Abstract
We formulate a general analysis to determine the two-dimensional dissociation constant (2D Kd), and use this method to study the interaction of CD2-expressing T cells with glass-supported planar bilayers containing fluorescently labeled CD58, a CD2 counter-receptor. Both CD2 and CD58 are laterally mobile in their respective membranes. Adhesion is indicated by accumulation of CD2 and CD58 in the cell-bilayer contact area; adhesion molecule density and contact area size attain equilibrium within 40 min. The standard (Scatchard) analysis of solution-phase binding is not applicable to the case of laterally mobile adhesion molecules due to the dynamic nature of the interaction. We derive a new binding equation, B/F = [(Nt × f)/(Kd × Scell)] − [(B × p)/Kd], where B and F are bound and free CD58 density in the contact area, respectively; Nt is CD2 molecule number per cell; f is CD2 fractional mobility; Scell is cell surface area; and p is the ratio of contact area at equilibrium to Scell. We use this analysis to determine that the 2D Kd for CD2-CD58 is 5.4–7.6 molecules/μm2. 2D Kd analysis provides a general and quantitative measure of the mechanisms regulating cell-cell adhesion.
INTRODUCTION
Cell membranes exert a powerful organizing influence on biologically important chemical reactions, and this organization is critically important for adhesive interactions. Proteins that are wholly or partially embedded in cellular membranes are confined to diffuse laterally in two dimensions. Interactions between proteins at the interface between two apposing membranes are governed by two-dimensional reaction rates and two-dimensional dissociation constants (1). Although the chemical properties of these interactions are of great importance, little experimental work has been performed focusing on the chemical nature of these pro-adhesive molecular interactions in the native membrane environment. In the immune system, antigen-independent adhesive interactions between T cells and antigen-presenting cells (APCs) are required before the interaction between T cell antigen receptors and antigenic peptide-major histocompatibility complex (MHC) protein complexes. Among the adhesion molecules promoting the cell-cell interaction between T cells and APCs are the T cell surface molecule CD2 and its widely expressed counter-receptor CD58 (2,3). Other antigen-independent adhesion molecules at the T cell surface are LFA-1, CD28, the intercellular adhesion molecules (ICAMs), and CD80/CD86 (4–8). Antigen-independent interactions not only enhance the strength of cell-cell adhesion but also provide important costimulatory signals to the T cell (9–12). This study focuses on the chemical nature of the CD2-CD58 binding interaction in the near two-dimensional interface between a T cell and a model membrane. Importantly, we develop a general methodology for dissecting the contributions of receptor affinity and mobility to this dynamic interaction.
CD2 and CD58 are members of the immunoglobulin (Ig) superfamily of proteins. Each molecule consists of two N-terminal Ig-like domains and a C-terminal membrane-anchoring structure: CD2 is a transmembrane receptor with a cytoplasmic tail, and CD58 has either a glycosylphosphatidylinositol (GPI) anchor or a transmembrane anchoring domain (13). The extracellular domains of CD2 and CD58 are similar in size to those of the T cell antigen receptor and the MHC proteins; both the CD2-CD58 and the T cell receptor-MHC interactions are predicted to require an intermembrane separation of ∼15 nm (14). This observation suggests that adhesive interactions between CD2 and CD58 may be an ideal accessory for antigen recognition, thus increasing the sensitivity of the latter process. The context provided by coreceptors such as CD2-CD58 could be essential for the process by which a remarkably low number of MHC-peptide complexes is required to trigger a T cell response (15). Indeed, it is well known that the CD2-CD58 interaction increases the sensitivity of T cells to antigen (16). Developing a quantitative model of the CD2-CD58 binding interaction would be relevant to understanding the context of fundamental antigen recognition processes.
The CD2-CD58 interaction has been well defined in solution. The interaction is notable for its low affinity (Kd estimates range from 2 to 9–22 μM) and rapid dissociation rate (koff > 5 s−1) (17–19). The relationship between the three-dimensional Kd (3D Kd) measured in these studies and the two-dimensional Kd (2D Kd) that governs the interaction in cell-cell contact areas is not obvious, however, because both receptors and ligands are confined to translate in only two dimensions. We have therefore set out to analyze the 2D Kd for the CD2-CD58 interaction directly, using as a model system the interaction between T cells and a glass-supported planar bilayer reconstituted with purified fluorescently labeled CD58. Accumulation of fluorescent CD58 in the contact area between T cell and bilayer is visualized by fluorescence microscopy, and radiolabeled antibody binding is used to relate the fluorescence intensity to molecular surface densities. In previous studies, we have fitted such data using a simple Scatchard analysis (20,21). However, we now introduce an analysis that accounts for the lateral mobility of CD2 in the T cell membrane (22) and for the accumulation of CD2 in the contact area between the T cell and the bilayer. The dynamic nature of the CD2 receptor density in the contact area during the equilibrium binding measurement violates a fundamental assumption underlying the Scatchard analysis. Here, we derive a general analysis of the interaction between laterally mobile receptors at an interface of two membranes. This analysis, which we have termed the Zhu-Golan analysis, considers the accumulation of both receptors and ligands within the contact area. We apply this analysis to the CD2-CD58 system, and obtain results that are strikingly different from those obtained using the previous Scatchard analysis. Because many adhesion molecules exhibit lateral mobility in cell membranes, our general analysis provides a quantitative method for determining the physiologically relevant 2D Kd from dynamic measurements of receptor-ligand interactions in single cells.
