Abstract
We aimed to evaluate whether changes in maize (Zea mays) leaf expansion rate in response to environmental stimuli or developmental gradients are mediated by common or specific expansins, a class of proteins known to enhance cell wall extensibility. Among the 33 maize expansin or putative expansin genes analyzed, 19 were preferentially expressed at some point of the leaf elongation zone and these expansins could be organized into three clusters related to cell division, maximal leaf expansion, and cell wall differentiation. Further analysis of the spatial distribution of expression was carried out for three expansins in leaves displaying a large range of expansion rates due to water deficit, genotype, and leaf developmental stage. With most sources of variation, the three genes showed similar changes in expression and consistent association with changes in leaf expansion. Moreover, our analysis also suggested preferential association of each expansin with elongation, widening, or both of these processes. Finally, using in situ hybridization, expression of two of these genes was increased in load-bearing tissues such as the epidermis and differentiating xylem. Together, these results suggest that some expansins may be preferentially related to elongation and widening after integrating several spatial, environmental, genetic, and developmental cues.
Leaves of monocotyledons offer an interesting model of organ expansion because spatial gradients along the leaf reflect the age of tissues, allowing one to consider tissues of known ages at one single sampling date (Silk, 1992; Ben Haj Salah and Tardieu, 1995). Cells are continuously produced in the meristematic region near the leaf base and expand in distal regions of the growing zone, while they are pushed forward by younger cells. Relative elongation rate (RER) usually has a bell shape in the growing zone, with a peak at about one-third of the zone and an end at a few centimeters from the leaf base (Schnyder et al., 1990; Muller et al., 2001). Local values of RER depend both on the water flux into expanding cells and on loosening of cell walls (Lockhart, 1965; Cosgrove, 1999). Because turgor pressure changes little along the growing zone (Tomos and Pritchard, 1994; Fricke, 2002; Bouchabké et al., 2006), the large spatial variability of elongation rates in this zone is most often attributed to spatial changes in cell wall mechanical properties (Passioura and Fry, 1992; Wu et al., 1996). Water deficit affects both the total elongation rate of the leaf and the spatial distribution of the RER (Tardieu et al., 2000), probably linked to local changes in cell wall mechanical properties (Chazen and Neumann, 1994; Wu et al., 1996).
Expansins are proteins involved in cell wall loosening (Cosgrove, 1999; Li et al., 2003a). They are expressed in all expanding parts of the plants or in organs that undergo cell wall modifications: root tips (Wu et al., 2001b), internodes (Lee and Kende, 2001), shoot apical meristems (Vogler et al., 2003), flowers (Gookin et al., 2003), pollen (Cosgrove et al., 1997), root hairs (Cho and Cosgrove, 2002), and ripening fruits (Brummell et al., 1999). The presence of expansins near primary or secondary xylem vessels suggests a role in xylem differentiation (Im et al., 2000; Gray-Mitsumune et al., 2004). Over- or underexpression of expansin genes affects expansion of Arabidopsis (Arabidopsis thaliana) leaves (Cho and Cosgrove, 2000), soybean (Glycine max) roots (Lee et al., 2003), or rice (Oryza sativa) internodes (Choi et al., 2003), but these effects can be counterintuitive or unexpected (Rochange et al., 2001), possibly because expansins belong to a large gene family with partly redundant functions (Li et al., 2003b).
The expansin family counts 32 and 53 genes in Arabidopsis and rice, respectively (Sampedro and Cosgrove, 2005). They are classified in two major subfamilies, α-expansins (EXPAs) and β-expansins (EXPBs; Kende et al., 2004). EXPBs were originally discovered in pollen (Cosgrove et al., 1997), but EXPBs have since been identified in other vegetative tissues (Lee and Kende, 2001; Reidy et al., 2001) with putatively similar roles as EXPAs. Individual expansins are preferentially expressed in specific organs (Lee and Kende, 2001, 2002 [in rice]; Wu et al., 2001a [in maize]; Shin et al., 2005). Several reports suggest that each expansin may have a specific temporal pattern of expression related to environmental stimuli (Brummell et al., 1999). For example, one expansin gene of Rumex is up-regulated by ethylene and flooding (Vreeburg et al., 2005), or one EXPA gene of rice is induced by submergence and gibberellin (Lee and Kende, 2001). In contrast, other reports suggest common regulations of several expansins by environmental or hormonal stimuli. In pear fruit (Pyrus communis) during ripening, six expansin genes show overlapping expression and similar regulation by ethylene (Hisawa et al., 2003). In common elderberry (Sambucus nigra), two distinct expansins related to leaf abscission are coordinately regulated by ethylene (Belfield et al., 2005). In chick pea (Cicer arietum), four EXPAs have coordinated overexpression by indole acetic acid or brassinolides (Sánchez et al., 2004).
The aim of this work was to evaluate to what extent changes in leaf expansion rate are mediated by individual expansin genes in response to several developmental, environmental, and genetic sources of variation. For that, we analyzed the spatial pattern of both local tissue expansion rate and the abundances of expansin transcripts, when leaf expansion rate underwent changes: (1) between genotypes with contrasting patterns of distribution of growth; (2) in response to water deficit, which affects this distribution; or (3) in leaves of different ages, which also affect the distribution of growth (Muller et al., 2001). We first performed an analysis of the expression of 33 expansin genes in the growing zone of leaves. Twenty expansin genes were preferentially expressed in this zone and were classified according to their spatial pattern of expression. We then evaluated the degree of association of the expression of three of these genes with leaf elongation and widening, as altered by water deficit, developmental stage, or spatial location within the leaf of different genotypes. Finally, we evaluated the tissue localization of expression of two of these expansins using in situ hybridization.
RESULTS
A Wide Range of Spatial Distributions of Leaf Expansion Rate Was Obtained
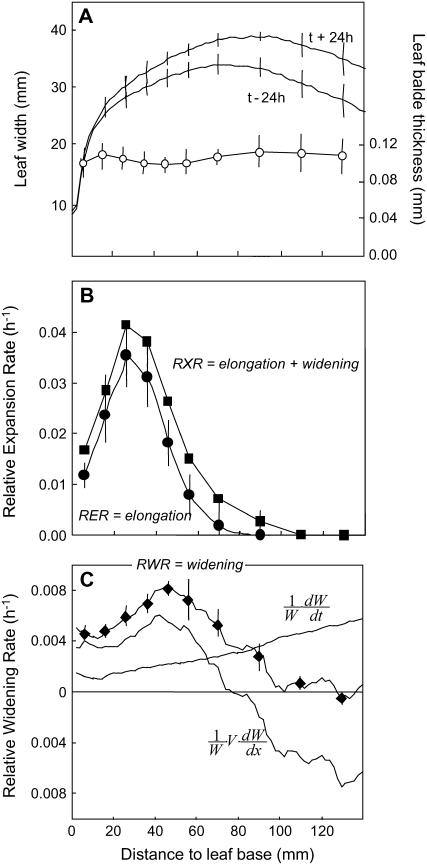
In well-watered plants of the inbred F2 line, the relative elongation rate (RER) in a longitudinal direction spanned over 70 mm, with a peak at 25 mm from the leaf base (Fig. 1B). Leaf width increased with distance from the leaf base in the first 70 mm and decreased at further distances (Fig. 1A). It also increased with time (Fig. 1A). The relative widening rate (RWR) was therefore computed as the sum of a spatial term due to local changes in width with distance from the base, and of a temporal term due to local changes in width with time (Fig. 1C). It showed a more flattened pattern than the RER and the widening zone extended beyond the elongating zone (Fig. 1B). The total relative rate of expansion (RXR) was estimated as the sum of the elongation and widening rates (Eq. 4) because of the absence of a consistent change in blade thickness with age and distance to the leaf base (Fig. 1A). Its pattern was similar to that of relative elongation rate, but over a longer distance in the leaf.
Figure 1.
Spatial distributions along the maize leaf base of leaf thickness, width, relative elongation rate (RER), and relative widening rate (RWR) and their sum (RXR). Data from rapidly elongating leaves of well-watered plants. A, Leaf blade thickness (○) and leaf width (line) along the maize leaf base at two successive times at 48-h interval. B, RER (•) and RXR (▪). C, RWR (♦). RWR is the result of the sum of a spatial term 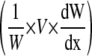 that depends on both the local elongation rate and the gradient of width with distance, and a temporal term
that depends on both the local elongation rate and the gradient of width with distance, and a temporal term 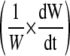 that depends on the temporal evolution of width at each position from the leaf base. Width and thickness data are means and sds of four to 12 plants. RER data are means and sds of three replicates of six plants each. RWR data are means and sds of independent calculations on three sets of plants.
that depends on the temporal evolution of width at each position from the leaf base. Width and thickness data are means and sds of four to 12 plants. RER data are means and sds of three replicates of six plants each. RWR data are means and sds of independent calculations on three sets of plants.
