Abstract
Background and aims: Gastric extranodal marginal zone B cell lymphoma of the mucosa associated lymphoid tissue (MALT)-type (MZBL) is a rare complication of Helicobacter pylori infection. Currently, no bacterial factor has been associated with the development of this disease. Our aim was to identify genes associated with lymphoma development.
Methods: We used subtractive hybridisation as a tool for comparative genomics between H pylori strains isolated from a patient with gastric MZBL and from a patient with gastritis only.
Results: When gastric MZBL strains were compared with gastritis strains, two open reading frames (ORFs) were significantly associated with gastric MZBL: JHP950 (74.4% v 48.7%, respectively; p = 0.023) and JHP1462 (25.6% v 2.6%, respectively; p = 0.004). The prevalence of JHP950 was 48.8% (p = 0.024) in duodenal ulcer strains and 39.3% (p = 0.006) in gastric adenocarcinoma strains, which makes this ORF a specific marker for gastric MZBL strains. In contrast, the prevalence of JHP1462 was 16% (p = 0.545) and 35.7% (p = 0.429) in duodenal ulcer and adenocarcinoma strains, respectively. These ORFs were present in reference strain J99 but not in reference strain 26695. JHP950 is located in the plasticity zone whereas the other, JHP1462, is located outside. Both encode for H pylori putative proteins with unknown functions.
Conclusion: Despite its low prevalence, the ORF JHP1462 can be considered a candidate marker for H pylori strains involved in severe gastroduodenal diseases. In contrast, the ORF JHP950 has a high prevalence, and is the first candidate marker for strains giving rise to an increased risk of gastric MZBL strains. Further confirmation in other studies is needed.
Keywords: Helicobacter pylori, MZBL, MALT lymphoma, genetic marker
Helicobacter pylori is the main cause of gastritis and is involved in the pathogenesis of several gastrointestinal diseases—for example, duodenal ulcer and gastric cancer.1,2H pylori is the first bacterium identified as being carcinogenic in humans.3 It has been related to two types of stomach neoplasia: gastric adenocarcinoma and mucosa associated lymphoid tissue (MALT) lymphoma.4–6 The relationship of H pylori infection and extranodal marginal zone B cell lymphoma of the MALT-type (MZBL) is now well established. Indeed, H pylori is the cause of approximately 80% of gastric MZBL.7–9 Gastric MZBL arises from a H pylori associated chronic gastritis which consists of polymorph and lymphocyte infiltration, and evolves from an initially polyclonal lymphoid proliferation towards a monoclonal lymphoid population.10,11 This evolution is accompanied by genetic abnormalities.12,13 Gastric MZBL induction and growth is probably antigen driven, as studies have shown that H pylori strains isolated from patients with MZBL can induce lymphoid proliferation.14–16 Moreover, a causal association between H pylori infection and gastric MZBL has been proven by demonstrating that eradication of H pylori leads to regression of early gastric MZBL.9,17,18 These findings suggest that bacterial virulence factors may exist and could be associated with disease development, but they remain to be identified.
In a previous study, we investigated the prevalence of seven H pylori virulence factors (cagA, cagE, vacA, iceA, babA, hopQ, and oipA) and two novel adhesins sabA and hopZ in a large collection of H pylori gastric MZBL strains in comparison with gastritis strains. Our conclusion was that none of these genes, considered individually, was associated with MZBL.19 Nevertheless, a gene cluster comprised of iceA1 allele, sabA “on” status, and hopZ “off” status was identified in strains for which the odds of having a gastric MZBL was 10 times higher than that of the gastritis strains.19
Following sequencing of the whole genome of two H pylori strains (J99 and 26695), DNA arrays were used on various strains and the results showed that 22% of the genes were indeed strain specific.20–23 We postulated that among these inconstant genes, operating alone or clustered together, some could be involved in gastric MZBL development.
The aim of our study was to identify genetic markers associated with H pylori strains isolated from patients with gastric MZBL in comparison with strains obtained from age matched dyspeptic patients with gastritis only, using subtractive hybridisation, a technique which allows identification of genes or sequences present in a particular strain in comparison with a chosen control strain.
EXPERIMENTAL PROCEDURES
Bacterial strains
The H pylori strains used for subtractive hybridisation were: (1) a strain called “tester”, isolated from a 29 year old male patient with gastric MZBL; and (2) a strain chosen as a control, called “driver”, from a 74 year old male patient with gastritis but no lymphoid infiltration, atrophy, or intestinal metaplasia. These two strains presented a common profile for six known H pylori virulence genes: that is, cagA+, cagE+, vacAs1m1, babA2, iceA1, and oipA “on”, and one adhesin sabA “on”.
To evaluate the prevalence of the open reading frames (ORFs) identified from the “tester” strain in the subtractive library, a collection of 43 H pylori strains obtained from gastric MZBL patients (27 men and 16 women, mean age 48.2 (13.4) years were used. These strains were isolated from patients included in two French multicentre studies by the Groupe d’Etude Français des Lymphomes Digestifs (GELD) of the Fédération Francophone de Cancérologie Digestive and the Groupe d’Etude des Lymphomes de l’Adulte (GELA).9,24 In addition, 39 H pylori strains isolated from age matched dyspeptic patients with histological gastritis only (29 men and 10 women; mean age 48.3 (14.7) years) and 41 H pylori strains isolated from age matched duodenal ulcer patients (34 men and seven women; mean age 49.7 (14.2) years) were also included. Isolation of H pylori from gastric biopsy specimens was carried out using standard culture methods, as previously described.25
Genomic DNA was extracted from isolated colonies using a commercial kit (Qiagen SA, Courtaboeuf, France). The integrity of the genomic DNA template was controlled by polymerase chain reaction (PCR) with primers specific to a region of the 16S rDNA of H pylori.