EXPERIMENTAL PROCEDURES
Cells and monoclonal antibodies
The Jurkat E6.1 T leukemia cell line (American Type Culture Collection, Rockville, MD) was maintained in RPMI 1640 medium (Sigma, St. Louis, MO) supplemented with 10% heat-inactivated fetal bovine serum (Sigma) and 2 mM L-glutamine/100 units/mL penicillin/0.1 mg/mL streptomycin (Sigma) at 37°C in a humidified atmosphere of 5% CO2. Human peripheral blood lymphocytes were purified as follows: Whole blood was layered in Accuspin System-Histopaque-1077 centrifuge tubes (Sigma) and centrifuged at 2450 rpm (1000 × g) for 10 min at 21°C. Mononuclear cells (MNCs) formed a single band at the Histopaque-plasma interface, while red cells and granulocytes passed through the filter barrier and formed a pellet at the bottom of the tube. MNCs were carefully aspirated using a Pasteur pipette; this fraction contained >87% lymphocytes as assessed by Wright's staining and differential counting. MNCs were suspended in 20 mL of RPMI 1640 supplemented with 10% autologous plasma and the L-glutamine-penicillin-streptomycin solution described above. Ten milliliters of cell suspension was transferred to a 100 × 20 mm culture dish and incubated for 24 h at 37°C in a humidified incubator. The nonadherent cells were collected by gently rocking the plate and centrifuged for 10 min at 1500 rpm (100 × g); this procedure resulted in recovery of a 98% pure population of lymphocytes in the cell pellet as assessed by Wright's staining and confirmed by immunophenotyping.
Anti-CD2 and anti-CD58 specific monoclonal antibodies (mAbs) were produced from hybridoma culture supernatants. TS2/18 and 35.1 are adhesion-blocking anti-CD2 mAbs (23); CD2.1 is a nonblocking anti-CD2 mAb (24); and TS2/9 is an adhesion-blocking anti-CD58 mAb (23).
Preparation of planar bilayer reconstituted with FITC-CD58
GPI-CD58 was conjugated to fluorescein isothiocyanate (FITC) as previously described (20). To remove additional free FITC, the FITC-CD58 conjugate was subjected to ultrafiltration using a 30,000 MW cutoff membrane (Centricon, Amicon, Beverly, MA) immediately after elution from the affinity column.
Egg phosphatidylcholine (PC) (Avanti Polar Lipids, Alabaster, AL) unilamellar liposomes were reconstituted with FITC-CD58, as described (20). Glass-supported planar bilayers were formed by fusing egg PC liposomes onto round glass coverslips (25 mm diameter, Fisher, Pittsburgh, PA). Coverslips were boiled in detergent solution (Linbro 7×, ICN Pharmaceuticals, Costa Mesa, CA) for 30 min, washed with distilled water for 30 min, and stored in 100% ethanol. For experiments, a coverslip was removed from the ethanol and dried using argon, then a scratch was placed by a diamond pencil at the center of the coverslip. This scratch helped to localize the focal plane of the microscope at the bilayer. A liposome droplet (10–15 μL) was deposited at the bottom of a plastic petri dish (50 × 9 mm), and the coverslip was placed on top of the droplet with its scratched surface down. After a 30-min incubation, 3 mL of binding buffer containing 25 mM HEPES (pH 7.4), 147 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, 1.8 mM CaCl2, 5 mM D-glucose, and 1% bovine serum albumin (BSA) (Calbiochem, San Diego, CA) was added to the dish. The coverslip was released from the bottom of the dish by gentle shaking. The coverslip was washed in binding buffer five times, then the dish containing the coverslip was immersed in a glass tank containing 1 liter of binding buffer. A stainless steel slide with a 15-mm diameter hole at the center was engineered to mount the bilayer-bearing coverslip. The slide was coated with vacuum grease on both sides around the hole, and a coverslip was gently pressed on the grease to seal one side of the hole. The slide was then placed into the tank with the opening of the hole facing up. The bilayer-bearing coverslip was carefully placed on the other side of the metal slide, with the bilayer face-down, and gently pressed on the grease to a firm seal. The sandwich-like slide assembly was removed from the tank and rinsed with distilled water. The nonbilayer coverslip was carefully slid off the opening to add cells and/or reagents to the sample; this coverslip was then slid back and pressed to seal the chamber again. Planar bilayers were never exposed to air.