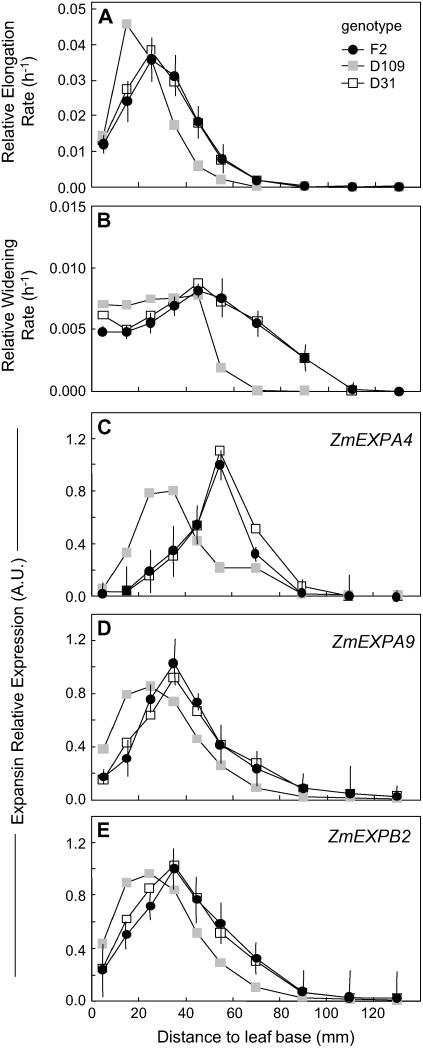
The three studied inbred lines presented appreciable differences in spatial patterns of expansion rate. The growing zone was shorter in the D109 line than in the F2 line (60 versus 70 mm), and the peak of RER shifted 10 mm basipetally in this line (Fig. 2A). In contrast, the F2 and D31 lines had similar spatial patterns. Differences between genotypes in relative widening rate were consistent with those of relative elongation rate (Fig. 2B).
Figure 2.
Comparison of spatial distributions of relative elongation rate (A) and relative widening rate (B) and of expressions of three expansin genes (C, D, and E for ZmEXPA4, ZmEXPA9, and ZmEXPB2, respectively) at the leaf base of well-watered plants from the F2 line (•) or two maize lines with either short (D9 line; gray square) or long (D31 line; □) elongation zones. Maximal expression from the F2 line was set to 1. RER data are means and sds of three replicates of six plants each. RWR data are means and sds of three independent sets of plants. Expression data are means of two technical replicates and two independent samples. Vertical bars only show sd for the F2 line for better legibility. RER and RWR data for F2 plants are the same as those shown in Figure 1, B and C.
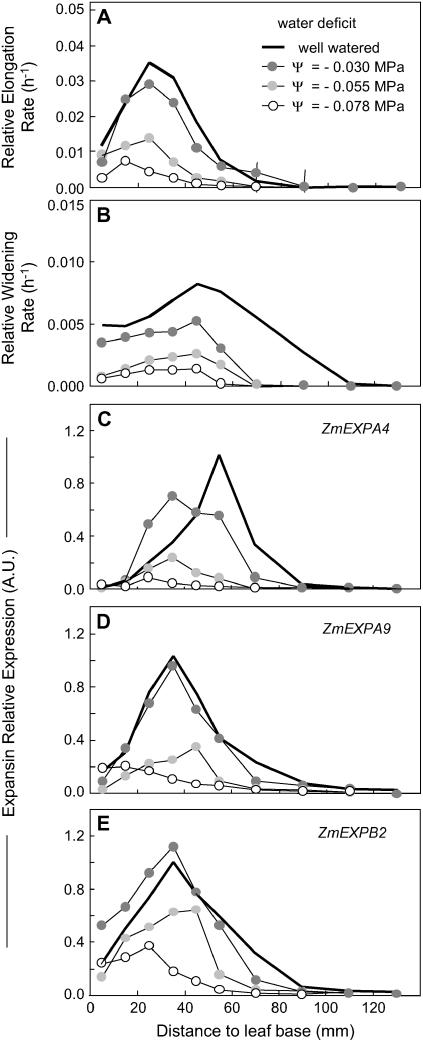
Water deficit affected expansion rate with a dose-response effect in the F2 line. Moderate water deficit (Ψw = −0.03 MPa) decreased the relative elongation rate at all positions beyond 20 mm from the leaf base, whereas the first 20 mm were almost unaffected (Fig. 3A). Values of relative elongation rate were still lower in the whole growing zone at further water deficit (Ψw = −0.055 and −0.078 MPa). The relative widening rate was reduced by the three levels of water deficit over the whole growing zone (Fig. 3B).
Figure 3.
Comparison of spatial distributions of relative elongation rate (A) and relative widening rate (B) and of expressions of three expansin genes (C, D, and E for ZmEXPA4, ZmEXPA9, and ZmEXPB2, respectively) at the leaf base of well-watered plants from the F2 line (same data as in Fig. 2; black line) or in plants exposed to increasing soil-water deficits, from moderate (Ψw = −0.3 MPa; dark gray circle), intermediate (Ψw = −0.55 MPa; light gray circle), to severe (Ψw = −0.78 MPa; ○). Maximal expression from the well-watered F2 line was set to 1. Same number of replicates as in Figure 2. Error bars are omitted for better legibility.
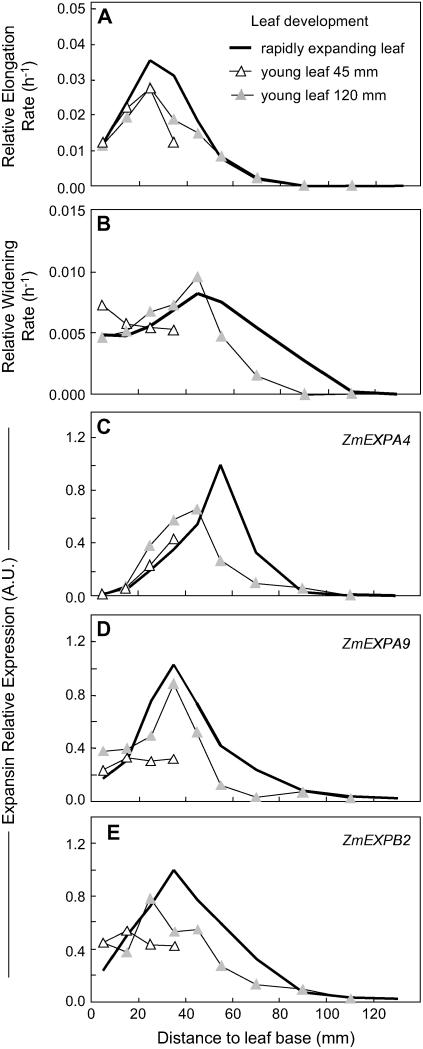
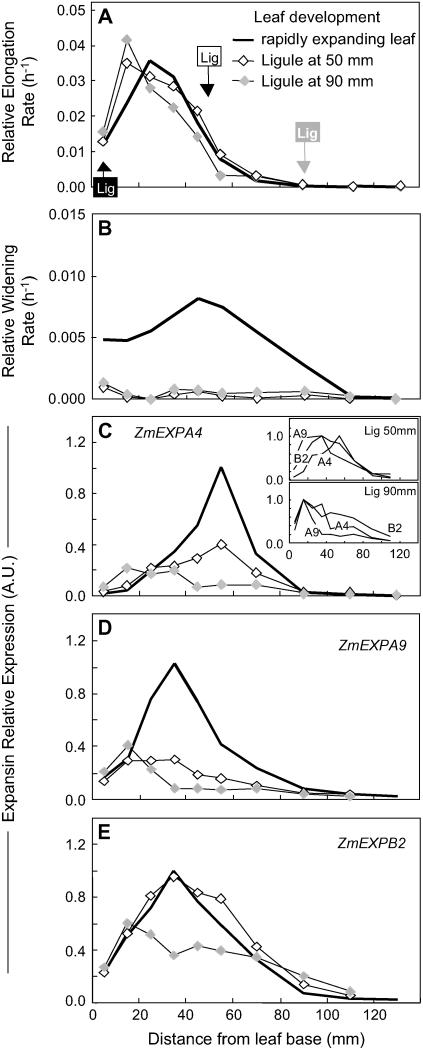
Leaf age also affected the spatial pattern of leaf expansion rate. In young leaves still hidden in the whorl (45 mm long, 17 d after leaf initiation), the relative elongation rate was similar to that of older leaves in the first 20 mm, but was truncated at further distances with a peak at 25 mm (Fig. 4A). Slightly older leaves (120 mm, 19 d after leaf initiation) also had a peak at 25 mm, but with an elongation zone of 70 mm, similar to that of emerged leaves, but with lower values (Fig. 4A). The relative widening rate was essentially similar in young leaves as compared to older, rapidly elongating leaves (Fig. 4B). The pattern of leaf elongation rate was more skewed in growing leaves, which were close to cessation of elongation, in which the ligule (limit between the lamina and the sheath) was located inside the growing zone. In these leaves, the growing zone progressively shortened, with peaks at 15 mm from the leaf base (Fig. 5A). Analysis of leaves of different ages was the only case where the patterns of RWR were independent of that of the relative elongation rate. In particular, the relative widening rate was near zero in leaves close to cessation of elongation (Fig. 5B). Overall, multiple comparisons of the effect of genotype, water deficit, and leaf age provided a series of patterns of relative elongation rate and relative widening rate to which we have compared the pattern of expression of expansin genes.