Twenty eight DNA samples extracted from H pylori strains isolated from German patients with gastric adenocarcinoma (12 men and 16 women; mean age 64.3 (9.4) years) were also included in this study.
The two H pylori strains for which the genome has been sequenced, 26695 and J99, were used as reference strains.20,21
Subtractive hybridisation
The PCR select bacterial genome subtraction kit (Clontech, Palo Alto, California, USA) was used with the “tester” (gastric MZBL) and “driver” (gastritis) strains. The manufacturer’s recommendations were followed with the exception of the choice of the restriction enzyme and the method of evaluation of the subtractive efficacy. The restriction enzyme AluI was chosen because of its ability to generate DNA fragments, from “tester” and “driver” strains, in the same molecular weight range (from 2.5 kb to 0.25 kb). The efficacy of subtractive hybridisation was evaluated using amplification of a fragment of H pylori 16S rDNA in a real time PCR experiment in a LightCycler thermocycler (Roche Diagnostics, Neuilly sur Seine, France). Results were plotted versus time, represented by cycle number, to produce a measure of PCR amplification at the end of each PCR cycle. A difference of 20 threshold cycles was observed between subtracted and unsubtracted products, indicating an important reduction in 16S rDNA abundance in the subtracted sample and therefore very good subtractive hybridisation efficacy (data not shown).
After the last step, the subtractive experiment products were cloned in the pTAdv plasmid (Clontech) according to the manufacturer’s recommendations. A total of 139 recombinant bacteria were inoculated into LB medium with 50 mg/l kanamycin and grown overnight at 37°C under agitation. Inserts were amplified by PCR using the following protocol: 1 ml of incubated LB medium was heated for 10 minutes at 95°C and then centrifuged for five minutes at 14 000 rpm. The boiling lysis supernatants were collected and the insert size from each clone of the library was determined by PCR amplification using 5 µl of the supernatant and vector based primers. The average sizes of subtracted fragments varied from 300 to 1850 bp, in agreement with the initial digestive DNA library. DNA fragments were purified using Microspin S-400 HR columns (Amersham Pharmacia Biotech Inc., Uppsala, Sweden), and direct sequencing was performed with the ABI PRISM BigDye Terminators v3.0 Cycle Sequencing Kit (PE Applied Biosystems, Foster City, California, USA) using an ABI 3700 Analyzer DNA Sequencer (PE Applied Biosystems).
Screening of the subtractive library
To determine if the subtractive clones were really “tester” specific, primers for each clone were designed using the conserved sequence between the sequence identified and the corresponding gene of H pylori in database sequences (http:/www.probes.toulouse.inra.fr/multialin/multialin.hlml).26 The specificity of our primers was tested on both H pylori reference DNAs. For the sequences with no homology found in the databases, the specificity of the amplified fragment was determined by direct sequencing and dot blot hybridisation between “tester” (gastric MZBL) and “driver” (gastritis) DNAs, as described below.
If a PCR product was amplified from the “tester” DNA and not from the “driver” DNA, the sequence was considered to be “tester” specific. PCR amplifications were carried out in a 25 μl volume containing 2.5 μl of 10× PCR buffer (Eurobio, Les Ulis, France), 1.5 mM MgCl2 (Eurobio), 200 μM (each) of the deoxynucleoside triphosphates (Eurobio), 2 U of Taq DNA polymerase (Eurobio), 1 μM (each) of the primers (Q BIOgen, Strasbourg, France) and 10 ng of H pylori DNA. After four minutes of initial denaturation at 94°C, each reaction mixture was amplified for 35 cycles as follows: 30 seconds at 94°C, 30 seconds of annealing at 60°C, and extension at 72°C (the time depending on the length of the sequence to be amplified and the processivity of the Taq polymerase). After the last cycle, an extension was continued for another 7–10 minutes. PCR products were separated eletrophoretically on agarose gels and stained with ethidium bromide.
Dot blot hybridisation
Screening of the PCR sequences specific for the “tester” strain was performed using our collection of H pylori gastric MZBL and gastritis strains. Dot blot analyses were carried out using the same batch of membranes, prepared with the same dilution of DNA. Genomic DNA (100 ng) of each strain (MZBL, gastritis, J99, and 26695 reference strains) were spotted on a Hybond N+ membrane (Amersham Pharmacia Biotech) by means of a Bio-Rad dot blot 96 well filtration system apparatus (Bio-Rad, Ivry-Sur-Seine, France). Hybridisation was performed using digoxigenin labelled probes of each ORF identified, as described previously.27 The probes were prepared by PCR after primer design in a conserved area between the “tester” DNA sequence and the J99 reference strain or between both the J99, 26695, and “tester” strain when the DNA sequence was present in all.
Reverse transcription-PCR
RNA was extracted from the “tester” strain and the reference strain J99 during the exponential growth phase using the RNAeasy kit (Qiagen). RNA samples were treated with Rnase-Free Dnase (Promega, Madison, Wisconsin, USA) and quantified spectrometrically. Each reverse transcription PCR (RT-PCR) was carried out in a 10 µl volume containing 250 ng of RNA template, 0.6 µM of each oligonucleotide primer, 0.125 µl of RNAsine (Promega), 2 µl of 5× Q buffer, and 2 µl of RT-Taq mix. Complementary DNA was synthesised at 45°C for 30 minutes followed by 95°C heat inactivation of the RT Taq polymerase for 15 minutes, and then by the amplification program used for PCR amplification. Absence of DNA in RNA preparations was verified by omitting the RT amplification step. After 40 PCR cycles, products were separated electrophoretically on agarose gels.