The uniformity and continuity of the planar bilayer was confirmed as follows. When bilayers containing fluorescein-labeled diacylphosphatidylethanolamine (Avanti Polar Lipids) were incubated with anti-fluorescein mAb (Molecular Probes, Eugene, OR), 48% of the fluorescence was quenched by the antibody (data not shown). This observation, which indicated that approximately half of the fluorescent lipid molecules were capable of interaction with the antibody while the other half were not, suggested that the membrane was a unilamellar bilayer rather than a monolayer or a multilamellar structure. Interestingly, FITC-labeled GPI-CD58 in the bilayer was completely quenched by anti-FITC mAb (Sigma), suggesting that all of the GPI-CD58 (hereafter called CD58) molecules were re-oriented to the upper leaflet of the bilayer (i.e., to the leaflet facing the aqueous solution) upon fusion of the liposomes into a planar bilayer, and that all of the FITC-GPI-CD58 (hereafter called FITC-CD58) in the bilayer would be capable of interaction with CD2 molecules expressed on the surface of T cells. This information was important for accurate determination of the effective free CD58 density in the bilayer.
Image analysis
Two-dimensional 180 × 180 μm fluorescence images of FITC-CD58 redistribution in the egg PC bilayer were scanned using a step size of 0.6 μm. A scratch on the coverslip was used to focus the microscope coarsely at the surface of the coverslip; the focus was then finely adjusted to obtain the maximum fluorescence intensity. Cellular autofluorescence and background fluorescence was obtained by scanning the contact area between cells and a bilayer containing unlabeled CD58. To define the contact area, the fluorescence intensity threshold for each image was set equal to the sum of the cellular autofluorescence/background fluorescence and the free FITC-CD58 fluorescence intensity (F), where F was measured from a cell-free area of the bilayer. Contact area determinations using this method were generally in good agreement with those determined using interference reflection microscopy (25). The fluorescence intensity was converted to an absolute surface density of CD58 in the bilayer by using a standard curve of fluorescence intensity versus FITC-CD58 density. The standard curve was obtained with bilayers prepared from vesicles containing FITC-CD58 at a density that was measured using the 125I-labeled anti-CD58 mAb TS2/9. Cellular autofluorescence/background fluorescence and free FITC-CD58 fluorescence (F) were subtracted from the fluorescence intensity in the cell-bilayer contact area to yield the surface density of bound CD2-CD58 complexes (B). There was no detectable nonspecific binding of cells to CD58-containing bilayers when the CD2-CD58 interaction was inhibited by the adhesion-blocking anti-CD2 mAb TS2/18.
The assumption that, at equilibrium, the free FITC-CD58 density in the contact area was equal to the (free) FITC-CD58 density outside the contact area is an approximation that does not take into account the possible reduction in free ligand density due to steric effects in the contact area. In experiments on CD28-CD80 and CD2-CD58 interactions (26) (T. Starr and M. L. Dustin, unpublished), a nonbinding GPI-anchored glycoprotein of similar size to the ligand, but labeled with a different fluorophore, was included in the bilayer as a control for this effect. Imaging of the control glycoprotein revealed a 20–40% reduction in free ligand in the contact area compared to the surrounding bilayer. This correction was particularly important at very high ligand density, where the amount of free ligand excluded from the contact area could exceed the amount of ligand bound. In this study, this correction would be very small due to the low density of free ligand compared to the density of bound ligand, and it was therefore unnecessary to apply the correction to the present data.
Confocal imaging of CD2 redistribution
Jurkat cells were incubated with the nonblocking, FITC-conjugated anti-CD2 mAb, FITC-CD2.1 (100 μg/ml), in binding buffer for 30 min. After the free antibody was removed by three washes, the cells were incubated for 40 min with a control bilayer (egg PC only) or a bilayer containing a high density of CD58 (270 molecules/μm2). Confocal images were then taken, at 1-μm intervals in the z-axis, using an interactive laser cytometer with confocal optics (Ultima, Meridian Instruments, Okemos, MI).
Fluorescence photobleaching recovery (FPR)
FPR was used to measure the fraction of laterally mobile CD2 molecules (f value) and the lateral diffusion coefficient of the mobile CD2 molecules. Cell surface CD2 molecules were labeled with FITC-CD2.1 on Jurkat T cell and peripheral blood T cell membranes. FPR experiments were performed using an interactive laser cytometer with confocal optics (Ultima, Meridian). The protocol for FPR measurements has been described in detail (20,22); here, the 250-μm emission pinhole was used to optimize z-axis resolution of the upper and lower T cell membranes without losing signal intensity. CD2 mobility was measured in the cell-bilayer contact area by focusing on the plane of most intense fluorescence, as described above. CD2 mobility in the membrane outside the contact area was measured by translating the focal plane ∼10 μm up from the plane of the bilayer to a region at the upper surface of the cell. The average diameters of Jurkat T cells and peripheral blood T cells were 11.1 and 7.0 μm, respectively.