Figure 4.
Comparison of spatial distributions of relative elongation rate (A) and relative widening rate (B) and of expressions of three expansin genes (C, D, and E for ZmEXPA4, ZmEXPA9, and ZmEXPB2, respectively) at the leaf base of well-watered plants of the F2 line when the leaf is rapidly elongating (25 d after leaf initiation, mean length = 300 mm; same data as in Fig. 2; black line) or in young leaves either 45-mm-long (▵, 17 d after leaf initiation) or 120-mm-long (light gray triangle, 19 d after leaf initiation). Maximal expression among the samples from the rapidly elongating leaf was set to 1. Error bars are omitted for better legibility.
Figure 5.
Comparison of spatial distributions of relative elongation rate (A) and relative widening rate (B) and of expressions of three expansin genes (C, D, and E for ZmEXPA4, ZmEXPA9, and ZmEXPB2, respectively) at the leaf base of well-watered plants of the F2 line when the leaf was rapidly elongating (mean length = 300 mm; same data as in Fig. 2; black line) or in older leaves (length > 500 mm) 2 d before cessation of elongation. Two independent sets of samples were collected with the ligule (sheath-to-blade junction indicated by Lig on the graph) located at 50 mm (⋄) or 90 mm (light gray diamond) from the leaf base. Maximal expression from the rapidly elongating leaf was set to 1, except in the inset in C, where the expression of the three genes is shown relative to their maximum in the older leaves. Error bars are omitted for better legibility.
Identification of Expansin Genes Whose Spatial Patterns of Expression Are Similar to That of Leaf Expansion Rate
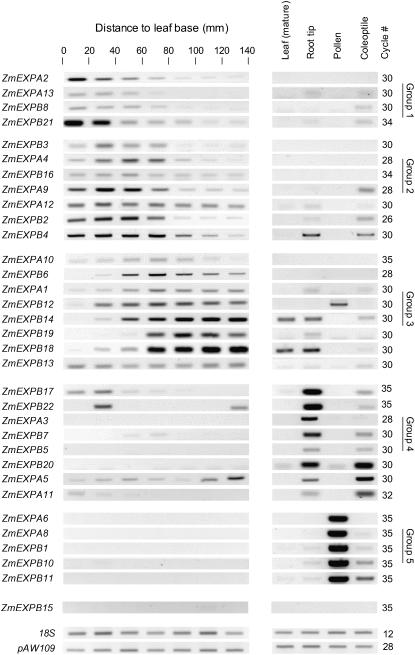
Forty-three sequences for expansins genes or putative genes were identified in the available cDNA libraries using the The Institute for Genomic Research maize contig database (http://tigrblast.tigr.org/tgi) and specific primer pairs were designed for 33 of them (Table I). To identify the expansin genes that are expressed in the leaf-growing zone, we evaluated the relative transcript abundance from gel-PCR images in successive 20-mm sections of the elongating zone. This was carried out in well-watered plants of the inbred F2 line during the period of maximal leaf elongation. We also evaluated transcript abundance in other growing tissues (root tips and coleoptiles) in nongrowing zones of the leaf and in pollen (Fig. 6). For each gene, a fixed cycle number (26–35) was used for all tissue types to allow comparison of transcript abundance. The specificity of amplification was checked by sequencing the 33 PCR products and the robustness of this analysis was verified by visually comparing images of gels with real-time PCR data (Supplemental Fig. S1). The repeatability of the patterns was verified by comparing results from three independent experiments (Supplemental Fig. S1).
Table I.
Primer sets used for gene-specific PCR amplifications of EXPA and EXPB sequences of maize
Forward primer positions are given relative to either the start or the stop codon (except when unavailable). GenBank numbers refer to either the full-length cloned sequence when available (underlined) or the TC of EST assembly.
| Gene Name | GenBank No. | Length | Primer Sequence (5′–3′) Forward / Reverse | Position from Start | From Stop |
|---|---|---|---|---|---|
| ZmEXPA1 | AF332169 | 1,090 | CGAGTGTGACTGTGAGCAAGAGA/GCCTTGCAGGTTTTTGTTTTG | +21 | |
| ZmEXPA2 | AF332170 | 1,294 | GCGTTAGCATGGTGTTTTAGTGT/GAGGCCCTAAACGCATAATTG | +181 | |
| ZmEXPA3 | AF332171 | 1,031 | TGGCTCAAGATCGGCATCTA/ACAAGCGAACGAGGCAGAAA | −360 | |
| ZmEXPA4 | AF332172 | 929 | CTACTCACTCCTGCTACCGTACTA/GAATCCCATCCACACATCTT | +95 | |
| ZmEXPA5 | AF332173 | 1,029 | GCCAGTTCTGAGGATGAACAGC/TGGTCCGATCCAGTCCGTAA | −8 | |
| ZmEXPA7 | TC310903 | 1,178 | GACGTTCCGGGTGATGACTA/GTGCATGAGCGAAAGAATGA | −107 | |
| ZmEXPA8 | TC287543 | 1,048 | TGGTCGTTCGTGGACAATAA/GCTGTCCGTGGTGTCAGTTA | +5 | |
| ZmEXPA9 | TC292888 | 1,066 | GAGGCGCCATGGTTTTATGA/CCTACCTCCGTTAATCTGCAAGC | +116 | |
| ZmEXPA10 | TC298364 | 1,226 | TTCTGTAGAGAATGGAGTGCCG/CCGGACATACCCGAATCC | −5 | |
| ZmEXPA11 | TC292885 | 1,349 | GCCGGCGAGGAAGAGCT/GTACACGCCGGGAATGCG | −86 | |
| ZmEXPA12 | TC298359 | 1,317 | GTTGGGCTGTGGTGTACATTCTC/GGAAAACCATTTATGACCAAGAGC | +60 | |
| ZmEXPA13 | TC291600 | 1,159 | CGGCGGTGGTGATGTAAGTAGA/GGAAATCCTTTCGCCATTCAAT | +28 | |
| ZmEXPB1 | AF332174 | 1,080 | CTACACTTCCAACGTCCAATTCTACT/TTCGATCATGAACCCGAACA | −25 | |
| ZmEXPB2 | AF332175 | 907 | CACCACCCACCACTACTACCA/AACGACTCAAAGGACCATGACAA | +30 | |
| ZmEXPB3 | AF332176 | 788 | GTATATGCCATGCGTCGTAC/TATCATCGATCCGATCCGTCC | +5 | |
| ZmEXPB4 | AF332177 | 1,273 | ACCATCGTAATCACCGACCA/CTTCAACCTCCTCTTTCATTCTCT | −474 | |
| ZmEXPB5 | AF332178 | 650 | GCCAACCACAGGCTACAG/CCAAATGACACCGAATCG | −129 | |
| ZmEXPB6 | AF332179 | 1,142 | CTCGCAGGACAGATCCTTGT/GGCACCGTATCTGGTAGCAT | +113 | |
| ZmEXPB7 | AF332180 | 1,173 | GGCCAACTCTGCGTCGTG/GGTCAGCCACCAGCGTC | −190 | |
| ZmEXPB8 | AF332181 | 1,479 | TACAAGCTACGCTCTCCCGC/GAAAAAATGCACTGACGAAAGG | +401 | |
| ZmEXPB10 | AY104999 | 860 | GGTTGTGGATCCTGCTTCGAGAT/GACCCTTCGAAATTGCATGT | −513 | |
| ZmEXPB11 | AY104125 | 1,191 | TCAAGTGCGATAAGCCTGTG/TGCACTTAACCCTTCGGAAC | +319 | −491 |
| ZmEXPB12 | TC287648 | 1,438 | GTGTAAGAAGAGGAGGAGGACA/GAAAAAAGAAACCGACACCGAC | +175 | |
| ZmEXPB13 | TC280521 | 1,720 | GGCTTGATCTGTTTGGTATTGTTG/TGAAACTCTGGACCGACGAC | −51 | |
| ZmEXPB14 | TC280520 | 1,191 | CGTCGTTGTGCTTGATTAGTTTG/GCCTTCCTCCATTTCGTTCC | +40 | |
| ZmEXPB15 | TC311993 | 1,172 | CGCCTGCTATCAGGTGAAAT/CCCGAGTGTACTGGATCTTGA | +248 | |
| ZmEXPB16 | TC289097 | 1,568 | CGTCGTGTTGTTGTTACTGGTGTT/CGATGCGATGCAAGAAAAATAC | −34 | |
| ZmEXPB17 | TC311991 | 650 | TAACGATGGCGGAATAACAGAA/GGTACGGTATAGGCACACGCAC | +0 | |
| ZmEXPB18 | TC310578 | 1,216 | ATGCAGAACAACTCCGGGTA/TACTGGACGAAGGAGCGGTA | −20 | |
| ZmEXPB19 | TC287642 | 1,608 | TGCATGCATCCTGGCTTTT/AAATCAACAGCACGTTCAGACA | +60 | |
| ZmEXPB20 | TC310589 | 809 | ATCCCACAATTCATCTCCTGCA/GATCTACCCGATCGAGCTGAG | +3 | |
| ZmEXPB21 | TC308480 | 888 | ATGTTTGTGGTATTGGCGTTGTG/ACATCACTCCCCCCTCTCAG | −21 | |
| ZmEXPB22 | BG873846 | 543 | ACCAGATACGGTGCCTCAAC/CACCCTCCTGAACTGGATGT | + | − |
Figure 6.