Statistical analysis
Statistical analysis was performed using STATA 7.0 statistical software (Stata Corporation, College Station, Texas, USA). A first comparison of the characteristics between MZBL and gastritis strains was performed using Fisher’s exact test.
In order to create a variable describing how the different strain characteristics were associated, a multiple correspondence analysis (MCA) was performed followed by a cluster analysis using the average aggregation method and Euclidian distance.28 Both methods used the Statbox 2.5 program (Grimmer Logiciels, Paris, France).
The odds of an association with MZBL and its 95% confidence interval (CI) were compared within groups of strains and between the different strata of the summary variables obtained by the MCA.
RESULTS
PCR analysis of the subtractive clones
We sequenced the inserts of 132 clones on both strands, of which 81 were non-redundant. PCR screening between “tester” (gastric MZBL) and “driver” (gastritis) DNAs of these 81 clones showed that 39 (48.2%) contained sequences which were specific to the “tester” strain used in the experiment. Homologies of these clones with the H pylori databases (http://www.tigrblast.tigr.org/cmr-blast) and the NCBI blast program (http://www.ncbi.nlm.nih.gov/BLAST) were analysed. The following classification was made: 26 matched with sequences from the two reference strains (J99 and 26695), 10 matched with only J99 specific sequences, none matched with only strain 26695, and three presented no homology with H pylori reference strains.
For 10 of the 26 sequences which matched with the two reference strains, PCR products obtained from the “tester” DNA were different from the expected size when compared with the “driver” DNA and H pylori J99 and 26695 reference DNAs. Direct sequencing of these products showed significant variations, such as insertions or deletions, and therefore they were not included in the dot blot screening of our strain collections to avoid impairing the analysis. Nine other sequences where variation corresponded to intergenic regions were also not tested. The seven remaining sequences (clones A–G) (table 1 ▶) were distributed throughout the H pylori genome and were included in our dot blot experiments.
Table 1.
Summary of sequence analysis of “tester” specific DNA (gastric MZBL) fragments from Helicobacter pylori used for dot blot
| Clone | DNA match with | Function | MZBL strains (n = 43) | Gastritis strains (n = 39) | p Value | |||
| Strain J99 | Strain 26695 | n | % | n | % | |||
| A | JHP305 | HP322 | Putative poly E-rich protein† | 9 | 20.9 | 10 | 25.6 | 0.794 |
| B | JHP428 | HP476 | Predicted glutamyl-tRNA synthetase† | 41 | 95.4 | 38 | 97.4 | 1.000 |
| C | JHP1031 | HP1105 | Putative LPS biosynthesis protein | 30 | 69.8 | 32 | 82.1 | 0.212 |
| D | JHP1044 | HP1116 | Putative | 17 | 39.5 | 11 | 28.2 | 0.353 |
| E | JHP1285 | HP1371 | Putative | 39 | 90.7 | 34 | 87.2 | 0.730 |
| F | JHP1301 | HP1409 | Putative | 16 | 37.2 | 14 | 35.9 | 1.000 |
| G | JHP1424 | HP1402 | Type I RE (restriction subunit) | 40 | 93.0 | 33 | 84.6 | 0.297 |
| H | JHP820 | None | Predicted 1,2-glycosyltransferase† | 16 | 37.0 | 22 | 56.4 | 0.120 |
| I | JHP921* | None | Predicted DNA TC ComB8 homologue† | 21 | 48.8 | 14 | 35.9 | 0.270 |
| J | JHP922* | None | Predicted DNA TC ComB8 homologue† | 25 | 58.1 | 16 | 41.0 | 0.184 |
| K | JHP946* | None | Putative | 24 | 55.8 | 25 | 64.1 | 0.503 |
| L | JHP949* | None | Putative | 17 | 39.5 | 14 | 35.9 | 1.000 |
| M | JHP950* | None | Putative | 32 | 74.4 | 19 | 48.7 | 0.023 |
| N | JHP960* | None | Putative | 23 | 53.5 | 20 | 51.3 | 1.000 |
| JHP961* | None | Putative | 23 | 53.5 | 19 | 48.7 | 0.825 | |
| O | JHP1297 | None | Putative type III RE | 43 | 100.0 | 36 | 92.3 | 0.103 |
| P | JHP1409 | None | Putative type IIS DNA ME (methyltranferase) | 33 | 76.7 | 33 | 84.6 | 0.415 |
| Q | JHP1462 | None | Putative | 11 | 25.6 | 1 | 2.6 | 0.004 |
p values were determined by Fisher’s exact test.
*Open reading frame (ORF) localised in the plasticity zone.
†Revised annotation according to Boneca and colleagues.48
MZBL, extranodal marginal zone B cell lymphoma of the MALT-type; LPS, lipopolysaccharide; RE, restriction enzyme; ME, modification enzyme; TC, transformation competence.
Among the 10 sequences matching with only strain J99, six were localised in the plasticity zone (clones I–N) and one of these corresponded to a deletion of two ORFs (that is, JHP960 and JHP961). These two ORFs were therefore PCR amplified using J99 DNA and also tested by dot blot. The four remaining sequences (clones H, O, P, and Q) were distributed throughout the H pylori J99 genome.
