Abstract
Coat protein I (COPI) vesicles arise from Golgi cisternae and mediate the recycling of proteins from the Golgi back to the endoplasmic reticulum (ER) and the transport of Golgi resident proteins between cisternae. In vitro studies have produced evidence for two distinct types of COPI vesicles, but the in vivo sites of operation of these vesicles remain to be established. We have used a combination of electron tomography and immunolabeling techniques to examine Golgi stacks and associated vesicles in the cells of the scale-producing alga Scherffelia dubia and Arabidopsis preserved by high-pressure freezing/freeze-substitution methods. Five structurally distinct types of vesicles were distinguished. In Arabidopsis, COPI and COPII vesicle coat proteins as well as vesicle cargo molecules (mannosidase I and sialyltransferase–yellow fluorescent protein) were identified by immunogold labeling. In both organisms, the COPI-type vesicles were further characterized by a combination of six structural criteria: coat architecture, coat thickness, membrane structure, cargo staining, cisternal origin, and spatial distribution. Using this multiparameter structural approach, we can distinguish two types of COPI vesicles, COPIa and COPIb. COPIa vesicles bud exclusively from cis cisternae and occupy the space between cis cisternae and ER export sites, whereas the COPIb vesicles bud exclusively from medial- and trans-Golgi cisternae and are confined to the space around these latter cisternae. We conclude that COPIa vesicle-mediated recycling to the ER occurs only from cis cisternae, that retrograde transport of Golgi resident proteins by COPIb vesicles is limited to medial and trans cisternae, and that diffusion of periGolgi vesicles is restricted.
Keywords: tomography, electron tomography, immunolabeling
Trafficking between the organelles of the secretory pathway of eukaryotic cells, endoplasmic reticulum (ER), Golgi, trans-Golgi network (TGN), and plasma membrane is mediated by vesicles. Traffic within this pathway flows along organized and directional routes, and each step involves a unique type of vesicle, which originates on one compartment and is targeted to another. There is general agreement that coat protein (COP)II vesicles are the carriers involved in anterograde ER-to-Golgi transport (1, 2). In contrast, there is still much confusion surrounding the trafficking patterns of COPI vesicles (3). There is strong evidence to support the notion that COPI vesicles originating from cis-Golgi cisternae recycle membrane molecules back to the ER (4). The targets of the COPI vesicles that bud from medial- and trans-Golgi cisternae are less clear, in part because of conflicting data, and in part because of conflicting hypotheses of Golgi trafficking (5, 6). The vesicle shuttle model postulates that COPI vesicles are involved in both anterograde and retrograde transport between cisternae (7), whereas the cisterna progression/maturation model proposes that COPI vesicles are used in retrograde transport only (8, 9). The extent to which COPI vesicles derived from the medial- and trans-Golgi cisternae can recycle molecules directly back to the ER is also uncertain (10).
To address these questions, we have performed a comparative electron tomography analysis of high-pressure frozen/freeze-substituted Golgi stacks of the alga Scherffelia dubia and of the higher plant Arabidopsis thaliana and have used immunolabeling experiments to confirm the identity of the COPI and COPII-type vesicles and to identify cargo molecules within COPI vesicles. S. dubia was chosen as the lead organism for the following reasons. First, the Golgi of S. dubia, like those of other proteoglycan scale-producing algae, have always been viewed as prototypes of Golgi that traffic according to the cisternal progression/maturation model (11, 12), because their distinctive scales are always seen inside the Golgi cisternae and never in the vesicles surrounding the stacks (13–15). Second, the large size of the stacks (≈1.5 μm in diameter, 17–20 cisternae) allows for an unambiguous localization of specific cisternal and vesicle types to discrete Golgi domains. Third, the rapid rate of cisternal assembly and turnover (one cisterna per 20–30 sec) gives rise to large numbers of transport vesicles, which facilitates the production of statistically relevant data sets (16). The two principal drawbacks of S. dubia are the lack of genetics and of cross-reactivity of well defined secretory pathway antibodies with S. dubia proteins.
The reasons for choosing Arabidopsis as a complementary experimental system to S. dubia were the following. Previous structural studies of Arabidopsis Golgi stacks (17, 18) have provided evidence for a cisternal maturation mode of trafficking and for structural similarities between the Arabidopsis and S. dubia Golgi. In particular, in Arabidopsis cells preserved by high-pressure freezing/freeze-substitution techniques, cis-, medial-, and trans-Golgi cisternae can be distinguished based on their position in the stack, the staining of their luminal contents, and the geometry of the cisternae. In addition, differences in Golgi-associated vesicle types and the visualization of a distinct type of coat on vesicles budding from Golgi cisternae have been noted (17, 19). Finally, COPI and COPII vesicular coat components have been characterized, and antibodies for verifying the identities of the vesicles and for identifying vesicle cargoes are available (20, 21).
Interestingly, electron tomography and live-cell imaging studies of the ER and Golgi membranes of Pichia pastoris (22, 23) have shown that many of the structural features reported for Arabidopsis Golgi can also be observed in the yeast, and that the P. pastoris Golgi operates according to the cisternal maturation model. Thus, the finding of this study that one type of COPI vesicle traffics between Golgi and ER, and a second type of COPI vesicle is involved in intra-Golgi trafficking, may be a characteristic feature of all walled organisms and possibly also may apply to the animal Golgi.
Results
Cis-, Medial-, and Trans-Golgi Cisternae Can Be Distinguished by Means of Morphological Criteria.