Gel-PCR analysis of the expression of 33 expansin genes along the base of the maize leaf and in different tissues. Mature leaf tissues were collected from the emerged, nongrowing part of a growing leaf. Root tissues were collected from growing root tips. All tissues were collected from well-watered plants of the F2 line. The number of PCR cycles differed among genes, but was fixed among tissues. 18S rRNA and pAW109 RNA were used as controls. Genes were clustered according to spatial pattern of expression. Group 1 refers to genes most expressed in the first 20 or 40 mm at the leaf base; group 2 refers to genes most expressed in the 20- to 60-mm region. Group 3 refers to genes most expressed beyond the elongation zone (i.e. beyond 60 mm). Group 4 refers to genes most expressed in growing coleoptiles or root tips. Group 5 refers to genes most expressed in pollen grains.
Sequences were sorted (Fig. 6) according to both their expression patterns in the leaf elongation zone and in other tissues. (1) Only one gene (Fig. 6) had no signal in any tested tissues. (2) Five genes (group 5), among which two EXPAs, had exclusive expression in the pollen. (3) Eight genes (group 4) had very low expression in growing leaf tissues and were more expressed in growing coleoptiles or root tips. (4) The remaining 19 genes were expressed at some point of the leaf-growing zone. A second classification was carried out according to the spatial pattern of gene expression. Maximal expression was observed either in the first 20-mm section of the leaf elongation zone (group 1, four sequences in Fig. 6), in the 20- to 60-mm section where expansion rate is maximal (group 2, seven sequences), or in the 60- to 140-mm section where expansion is low or nil (group 3, eight sequences). Overall, only the 11 genes that belong to groups 1 and 2 are likely to be closely involved in leaf expansion in the F2 line studied.
Do the Patterns of Expression of Three Expansin Genes Coincide with Those of the Leaf Expansion Rate over the Collection of Patterns?
Among the genes expressed in the elongating zone, we chose three with high expression (ZmEXPA4, ZmEXPA9, and ZmEXPB2); i.e. those for which the minimal cycle number was necessary to get clear spatial patterns in Figure 6. Real-time PCR on fully elongating leaves from well-watered plants of the F2 line confirmed that these three genes presented a bell-shaped pattern of expression (Fig. 2, C–E) with a maximum located 50 to 60 mm, 30 to 40 mm, and 30 to 40 mm from the base in ZmEXPA4, ZmEXPA9, and ZmEXPB2, respectively, and no expression beyond 100 mm. These spatial patterns were similar to those of expansion rate. In particular, peaks of expression coincided with peak of elongation for ZmEXPA9 and ZmEXPB2 and with peak of widening for ZmEXPA4.
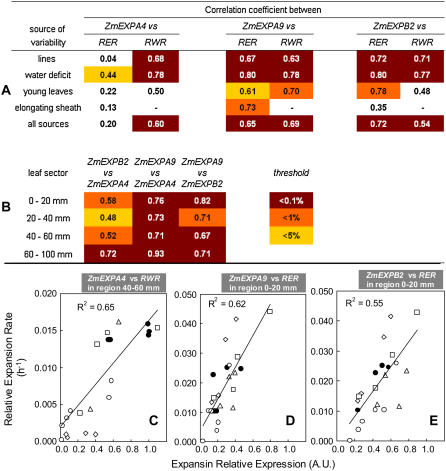
Changes in the patterns of expressions of the same three genes in the D31 and D109 lines closely mirrored those of leaf expansion rate (Fig. 2). In particular, patterns of expression were similar in the F2 and D31 lines and were shifted toward the leaf base in the D109 line, consistent with the changes of the patterns of leaf expansion rate. When data from the three lines were combined, strong correlation was observed for both ZmEXPA9 and ZmEXPB2 between, on one hand, the expression of the gene at all locations of the growing zone and, on the other hand, both relative elongation rate and relative widening rate in the same zones (Fig. 7A). In the same way, a strong correlation was observed for ZmEXPA4 and relative widening rate only.
Figure 7.
Analysis of correlation between expansin gene expression and relative elongation rate (RER) or relative widening rate (RWR) in maize leaves. A, Correlation coefficient corresponding to several data subsets for each of the three genes. B, Correlation coefficient linking the expression of ZmEXPA4, ZmEXPA9, and ZmEXPB2 along the maize leaf base. In both cases, the background indicates the probability level being either below 0.1%, 1%, or 5% thresholds using Fisher statistics. C, D, and E, Three clouds of points corresponding to correlations between expression and expansion for C, ZmEXPA4 expression, and RWR in the zone 40 to 60 mm from the leaf base. D and E, ZmEXPA9 and ZmEXPB2 expression and relative elongation rate in the zone 0 to 20 mm from the leaf base. •, Well-watered rapidly elongating F2 line; ○, same exposed to water deficit; □, D109 and D31 lines; ▵, young leaves before emergence; ⋄, leaves harvested at the time of sheath elongation. [See online article for color version of this figure.]
Soil-water deficit affected the expression of the same three genes (Fig. 3, C–E). At moderate water deficit, the expression patterns of ZmEXPA9 and ZmEXPB2 were similar to those of well-watered plants, whereas expression of ZmEXPA4 was shifted toward the leaf base. Increased water deficits resulted in reductions in expression of the three genes, with patterns consistent with those of local expansion rates. As a consequence, strong correlations were observed between local gene expression and expansion rate (both elongation and widening for ZmEXPA9 and ZmEXPB2; widening only for ZmEXPA4) over all positions and water deficit treatments (Fig. 7A).
In the same way, young leaves showed truncated distribution of the expressions of the three genes, consistent with patterns of relative elongation rate (Fig. 4, C–E). In these young leaves, correlation between expansion and expression was higher for ZmEXPA9 and ZmEXPB2 and looser for ZmEXPA4 (Fig. 7A). Leaf aging (Fig. 5, C–E) caused a marked decrease in the expressions of ZmEXPA4 and ZmEXPA9, whereas that of ZmEXPB2 was almost unaffected in leaves at the first stage of aging (ligule at 50 mm from the leaf base), and less affected than the other two genes at a further stage (ligule at 90 mm from the leaf base). This raises the possibility that the first two genes could be linked to widening rate, which was near zero at this stage, and that the third gene would be linked to the elongation rate, which was similar to that in fully expanding leaves. Leaf aging also caused a progressive basal shift in the peak of maximal expression consistent with basal shifts of patterns of relative elongation rate (Fig. 5C, inset).
When all sources of variability were combined, correlations between expansin expression and leaf expansion rate remained high, particularly between ZmEXPB2 and relative elongation rate, between ZmEXPA9 and both relative elongation rate and relative widening rate, as well as between ZmEXPA4 and relative widening rate (Fig. 7A). Correlations were not only due to coincidence of peaks of expression and expansion. When only subsets of data corresponding to a narrow 20-mm spatial window, but gathering of all other sources of variation was considered, correlations were most often observed with degree of association between expansin expression and growth depending on the spatial position of the leaf segment considered (data not shown). Examples of such correlations are given in Figure 7, C to E. Finally, correlation between expression of the three genes was also analyzed in different segments of the leaf base (Fig. 7B). Expression of ZmEXPA9 was well correlated with both ZmEXPA4 and ZmEXPB2 throughout the leaf base, whereas ZmEXPA4 and ZmEXPB2 showed much looser association, especially in the zone 0 to 60 mm from the leaf base.