Iodination of monoclonal antibodies
Protein A purified anti-CD2 mAbs TS2/18, 35.1 and CD2.1 were concentrated to >1 mg/mL as determined by BCA assays (Pierce, Rockford, IL), using bovine γ-globulin as the standard. Antibodies (100 μg) were iodinated using 1 mCi Na125I. Iodination was initiated with 1,3,4,6-tetrachloro-3α,6α-diphenylglycouril coated glass tubes (2 μg per tube) by the method of Fraker and Speck (27). Iodinated mAb was separated from free iodide by gel filtration, and active fractions were collected and pooled without a carrier. The concentration of the active pools was determined by BCA assay to allow accurate determination of specific activity.
Antibody binding assays
Jurkat T cells or peripheral blood T cells were washed twice with HEPES-buffered saline containing 2% BSA, and resuspended to 5 × 106 cells per mL. Cells were incubated with 2.3–230 nM iodinated mAb, in the presence and absence of 1.33 μM cold antibody, for 1 h at 4°C. TS2/18 and 35.1 mAbs displayed saturation binding to cells. Because this binding could have represented bivalent or monovalent binding to cell surface CD2 molecules, mAb molecular weights of 75,000 and 150,000 were used to calculate the limits of a range of specific activities. Bound and free counts were separated by centrifugation at 4000 × g for 3 min through an oil cushion (1.5 parts dibutylphthalate to 1 part dioctylphthalate (28)) in prechilled microsediment tubes (Sarstedt, Newton, NC). The cell pellet and supernatant were separated and counted. Scatchard analysis was used to calculate the average number of CD2 sites per cell, as described (25).
Unless otherwise indicated, all experimental measurements were performed at room temperature.
TWO-DIMENSIONAL ZHU-GOLAN ANALYSIS
Derivation of the two-dimensional Kd
The reaction of receptors (R) in a cell membrane with ligands (L) in a planar bilayer can be described by the general expression
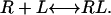 |
(1) |
At equilibrium,
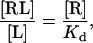 |
(2) |
where [RL], [L], and [R] are the densities of the receptor-ligand complex, the free ligand, and the free receptor, respectively, in the cell-bilayer contact area. Generally, in Eq. 2
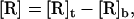 |
(3) |
where [R]t is the density of total receptors and [R]b is the density of bound receptors in the contact area. Because [R]b = [RL], we have
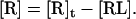 |
(4) |
Substitution for [R] in Eq. 2 results in
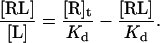 |
(5) |
If [R]t is a constant, then Eq. 5 can be written in the Scatchard form
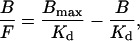 |
(6) |
and the Kd can be obtained from the negative reciprocal of the slope of a B/F vs. B plot.
Many cell surface receptors (such as CD2) are laterally mobile in the cell membrane, however, and the migration of these receptors into the contact area could result in variable levels of accumulation depending on the ligand density in the bilayer. In this case, [R]t in Eq. 5 is not a constant, but is instead a function of the free ligand density (F). Therefore, the Scatchard plot is not appropriate for analyzing the binding of laterally mobile molecules.
To establish an equation describing the two-dimensional binding equilibrium involving laterally mobile cell surface receptors, we assume that, at equilibrium, the free receptor density in the contact area is equal to the (free) receptor density outside the contact area. Thus, [R] in Eq. 2 can be expressed as
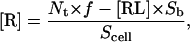 |
(7) |
where Nt is the total number of receptors on the cell surface, f is the fraction of laterally mobile receptors, Sb is the area of contact, and Scell is the total surface area of the cell. Note that, in this equation, only the laterally mobile receptors are considered to be capable of migration into the contact area to interact with the ligand in the apposing membrane.
Substitution of Eq. 7 into Eq. 2 yields
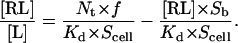 |
(8) |
Let
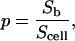 |
(9) |
and substitute B for [RL] and F for [L]. We then have the Zhu-Golan equation
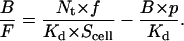 |
(10) |
Note, in the first term at the right side of Eq. 10, that the constant Nt × f/Scell represents the mean density of laterally mobile receptors. Thus, this term is not generally equivalent to the Bmax for two-dimensional binding. For the three-dimensional interaction (i.e., for the interaction between cell surface receptors and soluble ligands), however, this constant does reduce to Bmax, because for this case p = 1 (or Sb = Scell), and Eq. 10 reduces to Eq. 6. In other words, the Scatchard form in Eq. 6 is a special case of Eq. 10.