Analysis of the three original sequences allowed us to consider them as potentially coding but they did not present nucleic and protein homologies in databases to assign them a function (to be published elsewhere). These three sequences were included in our dot blot experiment.
Dot blot analysis of the selected clones (MALT v gastritis)
The distribution of the 24 selected sequences was tested by dot blot using our collection of H pylori gastric MZBL and gastritis strains. The specificity of the sequences found to be “tester” specific by PCR was verified by (1) the absence of hybridisation on the plot corresponding to the “driver” strain and (2) the absence of hybridisation for the original sequences on the plots corresponding to the “driver” DNA, strain J99, and strain 26695. Indeed, negative results by PCR may be due to either an absence of the tested gene (true negative) or a lack of primer annealing due to interstrain variation in the sequences targeted by the primers. Accordingly, three sequences, which matched with the two reference strains and were initially classified by PCR as “tester” specific, were removed from our panel because of a positive hybridisation with the “driver” plot DNA.
The 21 remaining sequences were then tested. The corresponding genes and results obtained are summarised in table 1 ▶. The proportion of each ORF was compared between strains from patients with gastric MZBL and with gastritis only. Two ORFs were significantly associated with gastric MZBL strains: JHP950 (74.4% v 48.7%, respectively; p = 0.023) and JHP1462 (25.6% v 2.6%, respectively; p = 0.004). JHP950 was located in the plasticity region of strain J99 whereas JHP1462 was located outside. These two ORFs encoded for two H pylori specific proteins of unknown function.
Study of the transcription of JHP950 and JHP1462 ORFs in gastric MZBL strain
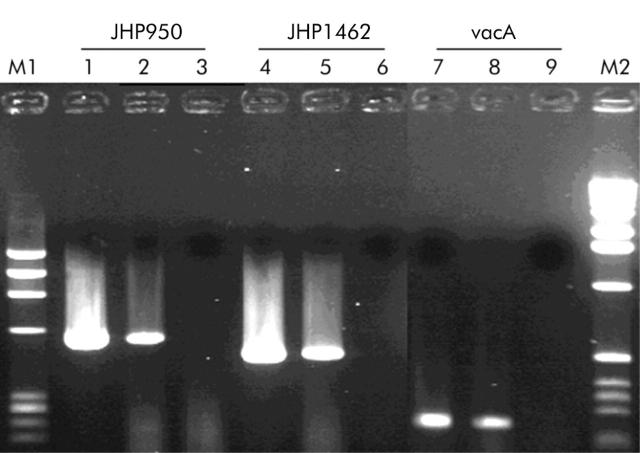
RT-PCR was performed for JHP950 and JHP1462 using the primers designed for PCR screening of our subtractive library in order to verify if they were transcribed in our “tester” strain. The vacA gene was included as a positive control of RT-PCR using primers previously published.29 The results showed that both were expressed (fig 1 ▶).
Figure 1.
Expression of the open reading frames (ORFs) JHP950 and JHP1462 of Helicobacter pylori determined by reverse transcription-polymerase chain reaction (RT-PCR) in the “tester” strain. Lanes M1 and M2 correspond to ΦX174 DNA/HaeIII markers (Eurobio) and to the 1 kb DNA ladder (Promega), respectively. Lanes 1, 4, and 7 correspond to PCR control on genomic DNA. Lanes 2, 5, and 8 correspond to RT-PCR product obtained on RNA extracted from the “tester” strain. Lanes 3, 6, and 9 correspond to PCR results on RNA preparations without the RT amplification step. Lanes 1–3, 4–6, and 7–9 correspond to templates amplified for JHP950, JHP1462, and vacA genes, respectively.
Multiple correspondence analysis
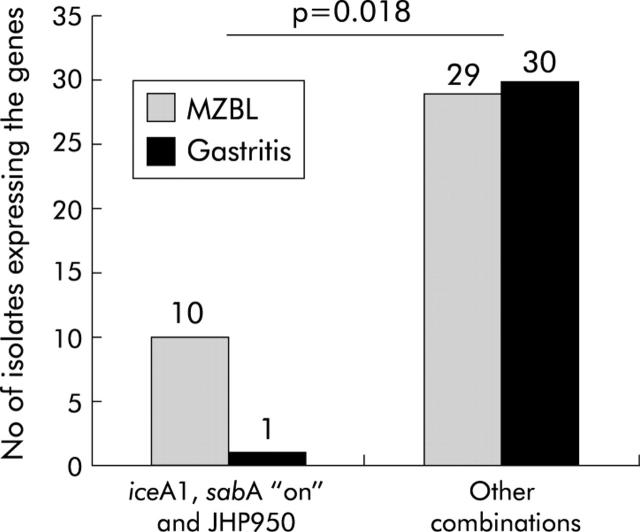
Firstly, an MCA was performed for all variables obtained from dot blot. We were unable to identify gene clusters which were significantly associated with gastric MZBL or gastritis strains. Thus the analysis was focused on the two ORFs found associated with the gastric MZBL strains. Because the prevalence of JHP1462 in gastric MZBL and gastritis strains was low, it was impossible to keep it in our final analysis. In contrast, the ORF JHP950 was included in the data from our previous study in which we identified a gene cluster in gastric MZBL strains, which is absent in gastritis strains, and containing iceA1, sabA “on”, and hopZ “off”.19 Interestingly, the dendrogram obtained from the results of the cluster analysis of the H pylori gastric MZBL strain population showed that the ORF JHP950 was included in the formally identified gene cluster (data not shown). The possible association between the presence of gastric MZBL and the characteristics of strains cultured from these patients was tested. On analysis of the summary variables according to the MCA results, it was remarkable that the odds of having gastric MZBL among patients harbouring JHP950, iceA1, and sabA “on” strains were 10 times higher than for the other defined groups (odds ratio 10.3 (95% CI 1.2–86.0)) (table 2 ▶); indeed among a total of 11 strains having these three markers, 10 (90.9%) corresponded to gastric MZBL strains (p = 0.018) (fig 2 ▶).