ZmEXPA4 and ZmEXPB2 Are More Expressed in Epidermal and Periveinal Regions
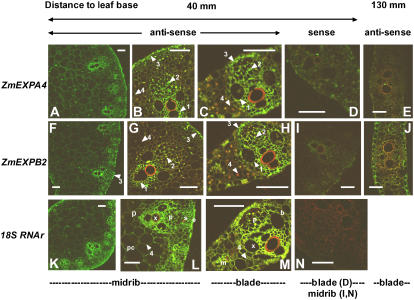
The locations of ZmEXPA4 and ZmEXPB2 expression were evaluated in leaf cross sections of regions with high (40 mm from the leaf base) or low (130 mm from the leaf base) expression. At 40 mm, broad views of the midrib (Fig. 8, A and F) using antisense probes showed a gradient of labeling for both genes with small cells at the periphery of the midrib, around xylem and phloem vessels, as well as in the epidermis strongly labeled in contrast with more loosely labeled highly vacuolated cells of the parenchyma. However, this pattern essentially mirrored that found for the control probe (18S rRNA; Fig. 8K), suggesting no tissue-specific expression of these two expansin genes. At higher magnification in the same region (Fig. 8, B, C, G, H, L, and M), subtle, but robust, differences between the expansin genes and the 18S control could be observed. Contrary to the 18S control probe (Fig. 8, L and M), little labeling was found with both expansin gene antisense probes in large parenchymal cells of the midrib and in most of the mesophyl cells of the leaf blades as well as in differentiating sclerenchyma (arrow 4 in Fig. 8, B, C, G, and H). Conversely, preferential association of ZmEXPA4 and ZmEXPB2 was detected in both leaf blade and midrib cross sections with (1) small, parenchymal cells boarding xylem vessels as well as the differentiating xylem cells themselves; (2) sieve elements and companion cells; and (3) the epidermis located above each vascular bundle (Fig. 8, arrows 1, 2, and 3). These patterns were remarkably conserved for both genes and no labeling was detected with the corresponding sense probes (Fig. 9, D, I, and N). Cross sections taken at 130 mm showed much fainter signals consistent with the strongly reduced expression found with reverse transcription (RT)-PCR in this region. In this region, no expression was found around the vascular bundles or in the sclerenchyma. However, little expression of ZmEXPB2 was consistently found in the epidermis in this distal region. Together, these results suggest that ZmEXPA4 and ZmEXPB2 have an undistinguishable pattern of expression at the tissue level. Their transcripts are essentially present in all cells of the blade or the midrib, but with an apparent preferential association with xylem and phloem cells, as well as with some epidermal cells.
Figure 8.
Localization of ZmEXPA4 and ZmEXPB2 expression by in situ hybridization at the base of a rapidly elongating maize leaf from well-watered plants. Localization of ZmEXPA4 RNA in the midrib and in the leaf blade (A–C and E) and localization of ZmEXPB2 RNA in the midrib and in the leaf blade (F–H and J) at two distances from the leaf base (40 and 130 mm). Controls, ZmEXPA4 sense probe (D) and ZmEXPB2 sense probe (I), 18S ribosomic RNA antisense probe (in the midrib, K and L; in leaf blade, M), 18S ribosomic RNA sense probe (in the midrib, N). x, Xylem vessels; p, phloem; s, sclerenchyma; e, epidermis; pc, parenchyma; b, vascular bundle sheath; m, mesophyl. Arrows 1 to 3 show strong labeling with ZmEXPA4 and ZmEXPB2 antisense probes in parenchymal cells around xylem vessels (1), in phloem sieve element, in companion cells (2), and in the epidermis located above vascular bundles (3). Arrow 4 points out the lack of labeling of mesophyl cells and large parenchymal cells with the ZmEXPA4 and ZmEXPB2 antisense probes by contrast with the 18S RNA antisense probe (M). All characteristics (laser power, gain) were set at identical values for images A and F, B to E, G to J, L, and N. Bars = 50 μm.
DISCUSSION
Clusters of Expansins Related to Cell Division, Rapid Tissue Expansion, and Cell Wall Differentiation at the Base of Maize Leaves
The results presented here suggest that 19 expansins were expressed at some point of the leaf (over 33 tested). This contrasts with a lower proportion (three expressed over 13 tested) observed by Wu et al. (2001a) in the whole leaf when no attempt was made to separate growing and nongrowing portions of the leaf. In particular, three of the genes reported as having no expression in the study of Wu et al. (2001a) were found expressed in a narrow leaf window in our study. These localized expressions may have been lost by dilution when gene expression in the whole leaf was analyzed.
A group of four expansins exhibited peak expression in the first 20-mm segment at the leaf base, where cell division is fastest at least in the epidermis (Ben Haj Salah and Tardieu, 1995; Tardieu et al., 2000). Cell wall synthesis is taking place during cell division to create new transverse cell wall separating daughter cells. Soybean EXPB was consistently found to be controlled by cytokines (Downes and Crowell, 1998), hormones that trigger cell division. This region is also the place for accelerating relative elongation rate (tissue must expand to maintain cell size between successive mitoses) and high widening rate. Expansins expressed in this region could also be involved in both processes.
Expression of several expansin genes in the region with maximal expansion rate is consistent with a role of expansins as wall-loosening agents (Lee and Kende, 2001; Wu et al., 2001b). Our analysis suggests that at least seven distinct expansins were involved in the region with maximal expansion rate. In the same way, five EXPA and five EXPB genes are expressed in the elongation zone of the rice internode (Lee and Kende, 2001, 2002). Two of the maize expansin genes identified in our study were more associated with the epidermis and with differentiating xylem vessels than with other leaf tissues in line with earlier analyses (Cho and Kende, 1998; Im et al., 2000; Reidy et al., 2001; Gray-Mitsumune et al., 2004). This localization fits well with the idea that these two tissues exert a load-bearing role and that they are the place where the cell wall must be strongly loosened during the expansion process (Kutschera, 1992; Cho and Cosgrove, 2000), although the inner tissue of organs may also actively participate in the control of growth rate (Peters and Tomos, 2000).
The presence of several expansins in the region where elongation has ceased is consistent with previous reports in fescue (Reidy et al., 2001) or rice (Lee and Kende, 2002). In this region, secondary cell wall deposition occurs (MacAdam and Nelson, 2002) and metaxylem elements mature (Paolillo, 1995; Evert et al., 1996). Our in situ localization suggests a role for expansins in xylem differentiation because some expansins are expressed around differentiating xylem vessels, but not around mature xylem elements. The presence of expansin expression in cells boarding xylem vessels was also reported in tall fescue (Reidy et al., 2001), zinnia (Im et al., 2000), and poplar (Populus spp.; Gray-Mitsumune et al., 2004).
Robust Associations between Expansin Expression and Leaf Elongation or Widening
Twelve expansins were preferentially expressed in the leaf elongating zone and expression of three of them strikingly mirrored the changes in local expansion rate in contrasting genotypes, in response to water deficit or developmental changes. In particular, basal shifts in expression were remarkably associated with those of elongation in all conditions or genotypes. These expansins were therefore probably involved in some processes associated with growth, most probably with cell wall loosening (Li et al., 2003b). However, our analysis identified conditions or regions with either lower or higher correlations between expansin expression and leaf expansion. In a study on submergence-induced EXPA expression in Rumex, Vreeburg et al. (2005), using high temporal resolution, also identified periods of high and low correlations of expansin activity or expression with petiole elongation. In the same way, Wu et al. (1996) identified regions of the root-growing zones with different susceptibility of tissues to expansins. These results and ours suggest that (1) other expansins than those analyzed in detail are involved in the control of expansion; and (2) other proteins are also likely to be involved. Peroxidases are known to stiffen the cell wall and could be involved in growth cessation (Bacon et al., 1997). Reactive oxygen species and reactive oxygen species scavenging enzymes are also proposed candidates for controlling the organ extension rate (Schopfer, 2001).
Recent literature has provided genetic evidence that the control of leaf elongation and leaf widening follow at least partially different routes. In maize leaves, quantitative trait loci controlling leaf length and leaf width are clearly distinct (Reymond et al., 2004). In Arabidopsis, distinct mutants involved in leaf or cell elongation and widening have been identified (Tsuge et al., 1996; Tsukaya, 2006). It is therefore reasonable to assume that some expansins may be associated more directly with either elongation or widening. Whereas our analysis with different genotypes or with water deficit could not distinguish these two processes, developmental analysis allowed identification of one gene (ZmEXPA4) preferentially associated with widening and showing coincidence of peaks of expression with peaks of widening rate in most cases. By contrast, ZmEXPB2 was more associated with elongation than with widening, whereas ZmEXPA9 was equally associated with both. In situ analysis showed that both genes more specifically associated with either elongation or widening had undistinguishable patterns of expression within leaf blade or midrib cross sections. Together, these results raise the hypothesis that both elongation and widening could partly occur through the action of distinct expansin genes on similar load-bearing tissues within the leaf, their concerted action thus contributing to some degree of coupling between both processes.
Are the Reductions in Leaf Expansion Rate in Response to Environmental Stimuli or Developmental Programs Mediated by Different Expansins?
A striking result was the similarity of the responses of the three studied expansins to all tested environmental stimuli, developmental changes, and genotypic differences. When all sources of variation were gathered, expression of the three genes was strongly correlated one to another throughout the leaf base. When working with two expansins involved in root hair initiation, Cho and Cosgrove (2002) showed that both hormonal (indole acetic acid, ethylene) and developmental (using a root hair-defective mutant) stimuli needed the same promoter region of both genes to trigger transcription. Our own results go in the same direction by suggesting that, when expressed in the same spatial window, at least some distinct expansins are downstream, unspecific targets of a range of converging developmental, genetic, and environmental cues.
Beyond this result, the question of the possible role of different expansin genes being similarly affected by a range of different stimuli is left open. In such a large multigene family, it has been suggested that functional redundancy could be the rule (Li et al., 2003b). This hypothesis is favored by results showing that most of the targeted knockout mutants in the moss Physcomitrella patens show no visible phenotype (Schipper et al., 2002). This question clearly deserves future study.