According to Eq. 10, the 2D Kd can be obtained from the negative reciprocal of the slope of a B/F vs. B × p plot. (Note that the Scatchard form plots B/F vs. B.) At the x-intercept (X), B/F = 0 and Eq. 10 reduces to
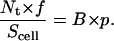 |
(11) |
Thus, Nt can be calculated from the experimentally determined values of f and Scell by using the expression
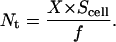 |
(12) |
The product Nt × f in Eq. 10 is equal to the total number of laterally mobile receptors, or the maximum number of receptors available for adhesion. Let
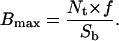 |
(13) |
Equation 10 can then be written as
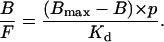 |
(14) |
When B = Bmax/2,
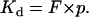 |
(15) |
This result is analogous to that in the three-dimensional analysis, in which Kd = F at half-maximal binding.
Role of immobile receptors in the contact area
Although laterally immobile receptors are not capable of migration into the cell-bilayer contact area, a fraction of these immobile receptors are present in the contact area from the beginning of the cell-bilayer interaction. To consider the potential contribution of these immobile receptors to ligand binding, we assume that the binding affinity of the immobile receptors is identical to that of the mobile receptors. From Eq. 5, the binding equilibrium can then be described by
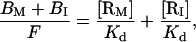 |
(16) |
where BM and BI are the densities of bound mobile receptors and bound immobile receptors in the contact area, respectively, and [RM] and [RI] are the densities of free mobile receptors and free immobile receptors in the contact area, respectively.
Assuming that the density of immobile receptors is a constant over the entire cell surface, [RI] can be calculated by
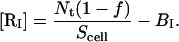 |
(17) |
Combining Eqs. 7, 16, and 17 yields the general equation for two-dimensional reactions involving both mobile and immobile receptor populations,
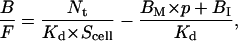 |
(18) |
where B is the sum of BM and BI.
If all of the cell surface receptors are mobile, or if the immobile receptors do not bind to ligands, then Eq. 10 provides an accurate description of the binding equilibrium. Otherwise, Eq. 18 is a more accurate description of the equilibrium when both mobile and immobile receptors contribute to ligand binding. However, since only total binding (B) can be measured experimentally, a plot of B/F vs. (BM × p + BI) is unachievable. Therefore, we use a plot of B/F vs. (BM + BI) × p, or more simply
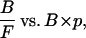 |
(19) |
to determine the apparent two-dimensional Kd (Kd,app). The potential error introduced by this approximation is addressed in Appendix B.
RESULTS
Adhesion of T cells to bilayers reconstituted with FITC-CD58
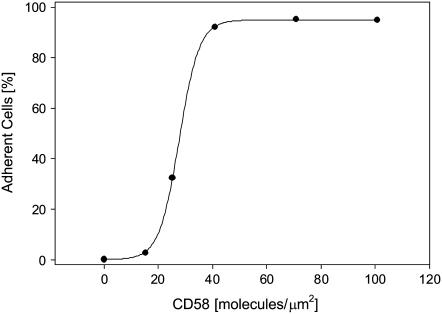
We used our analysis to measure the 2D Kd for binding between CD2 receptors on Jurkat T cells and purified CD58 ligands in egg PC planar bilayers. The bilayers were reconstituted with FITC-CD58 as described in Experimental Procedures. The formation of CD2-CD58 complexes was indicated by an increase in the fluorescence intensity under the cells and an increase in the cell-bilayer contact area, both of which were due to accumulation of FITC-CD58 molecules in the contact area (20,25). We first investigated the effect of CD58 density in the bilayer on cell adhesion (Fig. 1). Adherent cells could be differentiated from nonadherent cells by visual inspection: when the microscope was gently tapped, nonadherent cells drifted while adherent cells remained firmly seated. Cell adhesion was also confirmed by interference reflection microscopy (25). We found that a threshold CD58 density of 20 molecules/μm2 was required for stable cell adhesion. A CD58 density of 40 molecules/μm2 allowed >95% of cells to adhere to the bilayer (25).
FIGURE 1.
Jurkat T cell adhesion to CD58-containing bilayer, as a function of the initial density of CD58 in the bilayer. Adherent and nonadherent cells were scored as described in the text; at least 100 cells were used for each data point. A threshold density of ∼20 molecules/μm2 was required to support cell adhesion.