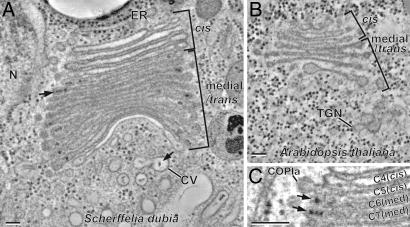
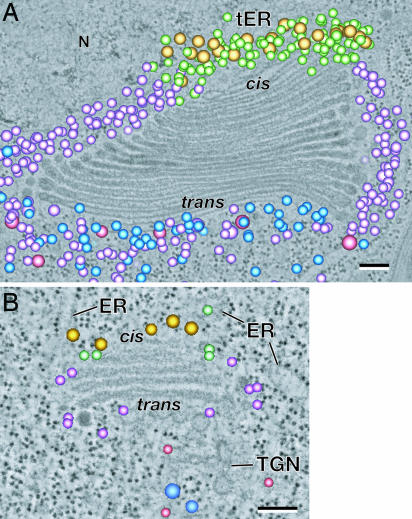
Cis-, medial-, and trans-Golgi cisternae can be positively identified by means of morphological criteria in thin-section and tomographic slice images of cryofixed and freeze-substituted plant [Arabidopsis (Fig. 1B) and Nicotiana sylvestris (17)] and P. pastoris yeast (23) cells. The same morphological criteria position in stack, luminal staining, and cisternal geometry can be used to identify the different cisternal types in the tomographic slice images of the S. dubia Golgi (Fig. 1 A and C). Thus, the luminal staining of the first four cisternae shown in Fig. 1A (first five to six cisternae in sections where the first one or two smaller cis cisternae are also seen) demonstrate that cis-type cisternae exhibit a much lighter luminal staining and possess a thicker lumen than the following medial cisternae. The abruptness of the increase in staining of the cisternal contents at the cis-to-medial cisterna boundary coincides with the sudden appearance of darkly stained scales in the lumen of the medial cisternae (Fig. 1 A and C). No darkly stained proteoglycan scales were ever observed in cis-Golgi cisternae. In Arabidopsis, too, the cis-Golgi cisternae were always very lightly stained (Fig. 1B).
Fig. 1.
Structural differentiation of Golgi cisternae of S. dubia and Arabidopsis. Tomogram slices of an S. dubia Golgi stack (A) and an Arabidopsis Golgi stack (B). In A, a proteoglycan scale within the cisternal lumen of the Golgi and another in the contractile vacuole (CV) are marked with arrows. In both species, the cis cisternae always exhibit a lightly stained and thicker lumen. A sudden transition to darkly stained luminal contents occurs at the cis-to-medial cisternal boundary in S. dubia (C). Simultaneous with the general increase of cisternal staining, the distinctive proteoglycan scales begin to be clearly discerned (C, arrows). A budding COPIa-type vesicle is indicated in C (COPIa). (Scale bars, 100 nm.)
Trans-Golgi cisternae can be distinguished by their position in the stack, collapsed central luminal domain, and frequently swollen margins. In higher plants, the trans-most Golgi cisternae typically show signs of becoming separated from the stacks during their transformation into completely independent TGN cisternae (Fig. 1B), which give rise to clathrin-coated and secretory vesicles. Because S. dubia secretes the Golgi-made scales by the contractile vacuole system to the cell surface (Fig. 1A) (24), the last lightly stained cisterna associated with each of its stacks (Fig. 1A) does not match the classical view of a TGN compartment in higher plants (17) or yeast (23). Thus, a strict comparison can only be made between the cis-, medial-, and trans-Golgi cisternae of these different organisms.
ER-, Cis-Golgi Cisternae, and Medial/Trans-Golgi Cisternae Give Rise to Three Types of Buds and Vesicles.
The excellent structural preservation of the different membrane systems in high-pressure-frozen S. dubia and Arabidopsis cells, together with the high resolution of the ≈2.3-nm-thick tomographic slices, has enabled us to distinguish COPII and two COPI-type vesicles by means of the following set of structural parameters: Site of vesicle origin, coat architecture, coat thickness, vesicle size, cargo staining, and spatial distribution around Golgi stacks (Figs. 2 and 3).
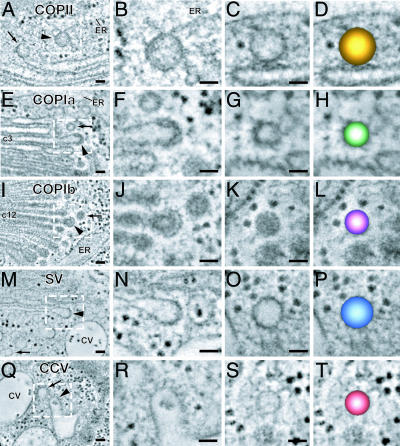
Fig. 2.
A gallery of tomogram slice images displaying examples of the five vesicle types associated with the transitional ER (A–D), cis cisternae (E–H), medial or trans cisternae (I–L), trans-most cisterna (M–P), and contractile vacuole (Q–T) of S. dubia. The budding profiles of the five types of vesicles display distinct coat morphologies (magnified in the second column). The COPII-type coat appears thin and is relatively tight against the ER membrane (B). The COPIa-type coat appears thicker and more diffuse (F), whereas the COPIb-type coat appears spaced out away from the cisternal membrane (J). The coats on the budding vesicles on the trans-most cisterna resemble COPII (N), and the clathrin coats on the CV system (R) have a characteristic clathrin structure and spacing. For each type of budding profile, vesicles with identical diameters and staining patterns are found nearby (arrows). Color-coded models of each type of vesicle are shown in the fourth column (D, H, L, P, and T). (Scale bars, 50 nm.)