MATERIALS AND METHODS
Plant Cultivation, Treatment Imposition, and Tissue Sampling
Three maize (Zea mays) inbred lines (F2, D31, and D109, the latter two being recombinant lines from a cross between F2 and Io) were used in this study (Causse et al., 1996). Plants were grown in a greenhouse in autumn and spring with average temperatures of approximately 22°C and 18°C during days and nights and additional lighting provided by rows of sodium lamps to an average photosynthetic photon flux density of 15 mol m−2 d−1. Plants were transferred for the last 72 h preceding harvest to a growth chamber with similar conditions to the greenhouse. Plants were grown in 7-L PVC columns filled with a sieved 40/60 mixture (v/v) of loamy soil and organic compost. The water content of the substrate was measured at the time of sowing by weighing several samples before and after 72-h drying at 105°C. Six seeds per column were sown in pairs and thinned to three by the emergence of leaf 3. Substrate was maintained at 0.40 g water/g dry soil by daily weighing the columns and watering with modified one-tenth-strength Hoagland solution. Predawn leaf water potential (pressure chamber; Soil Moisture Equipment) was close to zero in this condition (Ψw = −0.08 ± 0.02 MPa). This target humidity was preferred to nonlimited watering above the retention capacity of the substrate to avoid anoxia.
Upon emergence, the elongation rate of leaf 6 was continuously measured on each plant using rotative displacement transducers (full 360° smart position sensors; Vishay Electronic GmbH) connected to a datalogger (LTD-CR10 wiring panel; Campbell Scientific).
When leaf 5 was visible, irrigation was stopped for some columns to establish a range of drought treatments. The relative water content of the substrate in the selected columns reached 0.18 to 0.25 g water/g dry soil within 5 to 7 d, the time period when leaf 6 started to emerge. In these columns, substrate relative humidity was maintained by watering every other day at either 0.18, 0.20, or 0.25 g/g (severe, intermediate, and moderate water deficit, respectively), whereas control plants were kept in columns at a relative water content of 0.40 g water/g dry soil. Predawn leaf water potential was measured regularly during these periods. It reached on average −0.30 MPa (range −0.28 to −0.32 MPa), −0.55 MPa (range −0.49 to −0.62 MPa), and −0.78 MPa (range −0.62 to −0.89 MPa) in the moderate, intermediate, and severe water deficit treatment, respectively.
A set of 30 homogeneous plants was selected within each treatment and transferred to the growth chamber. Only those plants displaying similar and constant leaf elongation rates during the last two nights before sampling were used. Leaf 6 was collected after a period of 3 d in the growth chamber around the time of leaf 7 appearance, except for some plants, which were harvested either shortly after the appearance of leaf 5 (leaf 6 length was 30–140 mm long) or shortly after leaf 8 appearance. In the former case, samples were gathered according to leaf length (average 45 or 120 mm long). In the latter case, the elongating part of leaf 6 was almost entirely in the sheath and the ligule was located 30 to 110 mm from the leaf base. Samples were gathered according to the position of the ligule, being on average at 50 or 90 mm from the base. In all cases, six plants were chosen for determination of the spatial distribution of the relative elongation rate. Eight plants were selected for tissue harvesting and RNA extraction. Twelve plants were used for width and thickness determination. The later harvest was performed at three dates (four plants each) with a 24-h time lapse before, at, and after the principal harvest. All harvests were performed during the 2 h preceding dawn. Leaf 6 was carefully removed from the whorl. For RNA extraction, 10 consecutive sections, either 10 mm (0–60 mm from the leaf base) or 20 mm (60–140 mm from the leaf base) long, were cut, inserted into 10 cryotubes, and immediately frozen in liquid nitrogen. This operation was repeated two times (two biological repeats) with four leaves each whose sections were pooled. For width and thickness determination, leaf 6 width was measured every 5 mm until 140 mm from the leaf insertion point. Leaves were then cut in 1-cm-long sections and stored in pFA (3.5% p-formaldehyde, 50 mm Suc, 0.01% Silwet L-77, and 90 mm sodium phosphate, pH 6.8) and later used for thickness measurements on thick tissue slices (>100 μm) embedded in 4.5% agar and cut using a vibration microtome (HM 650 V; MICROM). Sections were observed using binoculars and numerical images were taken. Thickness was estimated by dividing section area by section width (Optimas 6.5; Media Cybernetics). This was performed on both the whole leaf (blade + midrib) or on the blade only. Because the proportion of the midrib within the leaf changed with distance, only blade data were later used and presented.
Spatial Distribution of Elongation
Spatial distribution of leaf elongation was measured using the pinning method (Muller et al., 2001) during the night. The leaf elongation zone was marked at dusk with 25 fine needles (0.2-mm diameter, 4-mm distance between consecutive needles). The position of marks (holes) was measured after a period during which pinned leaves elongated approximately 6 mm, but no longer than 12 h. Displacement velocity of each mark from the base of leaf 6 was obtained by subtracting initial (visible on nongrowing leaf 3 sheath) from final (visible on leaf 6) distances between each mark and the leaf 6 base. Distances were measured on scanned images of the leaf. Relative elongation rate of a point located midway from the ith to the i + 1th mark was calculated as the difference between displacement velocity of two neighboring marks corrected for the effect of piercing on elongation rate, which was known by leaving displacement transducers on both pinned and nonpinned plants. Calculation was performed on six replicate plants, interpolated at 5-mm intervals, and averaged.
Leaf Widening
A volume V is related to length (X), width (W), and thickness (T) by
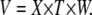 |
(1) |
The rate of volume change along the longitudinal axis (x) of the leaf is therefore
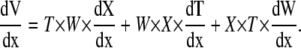 |
(2) |
Dividing by V, Equation 2 becomes
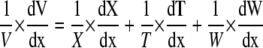 |
(3) |
or
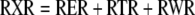 |
(4) |
where RXR, RER, RTR, and RWR are relative volumetric rate of expansion, relative elongation rate, relative thickening rate, and relative widening rate. Relative elongation rate was estimated as explained above. RWR and RTR were estimated using the continuity equation applied to width and thickness, respectively (Liang et al., 1997; Maurice et al., 1997), which can be simplified, in the case of width to:
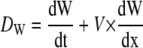 |
(5) |
where Dw is the width deposition rate, dW/dt is the temporal rate of width change (obtained by measuring width changes at each position of the leaf in the three consecutive harvests with a 24-h time lapse), V is the velocity of material point displacement (obtained by integration of RER profiles), and dW/dx is the longitudinal rate of width change. The latter was calculated as the local slope in point i of the relationship between width and distance to the leaf base. The RWR (width increase per unit width) could therefore be estimated as
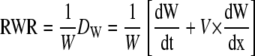 |
(6) |
For these calculations, each variable was interpolated at 2-mm intervals in the first 60 mm and at 5-mm intervals distally. It was then smoothed using a floating average procedure over five consecutive points. The same reasoning could apply to the determination of the RTR, but, because results revealed a lack of spatial gradient of thickness (Fig. 1A), RTR was neglected in the analysis.
Identification of Expansin Sequences in Maize
Identification of putative expansin genes was performed using BLAST algorithms (BLASTN and tBLASTX) on The Institute of Genomic Research maize contig database (http://tigrblast.tigr.org/tgi) with the whole set of 16 full-length cDNA sequences available for EXPA (5) and EXPB (11; Wu et al., 2001a; Li et al., 2003a). For that, singletons were excluded as well as tentative consensus (TC) sequences shorter than 500 bp or 150 amino acids. This resulted in the identification of 43 contigs corresponding to expansin genes. Most sequences (39) could be unambiguously aligned with genomic sequences (http://tigrblast.tigr.org/tgi_maize/index.cgi). Deduced amino acid sequences were then aligned (Supplemental Figs. S2 and S3) using ClustalW (http://www.ebi.ac.uk/clustalw) and Multalign (http://prodes.toulouse.inra.fr/multalin/multalin.html). Sequences displayed the typical expansin characteristics (Sampedro and Cosgrove, 2005), such as the series of Cys residues in the N-terminal region, the His-Phe-Asp motif in the central part of protein (although some EXPAs did not show the Phe-Asp motif as already pointed out by Li et al., 2003b), and the series of four Trp residues near the C terminus. Blocks of conserved sequences (Wu et al., 2001a; Sampedro and Cosgrove, 2005) allowed us to associate, in a nonambiguous way, the motifs (G)YG, (V)T(A)T(N), (CP)P(N), (A)GI(V)P, GG-R(F), and R(N)WG(Q) to the EXPA subfamily and the motifs G(K)GCG(S)CY, V(II)TD(M), RV(P)C, A(V)L, (S)WG, and (V)IP(A) to the EXPB subfamily. The 43 sequences could be organized in 15 EXPAs and 28 EXPBs. This list includes the 16 previously cloned sequences (underlined in Supplemental Figs. S2 and S3; Table I). Duplicate or near-duplicate genes are potentially present within this list, such as ZmEXPB9 to ZmEXPB11 (Li et al., 2003b).