Development of the cell-bilayer contact area
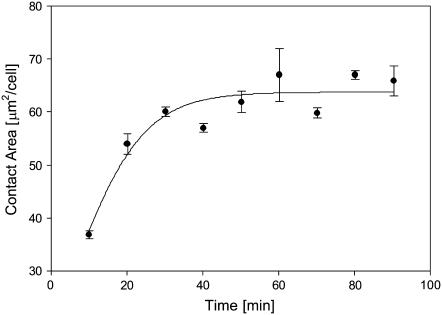
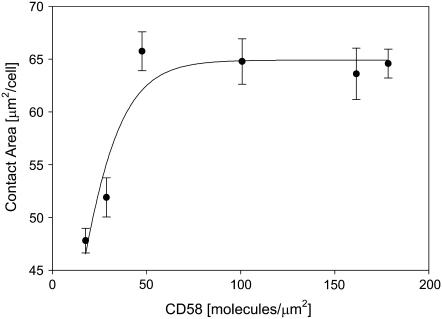
The size of the cell-bilayer contact area is one determinant of adhesion strength. The contact area size was quantified as described in Experimental Procedures. As shown in Fig. 2, incubation of Jurkat T cells with CD58-reconstituted bilayers resulted in a time-dependent increase in contact area, reaching a steady-state value at ∼40 min of incubation. The contact area at steady-state was also found to be a function of the initial density of FITC-CD58 molecules in the bilayer (Fig. 3). No definitive contact was formed at initial CD58 densities <20 molecules/μm2. At initial CD58 densities >40 molecules/μm2, the steady-state contact area size reached a plateau of 65 μm2/cell.
FIGURE 2.
Development of the cell-bilayer contact area as a function of incubation time. Jurkat T cells were incubated with bilayers containing FITC-CD58 at a density of 100 molecules/μm2. The size of the contact area was determined as described in the text; 30–100 cells were used for each data point. Data points represent mean ± SE.
FIGURE 3.
Size of the contact area as a function of initial CD58 density in the bilayer. Jurkat T cells were incubated for 40 min with bilayers containing different densities of FITC-CD58, and then contact area size was determined as described in the text; 30–170 cells were used for each data point. Data points represent mean ± SD. At initial CD58 densities <20 molecules/μm2, cells were not adherent to the bilayer.
Accumulation of CD58 in the contact area
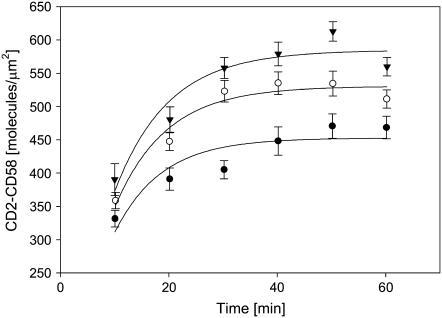
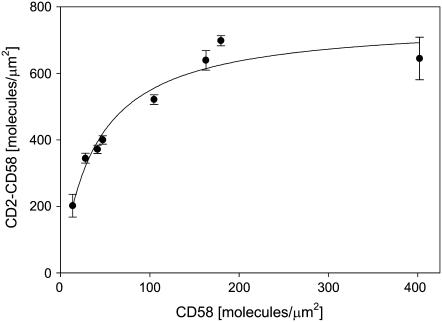
Laterally mobile CD58 molecules accumulated in the contact area due to binding interactions with CD2 on the T cell surface. Consistent with previous observations (20,25), the accumulation required ∼40 min to reach a steady-state value at all CD58 densities tested (Fig. 4). The time course of bound CD58 accumulation (Fig. 4) was identical to that of contact area development (Fig. 2), consistent with the interpretation that formation of CD2-CD58 complexes in the contact area provided the driving force for cell-bilayer adhesion. The steady-state level of bound CD58 accumulation depended on the initial density of CD58 in the bilayer (Fig. 5); nonlinear least-squares analysis showed that this relationship had the form of a saturable binding interaction with a maximum accumulation of 760 CD2-CD58 complexes/μm2.
FIGURE 4.
Time course of accumulation of bound CD58 molecules in the contact area. The initial densities of CD58 in the bilayer were 24 (solid circles), 50 (open circles), and 74 (solid triangles) molecules/μm2, respectively. Bound CD58 molecules were quantified as described in the text; 20–64 cells were used for each data point. Data points represent mean ± SD.
FIGURE 5.
Steady-state accumulation of bound CD58 molecules in the contact area as a function of initial CD58 density in the bilayer. Jurkat T cells were incubated for 40 min with bilayers containing different densities of FITC-CD58, and then the density of bound CD2-CD58 complexes was determined as described in the text. At least 50 cells were used for each data point. Data points represent mean ± SD.