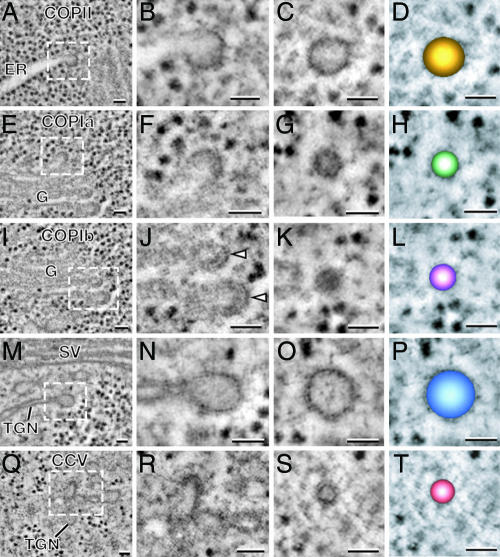
Fig. 3.
A gallery of tomogram slices showing types of Arabidopsis vesicles matching the five S. dubia vesicle types. (A–D) COPII-type vesicles. (E–H) COPIa-type vesicles. (I–L) COPIb-type vesicles. (M–P) Secretory vesicles. (Q–T) Clathrin-coated vesicles. Budding profiles and released vesicles are shown in the second and third columns, respectively. Color-coded models of each type of vesicle are shown in the fourth column. As in S. dubia, the cis- and medial-trans-Golgi cisternae give rise to two morphologically distinct COPI vesicles (COPIa and COPIb). The secretory and clathrin-coated vesicles are produced from the TGN cisternae (M–P and Q–T). (Scale bars, 50 nm.)
COPII-Type Vesicles.
These vesicles (Figs. 2 A–D and 3 A–D) are the only type of vesicle seen budding from ER cisternae. In S. dubia, ER vesicle budding sites, which are also known as transitional ER sites, are seen only adjacent to the cis-side of the Golgi stacks (Fig. 1A). In Arabidopsis and other higher plants, the spatial relationship between the ER budding sites and the Golgi stacks is less well defined, because the Golgi stacks are highly mobile (traveling along actin tracks at speeds of up to 4 μm/s) (25) and do not maintain a fixed spatial relationship to ER export sites (26). For these reasons, such associations are rarely observed in thin-section electron micrographs.
The COPII vesicles produced in S. dubia have a diameter of ≈70 nm and in Arabidopsis ≈60 nm (Figs. 2C and 3C). The COPII coat is not very distinct in either organism, but the presence of a coat layer is evidenced by the darker staining of the membrane around the buds/vesicles compared with the staining of the ER membrane (Figs. 2 A and B and 3 A and B). The COPII coat appears closely applied to the membrane, has a thickness of 5.6 nm ± 1.3 nm (Figs. 2B and 3B) and exhibits hints of a fine globular substructure. The contents of the vesicles are always lightly stained (Figs. 1 B and C and 2 B and C).
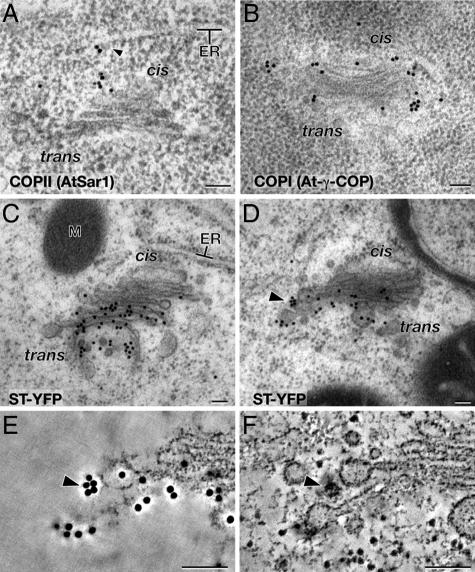
To confirm the identity of the COPII buds and vesicles, we have performed immunogold labeling of thin sections of high-pressure-frozen Arabidopsis root cells embedded in Lowicryl HM20 using anti-AtSar1 antibodies. AtSar1 is a homolog of the Sar1 GTPase that regulates the assembly/disassembly of the COPII coat and is required for ER-to-Golgi transport in plants (27). The anti-AtSar1 antibodies labeled both budding COPII vesicles and COPII vesicles associated with the cis-side of adjacent Golgi stacks (Fig. 5A).
Fig. 5.
Immunoelectron micrographs of COPII (A) and COPI (B) coat components in Arabidopsis meristem cells ST-YFP in BY2 tobacco suspension culture cells (C–F). (A) AtSar1-specific immunogold particles are seen on the ER surface and between the ER and the cis- side of Golgi. A budding vesicle labeled with AtSar1 immunogold particles is marked with an arrowhead. (B) At-γ-COPI-specific immunogold particles are seen along the periphery of the Golgi from the cis- to trans-most cisternae. (C and D) Immunoelectron micrographs of ST-YFP in BY2 cells. The ST-YFP-specific immunogold particles are confined to late medial and trans cisternae. The cluster of five immunogold particles marked with an arrowhead in D is shown at a higher magnification in E and F. (E) Tomographic slice image of the surface of the section in D showing the immunogold particle cluster (arrowhead). (F) Tomographic slice image ≈15 nm below the section surface depicted in E. A COPIb-type vesicle (arrowhead) is seen under the ST-YPF immunogold particles. (Scale bars, 100 nm.)