Specific PCR primers were designed for 33 genes using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi; Table I). Primer specificity was checked by BLASTN against maize TCs. Only primer sequences showing low identity (<60%) to other TCs were retained. When no primer could be found in this way, specific sequences were selected by hand on the basis of nucleotide alignments. In all cases, primers located in the 3′-untranslated region of putative sequences were given priority (Table I). The specificity of the amplification was checked by sequencing the 33 PCR products. New genes for which specific primer pairs could be designed and studied in this article were given a name (ZmEXPA6 to ZmEXPA13 and ZmEXPB12 to ZmEXPB22) and deposited at the expansin Web site (www.bio.psu.edu/expansins). Most consensus sequences were composed of expressed sequence tags (ESTs) originating from several source libraries, except a set of EXPA and EXPB sequences that exclusively originated from pollen/anther tissues.
RNA Isolation, Purification, RT, and Semiquantitative PCR
Tissue samples were ground in liquid nitrogen. Aliquots of approximately 100 mg were used for RNA extraction using TRIzol reagent (Invitrogen) according to the manufacturer's instructions followed by a DNAse treatment (RQ1 RNase free DNase I; Promega). Quality and quantity of RNA was checked by electrophoresis and OD260. A total of 2 μg RNA was used for RT (SuperScript II; Invitrogen) using a random hexamer primer. A total of 2.5 × 105 copies of GeneAmplimer pAW109 RNA (Applied Biosystems) were added to the RT reaction as an internal standard. RT products were diluted to 400 μL and stored.
PCR was performed using Taq (Hot Start master mix; Qiagen) according to the manufacturer's instructions. To get semiquantitative results, the number of cycles of the PCR reactions were adjusted for each gene to obtain barely visible bands in agarose gels. Aliquots of the PCR reactions were loaded on agarose gels and stained with ethidium bromide. All PCR products obtained with a given primer pair were visualized on the same gel to avoid differences due to staining or image acquisition. The constitutively expressed 18S rRNA gene (forward, 5′-CCATCCCTCCGTAGTTAGCTTCT-3′ and reverse, 5′-CCTGTCGGCCAAGGCTATATAC-3′) was used as an internal control of RNA quantity, whereas GeneAmplimer (Applied Biosystems) pAW109 RNA (forward, 5′-CATGTCAAATTTCACTGCTTCATC-3′ and reverse, 5′-TGACCACCCAGCCATCCTT-3′) was used as a positive control of the RT-PCR efficiency. 18S rRNA was chosen as a standard after preliminary experiments showing hardly visible variation in expression along the base of maize leaves, which was not the case with other possible standards, such as actin or glyceraldehyde-3-phosphate dehydrogenase (data not shown). Each PCR reaction was repeated at least once. The adequacy of the primer product size with the predicted product length was verified by using Smart ladder DNA markers. When more than one band was observed on the gel, the corresponding primer pair was discarded and a new one was designed.
Real-Time PCR
Real-time PCR was carried out using the 7900HT fast real-time PCR system (Applied Biosystems) and the SYBR green PCR master mix (Applied Biosystems) according to the manufacturer's instructions. Each 384 plate was split into four subsamples containing 2 μL of either the RT product (two repeats), or the same diluted at 1/10 or 1/100 to estimate PCR efficiencies. A similar setup was designed for 18S and pAW109 primers. Relative expression rates of target gene transcripts were then calculated according to Pfaffl (2001) using the following formula
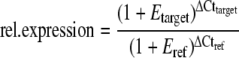 |
(7) |
where Etarget is the PCR efficiency for the target gene transcript (expansin gene), Eref is the PCR efficiency for the endogenous reference gene transcript (18S rRNA was used throughout because it always remained remarkably stable). ΔThreshold cycle (ΔCt) is the Ct difference between Ct of the sample and a reference Ct. In all cases, this reference Ct was chosen as the minimal one in the set of samples collected from well-watered F2 plants. The corresponding relative expression was therefore set to 1 in that sample with minimal Ct and maximal transcript abundance. All Ct values are means of two replicates. Most samples were present as duplicates so each relative abundance value is the mean of four Cts. Samples from control F2 plants were collected at three occasions to verify the robustness of the patterns (Supplemetal Fig. S1F).
In Situ Hybridization
Maize leaf blade or midrib samples were taken at several positions along the leaf base and fixed in paraformaldehyde 4%, potassium phosphate buffer (10 mm, pH 7.2) overnight at 4°C, dehydrated in ethanol series and butanol, and embedded in Paraplast X-TRA (Labonord). Cross sections (8 μm) were placed on silanized slides (DakoCytomation). Nucleic probes for ZmEXPA4 and ZmEXPB2 were specifically designed to reach a length >300 bp. A trial with ZmEXPA9 remained unsuccessful. Probes for ribosomal 18S were used as controls. Sense and antisense probes were labeled with UTP-digoxigenin during transcription. Labeling efficiency was estimated by dot blotting on membrane with a phosphatase alkaline conjugate antibody raised against digoxigenin (Roche Diagnostics). Slides were dewaxed, rehydrated, and then incubated for 40 min in proteinase K (2 μg mL−1) at 37°C. A probe concentration of 2 μg/mL was used in an overnight hybridization of tissue sections at 45°C in 10% dextran sulfate, 1× Denhart solution, 50% formamide, 1 μg μL−1 tRNA, and 2× SSC buffer. Posthybridization washing at graded stringency (2× SSC at 50°C and 1× SSC at 55°C) was followed by detection. Slides were first pretreated with a blocking solution (Roche Diagnostics) to prevent nonspecific binding of antibodies, then incubated with antidigoxigenin mouse antibody (Roche Diagnostics), rabbit anti-mouse FluoProbes 488 antibody IgG, and goat anti-rabbit FluoProbes 488 antibody IgG (Interchim) at room temperature. Sections were mounted in Mowiol. Fluorescence was detected using a confocal laser-scanning microscope (argon 488-nm laser; Zeiss 510 Meta). Specificity of labeling was checked by spectral analysis.
Robustness of the Data and Statistics
Significance of correlations was evaluated using Fisher statistics and probability threshold at 0.1%, 1%, and 5%. Robustness of expression data was evaluated by comparing Ct values of both technical replicates and mean Ct values of biological replicates. Ct value differences between technical replicates were < 0.5 in 80%, 79%, and 87% of the cases for ZmEXPA4, ZmEXPA9, and ZmEXPB2, respectively. When the Ct value was > 0.5, the value was not considered. Mean relative expression value differences between biological replicates showed a variation coefficient of 25% on average (range 1%–40%). Mean RER values between independent sets of plants showed a mean variation coefficient of 20% (range 10%–30%).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Comparison of gel-PCR with real-time PCR for a selection of six EXPAs or EXPBs in successive 2-cm-long segments at the base of maize leaves.
Supplemental Figure S2. List of maize EXPA genes and putative genes studied with corresponding names (GenBank no. of assembled ESTs and genomic DNA [top]). Alignment of translated sequences (bottom).
Supplemental Figure S3. List of maize EXPB genes and putative genes studied with corresponding names (GenBank no. of assembled ESTs and genomic DNA [top]). Alignment of translated sequences (bottom).
Supplementary Material
Acknowledgments
The authors thank Philippe Lessard for early help in primer design, Philippe Barrieu for technical input at the onset of this project, Jean-Luc Verdeil for help with the interpretation of in situ hybridization data, Bruno Moulia and François Gastal for fruitful discussions on the use of the continuity equation, and Xavier Sarda for access to and expertise on high-throughput real-time PCR technology.