Accumulation of CD2 in the contact area
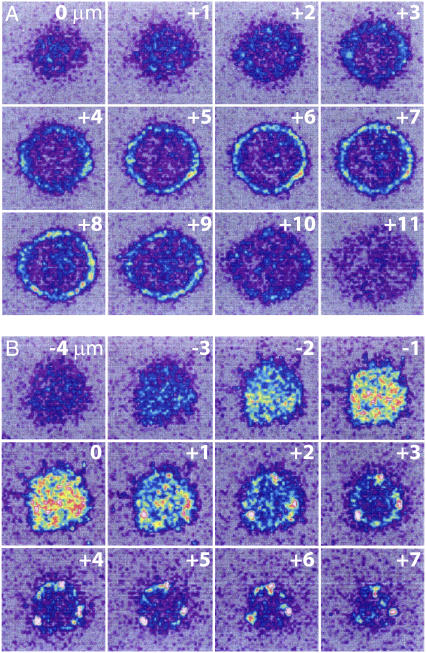
We also examined the formation of CD2-CD58 complexes by measuring the accumulation of labeled CD2 molecules in the contact area. In these experiments, the bilayers were reconstituted with unlabeled CD58, and CD2 on the T cells was labeled with FITC-CD2.1, a fluorescent anti-CD2 mAb that does not block CD2 binding to CD58. Confocal microscopy was used to determine the distribution of CD2 on cells interacting with control (egg PC) and CD58-containing bilayers. Cells resting on control bilayers showed a uniform distribution of CD2 in the plasma membrane, with no CD2 accumulation in the contact area. However, cells adherent to bilayers containing CD58 showed substantial redistribution of CD2 to the contact area (Fig. 6). The time course of CD2 accumulation in the contact area was similar to that of CD58 accumulation (not shown).
FIGURE 6.
Confocal images of CD2 distribution on the surface of Jurkat T cells. CD2 molecules were labeled using FITC-CD2.1, a fluorescent derivative of the nonblocking anti-CD2 mAb CD2.1. Cells were then incubated with an egg PC bilayer (A) or with an egg PC bilayer that had been reconstituted with 150 molecules/μm2 of unlabeled (i.e., nonfluorescent) CD58 (B). Confocal microscopy was used to image sections in 1-μm steps progressing from the bottom to the top of the cell, beginning from the glass surface (A) or from 4 μm below the bottom of the cell (B); z values are indicated relative to the plane of the coverslip in the upper right corner of each section. There was no accumulation of CD2 at the area of contact with bilayers lacking CD58 ((A), 0 to +3 μm), but cells interacting with bilayers containing CD58 showed significant redistribution of CD2 to the contact area ((B), −2 to +1 μm).
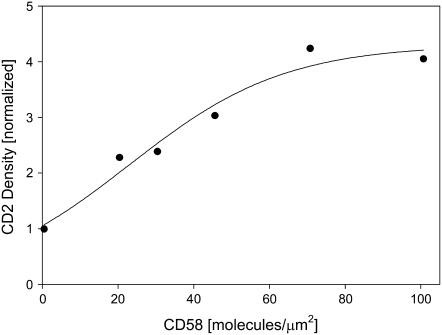
There was a direct relationship between the initial CD58 density in the bilayer and the steady-state level of CD2 accumulation in the contact area. The maximum density of accumulated CD2 was fourfold greater than the initial CD2 density on the T cell surface (Fig. 7). (Note that Fig. 7 shows the accumulation of total CD2 in the contact area, whereas Fig. 5 shows the accumulation of bound CD58 in the contact area. The accumulation of bound CD2 cannot be measured in this experimental system, because the density of free CD2 outside the contact area is difficult to quantify on the nonplanar membrane surface.) These important observations confirmed that CD2 density in the contact area was not a constant, but was rather a function of the CD58 density in the bilayer. These findings also supported the need for a new analytical method to analyze the two-dimensional dissociation constant for binding of laterally mobile cell adhesion molecules, since the standard Scatchard analysis assumes that the density of receptor binding sites is invariant.
FIGURE 7.
Steady-state accumulation of CD2 molecules in the contact area as a function of initial CD58 density in the bilayer. Jurkat T cells were labeled with FITC-CD2.1, and then incubated for 40 min with bilayers containing different densities of CD58. CD2 accumulation in the contact area was determined as described in the text; 20–130 cells were used for each data point. CD2 density in the contact area was normalized to the average CD2 density on the surface of Jurkat cells; according to the data in Table 2, this value was ∼136,000 molecules/cell ÷ 700 μm2, or 190 molecules/μm2.
Lateral mobility of CD2
As noted above, we derived the 2D Kd analysis to account for the lateral diffusion and accumulation of cell adhesion molecules in the contact area. We used fluorescence photobleaching recovery (FPR) and confocal microscopy techniques to measure directly the lateral mobility of CD2 in the plasma membranes of Jurkat T cells and peripheral blood T cells. As shown in Table 1, CD2 molecules on control Jurkat cells manifested a fractional mobility (f) of 0.75 ± 0.04 (mean ± SE) and a lateral diffusion coefficient of 7.9 ± 1.0 × 10−10 cm2 s−1. These values were consistent with previous reports (22). To measure CD2 mobility on adherent cells, Jurkat cells were incubated with planar bilayers containing CD58 at an initial density of 30 molecules/μm2. We chose a relatively low CD58 surface density for these experiments to allow cell adhesion to the bilayer while retaining a significant number of laterally mobile CD2 molecules outside the contact area. The f value of CD2 outside the contact area was 0.75 ± 0.06, which was identical to the f value of CD2 in control cells (Table 1). The fractional mobility of CD2 molecules in the contact area was 0.72 ± 0.04 and the lateral diffusion coefficient was 6.9 ± 0.7 × 10−10 cm2 s−1. These data indicated that ligation of CD2 by CD58 did not significantly alter the lateral mobility of CD2. The lateral mobility of CD2 on normal human peripheral blood T cells was similar to that of CD2 on Jurkat T cells, with an f value of 0.75 ± 0.03 and a lateral diffusion coefficient of 5.7 ± 0.5 × 10−10 cm2 s−1.