COPIa- and COPIb-Type Vesicles.
By definition, COPI-type vesicles bud from Golgi cisternae and are involved in both Golgi-to-ER and intraGolgi vesicle transport (4). To confirm that the vesicles budding from the Golgi stacks of Arabidopsis are indeed COPI-type vesicles, we have immunogold-labeled thin sections of Arabidopsis root tip cells with anti-At-γ-COP antibodies (21). These anti-COPI antibodies labeled both cisternal buds and vesicles around the edges of the cis-, medial-, and trans-type Golgi cisternae (Fig. 5B).
Both COPIa- and COPIb-type vesicles are produced from buds that arise on Golgi cisternae, but the two types of vesicles originate from different cisternae. In particular, the COPIa-type buds/vesicles arise from cis-Golgi cisternae (Figs. 2 E–G and 3 E–G), whereas the COPIb-type vesicles originate on medial- and trans-type cisternae [Figs. 2 I–K and 3 I–K; supporting information (SI) Figs. 7 and 8 provide 20 more COPIa- and COPIb-type vesicle images]. Another difference relates to the staining of the vesicle contents. As discussed above, the contents of the cis-Golgi cisternae stain much more lightly than those of the medial- and trans-Golgi cisternae (Fig. 1). Similarly, the COPIa-type buds and vesicles that originate from cis-Golgi cisternae have lightly stained contents and a readily discernable limiting membrane (Figs. 2 E–G and 3 E–G and SI Figs. 7 A–E and 8 A–E). In contrast, the contents of the COPIb-type buds and vesicles, which arise from the darkly stained medial- and trans-Golgi cisternae, are also darkly stained (Figs. 2 J and K, 3 J and K, SI Figs. 7 F–J and 8 F–J). This makes it difficult to distinguish the limiting membrane from the vesicle contents in the COPIb-type vesicles (Figs. 2K and 3K and SI Figs. 7 F–J and 8 F–J).
Budding COPIa- and COPIb-type vesicles also exhibit a different coat morphology, with the same differences seen in S. dubia and Arabidopsis (compare Figs. 2 E and F and 3 E and F for COPIa, Figs. 2 I and J and 3 I and J for COPIb; compare also SI Figs. 7 and 8). Most notably, the COPIa-type coats, while thicker than the COPII coats (11.4 ± 1.8 nm vs. 5.6 ± 1.3 nm), exhibit an irregular and less-defined architecture compared with the distinctive geometry of the COPIb-type coats. The COPIb-type budding vesicle coat consists of two layers, a compact inner layer that is closely applied to the membrane and a more pronounced concentric outer layer with thin bridging elements to the inner layer. The total thickness of the COPIb-type coat on the buds of the medial- and trans- types of cisternae is 17.9-2.5 nm. Interestingly, the dark outer layer of the COPIb-type coat of the buds does not persist on the vesicles that have been released (compare Figs. 2 J and K for S. dubia and 3 J and K for Arabidopsis; see also SI Figs. 7 and 8).
The diameter of the COPIa- and COPIb-type vesicles is the same in each cell type but differs between organisms. Thus, both COPIa- and COPIb-type vesicles average ≈55 nm in S. dubia but only ≈45 nm in Arabidopsis (SI Fig. 9).
COPII-, COPIa-, and COPIb-Type Vesicles Are Confined to Different Zones Around the Golgi Stacks.
To further characterize the differences between COPIa- and COPIb-type vesicles, we have mapped their distribution around Golgi stacks. The results of this analysis are illustrated in Fig. 4A and B, and the color coding of the different vesicle types is shown in Figs. 2 and 3. Most striking is the confinement of the two COPI vesicle types to two nonoverlapping zones around both the S. dubia and the Arabidopsis Golgi stacks. The S. dubia data are clearer because of the larger number of vesicles surrounding the stack (Fig. 4A), but the same spatial segregation is also evident in the Arabidopsis specimen (Fig. 4B). Detailed analysis of the S. dubia tomographic models has shown that the COPIa-type vesicles are confined to the space around the cis-type Golgi cisternae and between these cis cisternae and the adjacent ER membrane with the ER export site. In contrast, the COPIb-type vesicles are seen only around the margins of the medial- and trans-type cisternae and in the space beyond the trans-most Golgi cisterna but never in the space around the cis cisternae. Even when these tomographic models are examined in 3D space, no overlap between the COPIa- and COPIb-type vesicle-containing regions is seen. Thus, this segregation appears to be close to absolute. Although there are many fewer vesicles around the much smaller Arabidopsis Golgi stacks, the segregation of the COPIa-type vesicles to the space around the cis cisternae and of the COPIb-type vesicles to the space around the medial and trans cisternae, was always observed.
Fig. 4.
3D tomogram models of the Golgi-associated vesicles shown in Figs. 2 and 3 are superimposed on a single slice of an S. dubia (A) and of an Arabidopsis (B) Golgi stack. These images highlight the nonrandom distribution of the five different vesicle classes surrounding the Golgi stacks. The distribution of the COPII- (gold), COPIa- (light green), and COPIb-type vesicles (light purple) in S. dubia is duplicated in Arabidopsis. (Scale bars, 100 nm.)