This work was supported by the Genoplante-Maize program (grants to C.B. and A.M.) and the Swiss Science Foundation (grant to B.R.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Bertrand Muller (bertrand.muller@montpellier.inra.fr).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Bacon MA, Thompson DS, Davies WJ (1997) Can cell wall peroxidase activity explain the leaf growth response of Lolium temulentum L. during drought? J Exp Bot 48 2075–2085 [Google Scholar]
- Belfield EJ, Ruperti B, Roberts JA, McQueen-Mason S (2005) Changes in expansin activity and gene expression during ethylene-promoted leaflet abscission in Sambucus nigra. J Exp Bot 56 817–823 [DOI] [PubMed] [Google Scholar]
- Ben-Haj-Salah H, Tardieu F (1995) Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length. Plant Physiol 109 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchabké O, Tardieu F, Simonneau T (2006) Leaf growth and turgor in growing cells of maize (Zea mays L.) respond to evaporative demand under moderate irrigation but not in water-saturated soil. Plant Cell Environ 29 1138–1148 [DOI] [PubMed] [Google Scholar]
- Brummell DA, Harpster MH, Civello PM, Palys JM, Bennett AB, Dunsmuir P (1999) Modification of expansin protein abundance in tomato fruit alters softening and cell wall polymer metabolism during ripening. Plant Cell 11 2203–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse M, Santoni S, Damerval C, Maurice A, Charcosset A, Deatrick J, de Vienne D (1996) A composite map of expressed sequences in maize. Genome 39 418–432 [DOI] [PubMed] [Google Scholar]
- Chazen O, Neumann PM (1994) Hydraulic signals from the roots and rapid cell wall hardening in growing maize (Zea mays L.) leaves are primary responses to polyethylene glycol-induced water deficits. Plant Physiol 104 1385–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ (2000) Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc Natl Acad Sci USA 97 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Cosgrove DJ (2002) Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14 3237–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HT, Kende H (1998) Tissue localization of expansins in deepwater rice. Plant J 15 805–812 [DOI] [PubMed] [Google Scholar]
- Choi D, Lee Y, Cho HT, Kende H (2003) Regulation of expansin gene expression affects growth and development in transgenic rice plants. Plant Cell 15 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (1999) Enzymes and other agents that enhance cell wall extensibility. Annu Rev Plant Physiol Plant Mol Biol 50 391–417 [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ, Bedinger P, Durachko DM (1997) Group I allergens of grass pollen as cell wall-loosening agents. Proc Natl Acad Sci USA 94 6559–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Crowell DN (1998) Cytokinin regulates the expression of a soybean beta-expansin gene by a post-transcriptional mechanism. Plant Mol Biol 37 437–444 [DOI] [PubMed] [Google Scholar]
- Evert RF, Russin WA, Bosabalidis AM (1996) Anatomical and ultrastructural changes associated with sink-source transition in developing maize leaves. Int J Plant Sci 157 247–261 [Google Scholar]
- Fricke W (2002) Biophysical limitation of leaf cell elongation in source-reduced barley. Planta 215 327–338 [DOI] [PubMed] [Google Scholar]
- Gookin TE, Hunter DA, Reid MS (2003) Temporal analysis of alpha and beta-expansin expression during floral opening and senescence. Plant Sci 164 769–781 [Google Scholar]
- Gray-Mitsumune M, Mellerowicz EJ, Abe H, Schrader J, Winzell A, Sterky F, Blomqvist K, McQueen-Mason S, Teeri TT, Sundberg B (2004) Expansins abundant in secondary xylem belong to subgroup A of the alpha-expansin gene family. Plant Physiol 135 1552–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisawa K, Rose JKC, Nakani R, Inaba A, Kubo Y (2003) Differential expression of seven α-expansin genes during growth and ripening of pear fruit. Physiol Plant 117 564–572 [DOI] [PubMed] [Google Scholar]
- Im KH, Cosgrove DT, Jones AM (2000) Subcellular localization of expansin mRNA in xylem cells. Plant Physiol 123 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, Bradford K, Brummell D, Cho HT, Cosgrove D, Fleming A, Gehring C, Lee Y, McQueen-Mason S, Rose J, et al (2004) Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol Biol 55 311–314 [DOI] [PubMed] [Google Scholar]
- Kutschera U (1992) The role of the epidermis in the control of elongation growth in stems and coleoptiles. Bot Acta 105 246–252 [Google Scholar]
- Lee DK, Ahn JH, Song SK, Choi YD, Lee JS (2003) Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol 131 985–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kende H (2001) Expression of β-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol 127 645–654 [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kende H (2002) Expression of α-expansin and expansin-like genes in deepwater rice. Plant Physiol 130 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L-C, Bedinger PA, Volk C, Jones AD, Cosgrove DJ (2003. a) Purification and characterization of four β-expansins (Zea m1 isoforms) from maize pollen. Plant Physiol 132 2073–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Jones L, McQueen-Mason S (2003. b) Expansins and cell growth. Curr Opin Plant Biol 6 603–610 [DOI] [PubMed] [Google Scholar]
- Liang BM, Sharp RE, Baskin TI (1997) Regulation of growth anisotropy in well watered and water-stressed maize roots. I. Spatial distribution of longitudinal, radial and tangential expansion rates. Plant Physiol 115 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart JA (1965) An analysis of irreversible plant cell elongation. J Theor Biol 8 264–275 [DOI] [PubMed] [Google Scholar]
- MacAdam JW, Nelson CJ (2002) Secondary cell wall deposition causes radial growth of fibre cells in the maturation zone of elongating tall fescue leaf blades. Ann Bot (Lond) 89 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice I, Gastal F, Durand JL (1997) Generation of form and associated mass deposition during leaf development in grasses: a kinematic approach for non-steady growth. Ann Bot (Lond) 80 673–683 [Google Scholar]
- Muller B, Reymond M, Tardieu F (2001) The elongation rate at the base of a maize leaf shows an invariant pattern during both the steady-state elongation and the establishment of the elongation zone. J Exp Bot 52 1259–1268 [PubMed] [Google Scholar]
- Paolillo DJ (1995) Protoxylem maturation in the seedling wheat leaf. Am J Bot 82 337–345 [Google Scholar]
- Passioura JB, Fry SC (1992) Turgor and cell expansion: beyond the Lockhart equation. Aust J Plant Physiol 19 565–576 [Google Scholar]
- Peters WS, Tomos AD (2000) The mechanic state of “inner tissue” in the growing zone of sunflower hypocotyls and the regulation of its growth rate following excision. Plant Physiol 123 605–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidy B, McQueen-Mason S, Nösberger J, Fleming A (2001) Differential expression of α- and β-expansin genes in the elongation leaf of Festuca pratensis. Plant Mol Biol 46 491–504 [DOI] [PubMed] [Google Scholar]
- Reymond M, Muller B, Tardieu F (2004) Dealing with the genotype x environment interaction via a modelling approach: a comparison of QTLs of maize leaf length or width with QTLs of model parameters. J Exp Bot 55 2461–2472 [DOI] [PubMed] [Google Scholar]
- Rochange SF, Wenzel CL, McQueen-Mason SJ (2001) Impaired growth in transgenic plants over-expressing an expansin isoform. Plant Mol Biol 46 581–589 [DOI] [PubMed] [Google Scholar]
- Sampedro J, Cosgrove DJ (2005) The expansin superfamily. Genome Biol 6 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez MA, Mateos I, Labrador E, Dopico B (2004) Brassinolides and IAA induce the transcription of four alpha-expansin genes related to development in Cicer arietinum. Plant Physiol Biochem 42 709–716 [DOI] [PubMed] [Google Scholar]
- Schipper O, Schaefer D, Reski R, Fleming A (2002) Expansins in the bryophyte Physcomitrella patens. Plant Mol Biol 50 789–802 [DOI] [PubMed] [Google Scholar]
- Schnyder H, Seo S, Rademacher IF, Kühbauch W (1990) Spatial distribution of growth rates and epidermal cell lengths in the elongation zone during leaf development in Lolium perenne L. Planta 181 423–431 [DOI] [PubMed] [Google Scholar]
- Schopfer P (2001) Hydroxyl radical-induced cell-wall loosening in vitro and in vivo: implications for the control of elongation growth. Plant J 28 679–688 [DOI] [PubMed] [Google Scholar]
- Shin JH, Jeong DH, Park MC, An G (2005) Characterization and transcriptional expression of the α-expansin gene family in rice. Mol Cells 20 210–218 [PubMed] [Google Scholar]
- Silk WK (1992) Steady form from changing cells. Int J Plant Sci 153 49–58 [Google Scholar]
- Tardieu F, Reymond M, Hamard P, Granier C, Muller B (2000) Spatial distributions of expansion rate, cell division rate and cell size in maize leaves: a synthesis of the effects of soil water status, evaporative demand and temperature. J Exp Bot 51 1505–1514 [DOI] [PubMed] [Google Scholar]
- Tomos AD, Pritchard J (1994) Biophysical and biochemical control of cell expansion in roots and leaves. J Exp Bot 45 1721–1731 [Google Scholar]
- Tsuge T, Tsukaya H, Uchimiya H (1996) Two independent and polarized processes of cell elongation regulate leaf blade expansion in Arabidopsis thaliana. Development 122 1589–1600 [DOI] [PubMed] [Google Scholar]
- Tsukaya H (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57 477–496 [DOI] [PubMed] [Google Scholar]
- Vogler H, Caderas D, Mandel T, Kuhlemeier C (2003) Domains of expansin gene expression define growth regions in the shoot apex of tomato. Plant Mol Biol 53 267–272 [DOI] [PubMed] [Google Scholar]
- Vreeburg RA, Benschop JJ, Peeters AJ, Colmer TD, Ammerlaan AH, Staal M, Elzenga TM, Staals RH, Darley CP, McQueen-Mason SJ, et al (2005) Ethylene regulates fast apoplastic acidification and expansin A transcription during submergence-induced petiole elongation in Rumex palustris. Plant J 43 597–610 [DOI] [PubMed] [Google Scholar]
- Wu Y, Meeley RB, Cosgrove DJ (2001. a) Analysis and expression of the alpha-expansin and beta-expansin gene families in maize. Plant Physiol 126 222–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Thorne ET, Sharp RE, Cosgrove DJ (2001. b) Modification of expansin transcript levels in the maize primary root at low water potentials. Plant Physiol 126 1471–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.