Table 2.
Strength of the association of Helicobacter pylori genotypes and extranodal marginal zone B cell lymphoma of the MALT-type (gastric MZBL) status versus gastritis status
| Total No of patients | Patients with MZBL | Patients with gastritis | OR (95% CI) | |
| JHP950 | 82 | 43 | 39 | |
| Absent | 31 | 11 | 20 | |
| Present | 51 | 32 | 19 | 3.1 (1.2–7.8) |
| MCA summary variables | 70 | 39 | 31 | |
| Others | 59 | 29 | 30 | |
| iceA1, sabA “on”, and JHP950+ | 11 | 10 | 1 | 10.3 (1.2–86.0) |
OR, odds ratio, presented as univariate results; CI, exact binomial 95% confidence interval; MCA, multiple correspondence analysis.
Figure 2.
Distribution of the combination of iceA1 allele, sabA functional status, and the open reading frame (ORF) JHP950 according to Helicobacter pylori strains isolated from patients with gastric extranodal marginal zone B cell lymphoma of the MALT-type (gastric MZBL) or gastritis alone.
Verification of the specificity of the two putative markers
The specificity of the association of the ORFs JHP950 and JHP1462 with gastric MZBL strains was evaluated by determining their prevalence in a collection of duodenal ulcer strains (n = 41) by dot blot analysis, as described above. The prevalence in duodenal ulcer strains of OFRs JHP950 and JHP1462 was 48.8% (p = 0.024) and 16% (p = 0.545), respectively.
The prevalence of ORFs JHP950 and JHP1462 on strains isolated from 28 gastric adenocarcinoma was also determined. As DNA obtained from these strains was not concentrated enough to perform dot blot analysis, PCR screening was performed with the primers used for dot blot probe synthesis. A prevalence of 39.3% (p = 0.006) for JHP950 and of 35.7% (p = 0.429) for JHP1462 ORFs was found (table 3 ▶).
Table 3.
Prevalence of JHP950 and JHP1462 open reading frames (ORFs) in Helicobacter pylori strains isolated from patients with gastritis, duodenal ulcer, and gastric adenocarcinoma in comparison with strains isolated from patients with gastric MZBL
| Gastritis | Duodenal ulcer | Gastric adenocarcinoma | Gastric MZBL | |
| JHP950 | 48.7 (p = 0.023)* | 48.8 (p = 0.024)* | 39.3 (p = 0.006)* | 74.4 |
| JHP1462 | 2.6 (p = 0.004)* | 16.0 (p = 0.545) | 35.7 (p = 0.429) | 25.6 |
MZBL, marginal zone B cell lymphoma of the MALT-type.
p values were determined by Fisher’s exact test.
*Significant values versus gastric MZBL strains.
These results confirm that JHP950 is a specific marker for H pylori strains isolated from gastric MZBL (table 3 ▶).
DISCUSSION
Following sequencing of the whole genome of two strains of H pylori, the possibility arose to study the genetic diversity of H pylori strains using DNA arrays.23 Indeed, this approach is tempting but limits the possibilities to the genes already known. Our hypothesis was that H pylori pathogenesis in gastric MZBL could be linked to currently unidentified bacterial factors, and therefore that subtractive hybridisation represented the most accurate approach.
Subtractive hybridisation is now considered an important tool for comparative prokaryotic genomics.30 Most of the studies performed previously compared bacterial strains with apparent enhanced capacity for human infection and/or interpatient transmission and colonisation. For example, genomic differences have been identified in a virulent strain of Klebsiella pneumoniae as well as in the uropathogenic Escherichia coli strain 536 compared with the non-pathogenic strain MG1655, and more recently a genomic locus was identified and characterised in the Brazilian pupuric fever clonal group of Haemophilus influenzae biogroup aegyptius.31–33 Subtractive hybridisation has also been used to compare bacteria from different species in order to identify nucleotide specific sequences which could provide a basis for developing highly specific detection systems: for example, in Yersinia pestis, Salmonella enterica serovar Enteritidis, or Burkholderia cepacia genovar III strains.34–36 Concerning H pylori, representational difference analysis, a method similar to subtractive hybridisation, led to the to discovery of the cag pathogenicity island.37 Subtractive hybridisation was later validated by Akopyants et al and allowed identification of a 2 kb transposable element named IS607, as well as more recently a new type IV secretion system present in the plasticity zone of the H pylori genome.38–40
In a subtractive hybridisation experiment, the choice of strains to be compared is particularly important. Considering the fact that the natural evolution of H pylori infection begins with gastritis and moreover that host susceptibility factors of developing gastric MZBL are currently unknown, this choice is especially difficult. We based our choice on the pathology (gastric MZBL versus gastritis) and age of the patients from whom the strains were isolated (29 year old patient with gastric MZBL, 74 year old patient suffering from gastritis). The advanced age of the “driver” strain patient was chosen to minimise the possibility that gastritis would evolve to more severe gastrointestinal disease. As the mean age of patients suffering from gastric MZBL is close to 50 years, we could anticipate a particular pathogenic property in the tester strain issued from a much younger patient. Furthermore, the two strains were chosen because of their common profile for six known H pylori virulence genes—that is, cagA+, cagE+, vacAs1m1, babA2, iceA1, and oipA “on”, and one adhesin sabA “on”.