The spatial segregation of COPIa- and COPIb-type vesicles suggested that the two types of vesicles might traffic different sets of cargo molecules. To test this hypothesis, we used immunogold labeling of two Golgi resident proteins, sialyltransferase fused with yellow fluorescent protein (ST-YFP) and Arabidopsis mannosidase I (ManI), for determining whether these proteins localized to COPIa, COPIb, or both types of vesicles. ST-YFP-specific antibodies labeled late medial- and trans-Golgi cisternae, as well as COPIb- but not COPIa-type vesicles (Fig. 5C–F; see also SI Movie 1). Similarly, the ManI-specific antibodies, which are bound only to medial-Golgi cisternae, also labeled only COPIb-type vesicles (see SI Fig. 10).
The COPII-type vesicles are confined to the space around a given ER export site and the space between that site and the adjacent cis-Golgi cisternae, with the largest number of such vesicles seen in the vicinity of the cis-most cisterna (Fig. 4 A and B). Overall, the distribution of the COPII-type vesicles matches the distribution of the COPIa-type vesicles in both S. dubia and Arabidopsis.
Secretory-Type and Clathrin-Coated Vesicles Are Confined to Specific Regions Around Trans-Golgi and TGN Cisternae.
There are significant differences in the spatial and functional organization of the postGolgi membrane compartments of S. dubia and Arabidopsis, which relate to the different secretory mechanisms used by the two organisms. Thus, all of the products of the S. dubia Golgi appear to be secreted into a tubular membrane system that is part of the contractile vacuole apparatus (Fig. 1A) (24), whereas in Arabidopsis cells, all products appear to end up in the TGN (Figs. 1B and 4B) (28). Nevertheless, both systems produce two structurally distinct types of vesicles, secretory-type and clathrin-coated vesicles (Figs. 2 M–T and 3 M–T).
In S. dubia, the secretory-type vesicles (Fig. 2 M–O) are generated by the trans-most cisterna of the stack, which exhibits a conspicuous lack of staining of its cisternal contents except for the flagellar and thecal scales (Fig. 1A). These ≈72-nm-in-diameter vesicles possess a thin coat (compare staining of the bud and the adjacent cisternal membrane regions in Fig. 2N) and lightly stained contents (Fig. 2 N and O). The secretory-type vesicles of Arabidopsis bud from the TGN cisternae typically after they have separated from the Golgi stack (Figs. 1B and 3B). The diameter of these vesicles varies substantially (65–100 nm), as does the staining of their contents, although the majority possesses lightly stained cargo molecules (Fig. 3 N and O). The staining of the thin secretory vesicle coat is similar to the staining of the comparable vesicles in S. dubia.
In S. dubia, the clathrin-coated vesicles are seen on the tubular elements of the contractile vacuole system (Figs. 1A and 2 Q–S), whereas in Arabidopsis, they arise on the TGN cisternae (Figs. 1B, 3 Q–S, and 4B). Although the structure of the clathrin coats appears very similar in the tomographic images of both organisms, the diameters of the vesicles are quite different, ≈55 nm in S. dubia and ≈35 nm in Arabidopsis.
In both S. dubia and in Arabidopsis, the secretory and clathrin-coated vesicles are confined to the trans-side of the Golgi stacks and to the vicinity of the TGN cisternae (Fig. 4 A and B). Some intermingling of COPIb-type vesicles with the secretory and clathrin-coated vesicles is also seen in these regions, most notably in S. dubia.
Discussion
COPI vesicles mediate two types of transport, the recycling of molecules from Golgi to ER (4) and intraGolgi transport (29). The main purpose of this study was to investigate the structural basis of these two trafficking pathways using electron tomography of high-pressure-frozen algal and plant cells. We have identified two distinct classes of COPI vesicles, designated COPIa and COPIb vesicles, which can be distinguished based on a combination of six different morphological criteria. We have also demonstrated that COPIa and COPIb vesicles transport different types of cargo proteins. Together with the findings of others (10, 30, 31), our results suggest that the COPIa-type vesicles that traffic from Golgi to the ER constitute a subclass of COPI vesicles that are both structurally and compositionally distinct from the COPIb-type vesicles that mediate intraGolgi trafficking. The spatial segregation of these two vesicle types is consistent with the hypothesis that diffusion of periGolgi vesicles is restricted (7).
COPII and COPI Buds and Vesicles Can Be Efficiently Immunolabeled in High-Pressure-Frozen and Lowicryl HM20-Embedded Cells.
There is a general consensus that cells preserved by cryofixation methods provide more reliable structural information on dynamic membrane systems than cells preserved by chemical means (32, 33). Indeed, COPII buds can be visualized only in plant cells preserved by high-pressure freezing, never in chemically fixed cells (34–36). However, it has been claimed that in samples preserved by high-pressure-freezing, freeze-substitution, and resin-embedding techniques, the COPII and COPI vesicles lose their antigenicity, and that labeling of such vesicles can occur only in chemically fixed samples prepared by the Tokuyasu method (2, 21, 37). Here we demonstrate that COPII, COPI vesicles, and cargo proteins in the COPI vesicles can be efficiently labeled in high-pressure-frozen and freeze-substituted Arabidopsis root-tip cells embedded in Lowicryl HM20 at −60°C (Fig. 5 and SI Fig. 10). These immunolabeling experiments also prove that the structurally distinct buds and vesicles designated COPII and COPI in our electron tomography images of Arabidopsis (Fig. 3) contain the expected coat proteins.
