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 76.Riley M, Labedan B. J Mol Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- 77.Graham D E, Overbeek R, Olsen G J, Woese C R. Proc Natl Acad Sci USA. 2000;97:3304–3308. doi: 10.1073/pnas.050564797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.