MCA analysis, initially carried out on all of our dot blot data, failed to identify a gene cluster associated with gastric MZBL strains. However, despite the fact that our collection of gastric MZBL strains is the largest of those published, the number of strains included in this study may not be sufficient to efficiently perform MCA.
The first category of sequences identified presented significant homologies with genes present in strains J99 and 26695 but with important deletions or insertions. Such variations can modify the functional status of a gene or the functionality of the deduced protein. Moreover, it has been suggested that mixtures of matching and divergent sequences can facilitate quite dramatic changes in bacterial phenotype.38 We originally expected to find new H pylori genes and indeed we identified three sequences of unknown genomic localisation, corresponding to new putative coding regions. Nevertheless, none of these sequences was significantly associated with gastric MZBL. Such results could not have been found using a standard approach for detecting genetic diversity, and thus our findings provide more information for the future in investigating the particular properties of these strains.
The results of the dot blot experiments (MALT versus gastritis) highlighted two ORFs (JHP950 and JHP1462). Globally, repartition of both ORFs (JHP950 and JHP1462) was statistically different (p = 0.016 and p = 0.002, respectively) when comparing the four H pylori strain collections studied.
The prevalence of ORF JHP1462 (table 3 ▶) in MZBL strains was indeed low (25.6%) but when compared with gastritis strains the difference was highly significant (p = 0.004). However, no significant difference was found between gastric MZBL strains and duodenal ulcer and gastric adenocarcinoma strains (table 3 ▶), indicating that JHP1462 is not a specific genetic marker for H pylori gastric MZBL strains but could be considered more generally as a candidate marker for H pylori strains associated with severe gastroduodenal diseases. It is located outside the plasticity zone, in common with two thirds of H pylori strain specific genes.
In contrast, the prevalence of the JHP950 ORF in gastric MZBL strains was significantly higher than in strains isolated from duodenal ulcer and gastric adenocarcinoma. The difference in prevalence for this marker was in the same range as the difference between the prevalence of cagA in duodenal ulcer and gastric adenocarcinoma strains compared with gastritis strains. Indeed, in the past we looked for the cagA gene in relation to clinical presentation in a collection of 167 H pylori strains. The cagA gene was found in 82.6% of duodenal ulcer strains versus 63.5% of gastritis strains.41 These results were confirmed later in several European studies.42–45 The cagA gene was first considered as a marker of virulence for H pylori. Indeed, following the discovery of the cag pathogenicity island and of the interaction of the CagA protein with the epithelial cell, it is now considered to be a true virulence factor associated with severe clinical outcome (that is, duodenal ulceration and gastric adenocarcinoma).37,46,47 Similarly, the ORF JHP950 is part of the plasticity zone of strain J99. This region is approximately 45 kb long in strain J99 and 68 kb long in strain 26695, and contains one third of the strain specific genes. Occhialini et al studied several ORFs in the plasticity region of strain J99 by hybridisation and found that most were functional.27 We verified by RT-PCR that JHP950 and JHP1462 were transcribed. They were originally annotated as H pylori specific proteins with unknown functions (that is, they have no orthologs in other bacterial sequences). This classification is still accurate after publication of a revised annotation and comparative analysis of H pylori genomes.48 Our results are in line with the study of Salama et al who found that the most important class of strain specific genes was more frequent among strain specific genes (58%) than in the core of the genome (21%) which contains genes encoding, for example, metabolic functions.23
A limit of our study was the use of PCR instead of hybridisation for detection of the ORFs JHP950 and JHP1462 in adenocarcinoma strains. Indeed, there are very few collections of H pylori strains obtained from patients with gastric adenocarcinoma in Europe. The DNA material obtained did not allow us to perform dot blot hybridisation but only detection by PCR. The latter technique can had to false negative results because of a lack of primer binding due to limited differences in sequences. Therefore, we tested the accuracy of our method on our strain collections and found a sensitivity of 90% (JHP950) and 95% (JHP1462) versus dot blot hybridisation, which confirms the relevance of our approach.
MCA analysis allowed us to show that the JHP950 marker is part of a gene cluster significantly associated with gastric MZBL strains. This gene cluster, which is comprised of JHP950, iceA1 alleles, and a sabA “on” status, partially validates our initial findings.19 The sabA gene encodes for an adhesin. The association of JHP950 with iceA1, a gene activated by contact with the gastric epithelium, and an adhesin seems to be in line with the function of these genes.49,50 This underlines the fact that JHP950 might have functional importance during interaction of the bacterium with cells. We can hypothesise that if JHP950 corresponds to a new H pylori virulence factor, H pylori strains harbouring this triple association of genes could have a pathogenic advantage. Indeed, the odds of having gastric MZBL among patients harbouring JHP950, iceA1, and sabA “on” strains were 10 times higher than for the other defined groups. Koehler et al recently reported that the vacA m2 allele was the predominant subtype in MALT lymphoma.51 We also previously reported a slight association between m2 genotype and MALT strains (when considering the m genotype only) but we found no association between vacA s1m2 alleles and iceA1 genotypes.19 However, we share the similar conclusion (that is, certain H pylori subtype combinations have a predictive value for the development of gastric MZBL).