Cis-Golgi Cisternae Give Rise to COPI-Type Vesicles That Differ Structurally from COPI-Type Vesicles That Bud from Medial- and Trans-Golgi Cisternae.
Vesicles that bud from Golgi cisternae are known as COPI vesicles, because they share COPI-type coat proteins. Depending on the model of Golgi trafficking, cisternal maturation vs. vesicle shuttle, two or three functional types of COPI vesicles have been postulated to exist (29). Thus, in the scale-secreting alga S. dubia, whose Golgi serve as prototypes for Golgi trafficking according to the cisternal maturation model (13, 15), two types of COPI vesicles would be expected to be observed. Because plant Golgi stacks appear to move their cargo molecules by the same mechanism (18, 36, 38), they, too, should possess two functional types of COPI vesicles, those that recycle molecules from Golgi to ER, and those involved in retrograde transport within the stacks.
Both S. dubia and Arabidopsis Golgi give rise to two distinct types of COPI-type vesicles that display the same morphological features in the two organisms (Figs. 2 and 3 and SI Figs. 7 and 8). In particular, the vesicles designated COPIa possess a lightly stained core similar to the lightly stained lumen of the cis cisternae from which they bud, a clearly visible membrane, and a ≈11-nm-thick coat with a characteristic architecture. In contrast, all vesicles designated COPIb have dark cores similar to the dark luminal contents of the medial and trans cisternae from which they bud, a boundary membrane that is difficult to discern, and a characteristic double-layered coat, which has a thickness of ≈18 nm. In addition, the COPIa- and COPIb-type vesicles occupy the same subdomains around the Golgi stacks in both organisms. The only systematic difference between the COPI vesicles in the two organisms is their size, ≈55 nm in diameter in S. dubia and ≈45 nm in Arabidopsis. The advantage of using this multiparameter structural approach for identifying different types of periGolgi vesicles compared with the Tokuyasu immunolabeling method is that it allows for the positive identification of close to 100% of the vesicles (Figs. 2–4) vs. a much smaller percentage in the immunolabeled samples.
COPI vesicle budding can be reconstituted in vitro by using partly purified rat liver Golgi membranes (39). Analysis of the resulting vesicles has shown that they are of two types, vesicles enriched in p24 family proteins that cycle between the ER and the Golgi and vesicles that lack such proteins but that are enriched in Golgi resident enzymes (30, 31). In addition, the two vesicle types also display different affinities for Golgi-associated tethering factors (31). These differences are consistent with the idea that the p24-enriched vesicles correspond to the COPIa-type vesicles that localize to the region between ER export sites and cis-Golgi cisternae and the p24-depleted and Golgi enzyme-enriched vesicles to the COPIb-type vesicles around the medial- and trans-type cisternae. Martinez-Menarguez et al. (10) have provided immunolabeling evidence for two types of peri-Golgi vesicles being involved in the recycling of Golgi resident proteins in normal rat kidney epithelial cells, with one type mediating intraGolgi transport and the other recycling Golgi resident proteins to the ER. Our data support the first but not the second of these hypotheses (see below).
Retrograde Transport of Golgi Enzymes by COPIb-Type Vesicles Is Limited to Medial- and Trans-Golgi Cisternae.
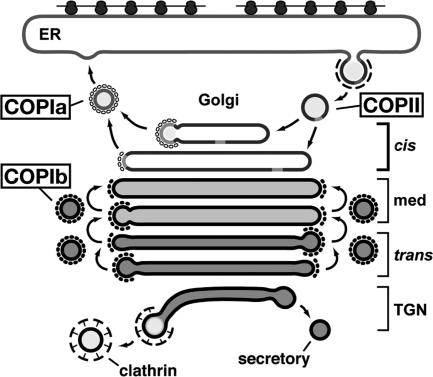
One of the most striking discoveries of this study is the nonoverlapping distribution of COPIa- and COPIb-type vesicles around the Golgi stacks as well as the correlation of this distribution with cisternal types and cargo trafficking patterns (Figs. 4–6). Although the cisternal types were originally identified by means of structural criteria (17), the cisternae identified in this manner have been shown to correspond to functionally distinct cisternal types (40), and similar findings have been reported for the yeast P. pastoris (23). The confinement of the COPIa-type vesicles to the region between the cis-type cisternae and the ER export sites confirms expectations for Golgi-to-ER recycling vesicles (4), whereas the exclusion of the COPIb-type vesicles from the same region and their confinement to the regions immediately adjacent to medial- and trans-type cisternae provides insights into intraGolgi trafficking. First, recycling from Golgi to ER must occur only from cis cisternae, never from medial and trans cisternae (Fig. 6), consistent with recycling data for Arabidopsis ER retention sequence-tagged proteins (41). Second, retrograde transport of Golgi resident enzymes is limited essentially to medial- and trans-Golgi cisternae (Fig. 6). Indeed, the retrograde transport of medial-Golgi enzymes into the trans-most cis cisterna may even define the transformation of a cis-Golgi cisterna into a medial cisterna, and a corresponding retrograde transport step at the trans-most medial cisterna would be expected to give rise to a new trans cisterna.
Fig. 6.
Model summarizing the sites of origin and trafficking patterns of the five ER/Golgi/TGN(CV)-associated vesicle classes described in this paper. Note the spatial segregation of COPIa and COPIb vesicles. The cis-Golgi cisternae are assembling cisternae that produce COPI vesicles that differ in their trafficking pattern from those originating on medial and trans cisternae.
Materials and Methods
High-Pressure Freezing and Freeze Substitution.
d-Manitol (Sigma, St. Louis, MO) was added to the S. dubia culture medium as an external cryoprotectant to a final concentration of 100 mM 1 h before freezing. Log-phase growth cultures were collected by centrifugation, and the cell pellets were rapidly frozen in a BAL-TEC HPM-010 high-pressure freezer (BalTec/RMC, Tucson, AZ). Freeze substitution was carried out in 2% OsO4/0.5% uranyl acetate in anhydrous acetone at approximately −80°C for 4 days. The samples were then gradually warmed to room temperature over the next 2 days. After bringing the samples to room temperature, they were rinsed three times in dry acetone, removed from the planchets, and slowly infiltrated with increasing concentrations of Epon resin (Ted Pella, Redding, CA) over 4 days (42). Resin-infiltrated samples were polymerized under vacuum at 60°C for 48 h.
Root tips from Arabidopsis seedlings were frozen and freeze-substituted as described in Segui-Simarro et al. (43). For protein immunolabeling, frozen root tips were freeze-substituted in acetone with 0.2% uranyl acetate and 0.25% glutaraldehyde at −90°C for 4 days and slowly warmed to −60°C for 6 h. After three acetone rinses, samples were infiltrated with Lowicryl HM20 (Electron Microscopy Sciences, Fort Washington, PA) at −60°C as follows: 1 day each in 25%, 50%, and 75% HM20 in acetone. After three changes of fresh 100% HM20 over 2 days, samples were polymerized at −60°C under UV light for 24 h.
Immunolabeling of AtSar1, At-γ-COP, ST-YFP, and ManI.
Anti-AtSar1 (Rose Biotech, Hayward, CA) (18), anti-At-γ-COP (21), and anti-ManI (44) antibodies were used to detect AtSar1, At-γ-COP, and ManI in Arabidopsis root meristem cells, respectively. ST-YFP was detected in a BY2 cell line expressing ST-YFP with an anti-GFP antibody. The sections of HM20-embedded samples were blocked with 2% nonfat milk in PBST (PBS plus 0.1% Tween 20). Antibodies were diluted 1:20 in 1% nonfat milk in PBST, and sections were incubated with antibody dilutions for 1 h at room temperature. After three rinses in 1% nonfat milk in PBST, sections were incubated with the secondary antibody (anti-rabbit IgG conjugated to 15-nm gold particles, diluted 1:20 in 1% nonfat milk in PBST) for 1 h. After thorough rinsing, the immunolabeled samples were poststained as described in Segui-Simarro et al. (43).
Other Methods.
Conditions for S. dubia culturing and Arabidopsis growth, general procedures of electron microscopy, dual-axis tilt series imaging, and tomogram reconstruction/modeling methods are provided in SI Text.
Supplementary Material
Acknowledgments
We thank Dr. David Robinson (University of Heidelberg, Heidelberg, Germany), Dr. Sebastian Y. Bednarek (University of Wisconsin, Madison, WI), and Dr. Andreas Nebenführ (University of Tennessee, Knoxville, TN) for the anti-At-γ-COP, anti-ManI antibodies, and BY2 cell line expressing ST-YFP, respectively. We also thank the members of the Staehelin laboratory and the Boulder Laboratory for 3D EM of cells, technical advice, and helpful discussions. This work was supported by National Institutes of Health Grant GM-61306 (to L.A.S.).
Abbreviations
- ER
endoplasmic reticulum
- COP
coat protein
- TGN
trans-Golgi network
- ST-YFP
sialyltransferase fused with yellow fluorescent protein
- ManI
mannosidase I.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609818104/DC1.
References
- 1.Palmer KJ, Stephens DJ. Trends Cell Biol. 2004;14:57–61. doi: 10.1016/j.tcb.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Zeuschner D, Geerts WJ, van Donselaar E, Humbel BM, Slot JW, Koster AJ, Klumperman J. Nat Cell Biol. 2006;8:377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- 3.Pelham HR. J Cell Biol. 2001;155:1099–1101. doi: 10.1083/jcb.200110160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 5.Altan-Bonnet N, Sougrat R, Lippincott-Schwartz J. Curr Opin Cell Biol. 2004;16:364–372. doi: 10.1016/j.ceb.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Orci L, Stamnes M, Ravazzola M, Amherdt M, Perrelet A, Sollner TH, Rothman JE. Cell. 1997;90:335–349. doi: 10.1016/s0092-8674(00)80341-4. [DOI] [PubMed] [Google Scholar]
- 7.Orci L, Perrelet A, Rothman JE. Proc Natl Acad Sci USA. 1998;95:2279–2283. doi: 10.1073/pnas.95.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra V, Mayor S. Nature. 2006;441:939–940. doi: 10.1038/441939a. [DOI] [PubMed] [Google Scholar]