In conclusion, if JHP1462 cannot be considered a specific genetic marker of gastric MZBL strains but rather as a new candidate marker of H pylori virulent strains in general, in contrast JHP950 can be considered as the first candidate marker of gastric MZBL H pylori strains. The association of JHP950 with gastric MZBL strains is similar to that between cagA and duodenal ulcer strains. Moreover, JHP950 forms a gene cluster with iceA1 and sabA “on” which is associated with gastric MZBL strains. Future studies are needed to confirm this association and to understand the biological function of this ORF in order to gain insight into the pathogenesis of H pylori and the development of gastric MZBL.
Acknowledgments
The authors thank the Groupe d’Etude Français des Lymphomes Digestifs (GELD) of the Fédération Français de Cancérologie Digestive, the Délégation de la Recherche Clinique of the Assistance Publique-Hôpitaux de Paris (AP-HP), the Groupe d’Etude des Lymphomes de l’Adulte (GELA) for supplying H pylori strains, and the Conseil Régional d’Aquitaine and the Fonds de Recherche of the Societé Nationale Française de Gastroentérologie for financially supporting the project.
Abbreviations
RT-PCR, reverse transcription-polymerase chain reaction
MALT, mucosa associated lymphoid tissue
gastric MZBL, gastric extranodal marginal zone B cell lymphoma of the MALT-type
MCA, multiple correspondence analysis
ORF, open reading frame
REFERENCES
- 1.Mégraud F , Lamouliatte H. Helicobacter pylori and duodenal ulcer—Evidence suggesting causation. Dig Dis Sci 1992;37:769–72. [DOI] [PubMed] [Google Scholar]
- 2.Uemura N , Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med 2001;345:784–9. [DOI] [PubMed] [Google Scholar]
- 3.IARC. Schistosomes, liver flukes and Helicobacter pylori. In: Monographs on the evaluation of carcinogenic risks to humans. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, vol 61. Lyon: IARC, 1994:1–241. [PMC free article] [PubMed]
- 4.Correa P , Fox J, Fontham E, et al. Helicobacter pylori and gastric carcinoma. Cancer 1990;66:2569–74. [DOI] [PubMed] [Google Scholar]
- 5.Forman D , Webb P, Parsonnet J. Helicobacter pylori and gastric cancer. Lancet 1994;34:243–4. [PubMed] [Google Scholar]
- 6.Parsonnet J , Hansen S, Rodriguez L, et al. Helicobacter pylori infection and gastric lymphoma. N Engl J Med 1994;330:1267–71. [DOI] [PubMed] [Google Scholar]
- 7.Seydel J , Ullrich A, Bender R, et al. Helicobacter pylori and carcinogenesis of gastric B-cell lymphomas. Int J Cancer 2003;104:646–49. [DOI] [PubMed] [Google Scholar]
- 8.Ullrich A , Fischbach W, Blettner M. Incidence of gastric B-cell lymphomas: a population-based study in Germany. Ann Oncol 2002;13:1120–7. [DOI] [PubMed] [Google Scholar]
- 9.Ruskoné Fourmestraux A , Lavergne A, Aegerter PH, et al. Predictive factors for regression of gastric MALT lymphoma after anti-Helicobacter pylori treatment. Gut 2001;48:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsushima Y , Kinoshita Y, Fukui H, et al. Immunological and molecular analysis of B lymphocytes in low-grade MALT lymphoma of the stomach. Are there any useful markers for predicting outcome after Helicobacter pylori eradication? J Gastroenterol 2002;37:428–33. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto M , Haruma K, Hiyama T, et al. High incidence of B-cell monoclonality in follicular gastritis: a possible association between follicular gastritis and MALT lymphoma. Virchows Arch 2002;440:376–80. [DOI] [PubMed] [Google Scholar]
- 12.Liu HX, Ruskone-Fourmestraux A, LavergneSlove A, et al. Resistance of t(11;18) positive gastric mucosa-associated lymphoid tissue lymphoma to Helicobacter pylori eradication therapy. Lancet 2001;357:39–40. [DOI] [PubMed] [Google Scholar]
- 13.Morgner A , Bayerdorffer E, Neubauer A, et al. Helicobacter pylori associated gastric B cell MALT lymphoma: predictive factors for regression. Gut 2001;48:290–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussell T , Issacson PG, Crabtree JE, et al. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 1993;342:571–4. [DOI] [PubMed] [Google Scholar]
- 15.Hussell T , Isaacson PG, Crabtree JE, et al. Helicobacter pylori-specific tumour-infiltrating T cells provide contact dependent help for the growth of malignant B cells in low-grade gastric lymphoma of mucosa-associated lymphoid tissue. J Pathol 1996;178:122–7. [DOI] [PubMed] [Google Scholar]
- 16.D’Elios MM, Amedei A, Manghetti M, et al. Impaired T-cell regulation of B-cell growth in Helicobacter pylori-related gastric low-grade MALT lymphoma. Gastroenterology 1999;117:1105–12. [DOI] [PubMed] [Google Scholar]
- 17.Wotherspoon AC, Doglioni C, Diss TC, et al. Regression of primary low-grade B-cell gastric lymphoma of mucosa-associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 1993;342:575–7. [DOI] [PubMed] [Google Scholar]
- 18.Ruskoné Fourmestraux A , Rambaud JC. Gastrointestinal lymphoma: prevention and treatment of early lesions. Best Pract Res Clin Gastroenterol 2001;15:337–54. [DOI] [PubMed] [Google Scholar]
- 19.Lehours P , Ménard A, Dupouy S, et al. Evaluation of the association of nine Helicobacter pylori virulence factors with strains involved in low grade gastric MALT lymphoma. Infect Immun 2004;72:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomb JF, White O, Kerlavage AR, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 1997;388:539–47. [DOI] [PubMed] [Google Scholar]
- 21.Alm RA, Ling LSL, Moir DT, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 1999;397:176–80. [DOI] [PubMed] [Google Scholar]
- 22.Alm RA, Trust TJ. Analysis of the genetic diversity of Helicobacter pylori: the tale of two genomes. J Mol Med 1999;77:834–46. [DOI] [PubMed] [Google Scholar]
- 23.Salama N , Guillemin K, McDaniel TK, et al. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc Natl Acad Sci U S A 2000;97:14668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy M , Copie Bergman C, Traulle C, et al. Conservative treatment of primary gastric low-grade B-cell lymphoma of mucosa-associated lymphoid tissue: Predictive factors of response and outcome. Am J Gastroenterol 2002;97:292–7. [DOI] [PubMed] [Google Scholar]
- 25.Lehours P , Ruskone Fourmestraux A, Lavergne A, et al. Which test to use to detect Helicobacter pylori infection in patients with low-grade gastric mucosa-associated lymphoid, tissue lymphoma? Am J Gastroenterol 2003;98:291–5. [DOI] [PubMed] [Google Scholar]
- 26.Corpet F . Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988;16:10881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Occhialini A , Marais A, Alm R, et al. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun 2000;68:6240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouanet H , Le Roux B. Analyse des données multidimensionnelles. Paris: Dunod, 1993.