- 9.Glick BS. Curr Opin Cell Biol. 2000;12:450–456. doi: 10.1016/s0955-0674(00)00116-2. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Menarguez JA, Prekeris R, Oorschot VM, Scheller R, Slot JW, Geuze HJ, Klumperman J. J Cell Biol. 2001;155:1213–1224. doi: 10.1083/jcb.200108029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown RM, Jr, Franke WW, Kleinig H, Falk H, Sitte P. J Cell Biol. 1970;45:246–271. doi: 10.1083/jcb.45.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farquhar MG, Palade GE. J Cell Biol. 1981;91:77s–103s. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Becker B, Bolinger B, Melkonian M. Trends Cell Biol. 1995;5:305–307. doi: 10.1016/s0962-8924(00)89047-9. [DOI] [PubMed] [Google Scholar]
- 14.Melkonian M, Preisig HR. Nordic J Bot. 1986;6:235–256. [Google Scholar]
- 15.Elsner M, Hashimoto H, Nilsson T. Mol Membr Biol. 2003;20:221–229. doi: 10.1080/0968768031000114024. [DOI] [PubMed] [Google Scholar]
- 16.McFadden GI, Melkonian M. Protoplasma. 1986;131:174–184. [Google Scholar]
- 17.Staehelin LA, Giddings TH, Jr, Kiss JZ, Sack FD. Protoplasma. 1990;157:75–91. doi: 10.1007/BF01322640. [DOI] [PubMed] [Google Scholar]
- 18.Nebenführ A. In: The Golgi Apparatus and the Plant Secretory Pathway. Robinson DG, editor. Vol 9. Oxford: Blackwell; 2003. pp. 76–89. [Google Scholar]
- 19.Driouich A, Staehelin LA, Faye L. Plant Physiol Biochem. 1994;32:731–749. [Google Scholar]
- 20.Bar-Peled M, Raikhel NV. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG. Plant Cell. 2000;12:2219–2236. doi: 10.1105/tpc.12.11.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bevis BJ, Hammond AT, Reinke CA, Glick BS. Nat Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- 23.Mogelsvang S, Gomez-Ospina N, Soderholm J, Glick BS, Staehelin LA. Mol Biol Cell. 2003;14:2277–2291. doi: 10.1091/mbc.E02-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donohoe BS. Boulder, CO: Univ of Colorado; 2005. pp. 75–84. PhD thesis. [Google Scholar]
- 25.Nebenführ A, Gallagher LA, Dunahay TG, Frohlick JA, Mazurkiewicz AM, Meehl JB, Staehelin LA. Plant Physiol. 1999;121:1127–1142. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG. Plant Cell. 2005;17:1513–1531. doi: 10.1105/tpc.104.026757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A. Plant J. 2000;23:517–525. doi: 10.1046/j.1365-313x.2000.00823.x. [DOI] [PubMed] [Google Scholar]
- 28.Jurgens G. Annu Rev Cell Dev Biol. 2004;20:481–504. doi: 10.1146/annurev.cellbio.20.082503.103057. [DOI] [PubMed] [Google Scholar]
- 29.Rabouille C, Klumperman J. Nat Rev Mol Cell Biol. 2005;6:812–817. doi: 10.1038/nrm1735. [DOI] [PubMed] [Google Scholar]
- 30.Lanoix J, Ouwendijk J, Stark A, Szafer E, Cassel D, Dejgaard K, Weiss M, Nilsson T. J Cell Biol. 2001;155:1199–1212. doi: 10.1083/jcb.200108017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malsam J, Satoh A, Pelletier L, Warren G. Science. 2005;307:1095–1098. doi: 10.1126/science.1108061. [DOI] [PubMed] [Google Scholar]
- 32.Donohoe BS, Mogelsvang S, Staehelin LA. Methods. 2006;39:154–162. doi: 10.1016/j.ymeth.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 33.McIntosh R, Nicastro D, Mastronarde D. Trends Cell Biol. 2005;15:43–51. doi: 10.1016/j.tcb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 34.Aniento F, Matsuoka K, Robinson DG. In: The Plant Endoplasmic Reticulum. Robinson DG, editor. New York: Springer; 2006. [Google Scholar]
- 35.Craig S, Staehelin LA. Eur J Cell Biol. 1988;46:81–93. [PubMed] [Google Scholar]
- 36.Ritzenthaler C, Nebenführ A, Movafeghi A, Stussi-Garaud C, Behnia L, Pimpl P, Staehelin LA, Robinson DG. Plant Cell. 2002;14:237–261. doi: 10.1105/tpc.010237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokuyasu KT. J Microsc. 1986;143:139–149. doi: 10.1111/j.1365-2818.1986.tb02772.x. [DOI] [PubMed] [Google Scholar]
- 38.Hillmer S, Movafeghi A, Robinson DG, Hinz G. J Cell Biol. 2001;152:41–50. doi: 10.1083/jcb.152.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostermann J, Orci L, Tani K, Amherdt M, Ravazzola M, Elazar Z, Rothman JE. Cell. 1993;75:1015–1025. doi: 10.1016/0092-8674(93)90545-2. [DOI] [PubMed] [Google Scholar]
- 40.Zhang GF, Staehelin LA. Plant Physiol. 1992;99:1070–1083. doi: 10.1104/pp.99.3.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Phillipson BA, Pimpl P, daSilva LL, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J. Plant Cell. 2001;13:2005–2020. doi: 10.1105/TPC.010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otegui MS, Mastronarde DN, Kang BH, Bednarek SY, Staehelin LA. Plant Cell. 2001;13:2033–2051. doi: 10.1105/TPC.010150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segui-Simarro JM, Austin JR, II, White EA, Staehelin LA. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preuss ML, Serna J, Falbel TG, Bednarek SY, Nielsen E. Plant Cell. 2004;16:1589–1603. doi: 10.1105/tpc.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.