- 29.Atherton JC, Cover TL, Twells RJ, et al. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J Clin Microbiol 1999;37:2979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Agron PG, Macht M, Radnedge L, et al. Use of subtractive hybridization for comprehensive surveys of prokaryotic genome differences. FEMS Microbiol Lett 2002;211:175–82. [DOI] [PubMed] [Google Scholar]
- 31.Lai YC, Yang SL, Peng HL, et al. Identification of genes present specifically in a virulent strain of Klebsiella pneumoniae. Infect Immun 2000;68:7149–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janke B , Hacker J, Blum-Oehler G. Genetic characterization of the uropathogenic E. coli strain 536—a subtractive hybridization analysis. Adv Exp Med Biol 2000;485:53–6. [DOI] [PubMed] [Google Scholar]
- 33.Li MS, Farrant JL, Langford PR, et al. Identification and characterization of genomic loci unique to the Brazilian purpuric fever clonal group of H. influenzae biogroup aegyptius: functionality explored using meningococcal homology, Mol Microbiol 2003;47:1101–11. [DOI] [PubMed] [Google Scholar]
- 34.Radnedge L , Gamez-Chin S, McCready PM, et al. Identification of nucleotide sequences for the specific and rapid detection of Yersinia pestis. Appl Environ Microbiol 2001;67:3759–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agron PG, Walker RL, Kinde H, et al. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl Environ Microbiol 2001;67:4984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L , Spilker T, Coenye T, et al. Identification by subtractive hybridization of a novel insertion element specific for two widespread Burkholderia cepacia genomovar III strains. J Clin Microbiol 2003;41:2471–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Censini S , Lange C, Xiang Z, et al.cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci U S A 1996;93:14648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akopyants NS, Fradkov A, Diatchenko L, et al. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc Natl Acad Sci U S A 1998;95:13108–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kersulyte D , Mukhopadhyay AK, Shirai M, et al. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J Bacteriol 2000;182:5300–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersulyte D , Velapatino B, Mukhopadhyay AK, et al. Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J Bacteriol 2003;185:3764–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labigne A , Lamouliatte H, Birac C, et al. Distribution of the cagA gene among Helicobacter pylori strains associated with peptic ulcer. Am J Gastroenterol 1994;89:1326. [Google Scholar]
- 42.Gerhard M , Lehn N, Neumayer N, et al. Clinical relevance of the Helicobacter pylori gene for blood-group antigen-binding adhesin. Proc Natl Acad Sci U S A 1999;96:12778–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bach S , Makristathis A, Pinto A, et al. Helicobacter pylori type I strains among Austrian and Portuguese patients with gastritis, peptic ulcer or gastric cancer. Eur J Clin Microbiol Infect Dis 1999;18:807–10. [DOI] [PubMed] [Google Scholar]
- 44.Palli D , Menegatti M, Masala G, et al. Helicobacter pylori infection, anti-cagA antibodies and peptic ulcer: a case-control study in Italy. Aliment Pharmacol Ther 2002;16:1015–20. [DOI] [PubMed] [Google Scholar]
- 45.van Doorn L - J, Figueiredo C, Sanna R, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology 1998;115:58–66. [DOI] [PubMed] [Google Scholar]
- 46.Covacci A , Censini S, Bugnoli M, et al. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Nat Acad Sci U S A 1993;90:5791–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akopyants NS, Clifton SW, Kersulyte D, et al. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol 1998;28:37–53. [DOI] [PubMed] [Google Scholar]
- 48.Boneca IG, de Reuse H, Epinat J-C, et al. A revised annotation and comparative analysis of Helicobacter pylori genomes. Nucleic Acids Res 2003;31:1704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peek RMJ, Thompson SA, Donahue JP, et al. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc Am Soc Phys 1998;110:531–44. [PubMed] [Google Scholar]
- 50.Mahdavi J , Sonden B, Hurtig M, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 2002;297:573–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koehler CI, Mues MB, Dienes HP, et al. Helicobacter pylori genotyping in gastric adenocarcinoma and MALT lymphoma by multiplex PCR analyses of paraffin wax embedded tissues. J Clin Pathol Mol Pathol 2003;56:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]