TABLE 1.
Lateral mobility of CD2
| Jurkat T cells | Peripheral blood T cells | |||||
|---|---|---|---|---|---|---|
| D* [× 10−10 cm2 s−1] | f† | N | D [× 10−10 cm2 s−1] | f | N | |
| Control cells‡ | 7.9 ± 1.0 | 0.75 ± 0.04 | 23 | 5.7 ± 0.5 | 0.75 ± 0.03 | 33 |
| Adherent cells§ | ||||||
| Top of cell¶ | 6.7 ± 0.7 | 0.75 ± 0.06 | 18 | |||
| Contact area | 6.9 ± 0.7 | 0.72 ± 0.04 | 35 | |||
The fluorescence photobleaching recovery (FPR) method is described in Experimental Procedures; data represent mean ± SE.
D, diffusion coefficient.
f, fractional mobility.
There was no CD58 in the bilayer.
Cells were incubated for 30 min with bilayers containing CD58 (at an initial density of 30 molecules/μm2), and then FPR experiments were performed.
The focus was first oriented at the plane of the contact area, and was then moved 10 μm “up” along the z-axis.
Determination of Nt by radiolabeled antibody binding
The number of cell surface CD2 molecules on Jurkat T cells and peripheral blood T cells (Nt) was determined by radiolabeled antibody binding, using the anti-CD2 mAbs TS2/18 and 35.1 (see Experimental Procedures). The two antibody assays yielded a range of Nt values from 43,000 to 86,000 CD2 molecules/cell for Jurkat T cells, and from 12,000 to 24,000 CD2 molecules/cell for peripheral blood T cells. Normalizing for the calculated surface area of each cell type, the average CD2 density was 60–120 molecules/μm2 on Jurkat T cells and 43–86 molecules/μm2 on peripheral blood T cells (see below).
Determination of 2D Kd and Nt by Zhu-Golan analysis
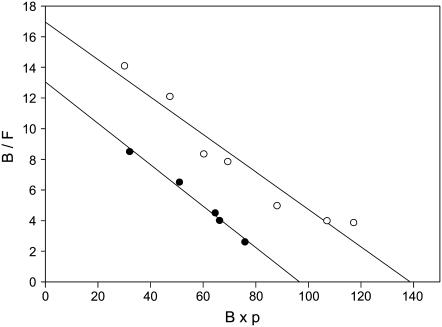
According to Eq. 10, the 2D Kd for the CD2-CD58 binding interaction can be obtained from plots of B/F vs. B × p. Experimental values of B, F, f, and p were obtained as described above. The surface area of the cells (Scell) was calculated using the diameter of projected cell images and a surface roughness factor of 1.8 for the T cell (25,29). We performed multiple measurements of CD2-CD58 binding affinity (see Fig. 8 for a representative Zhu-Golan plot), and determined that the 2D Kd for this interaction was 7.6 ± 0.9 molecules/μm2 for Jurkat T cells and 5.4 ± 1.0 molecules/μm2 for peripheral blood T cells (Table 2).
FIGURE 8.
Representative Zhu-Golan (B/F vs. B × p) plots (Eq. 10) describing interactions of Jurkat T cells (open circles) and peripheral blood T cells (solid circles) with bilayers containing CD58. Cells were incubated for 40 min with bilayers containing different densities of FITC-CD58. The densities of bound (B) and free (F) CD58 in the contact area, and the ratio of contact area size to surface area of the cell (p), were then determined as described in the text. Each point represents data from 27 to 176 cells. Curves were analyzed to determine the 2D Kd for the CD2-CD58 binding interaction and the total number (Nt) of CD2 molecules on the T cell surface. For Jurkat T cells, 2D Kd was 8.1 molecules/μm2 (r2 = 0.97) and Nt was 130,000 molecules. For peripheral blood T cells, 2D Kd was 7.4 molecules/μm2 (r2 = 0.99) and Nt was 36,000 molecules.
TABLE 2.
Results of binding analysis
| Cell type | Scell* [μm2] | pmax | X | Kd [mol/μm2] | Kd,ideal† [mol/μm2] | Nt‡ [mol/cell × 103] |
|---|---|---|---|---|---|---|
| Jurkat T cells¶ | ||||||
| Mean | 700 | 0.100 ± 0.005 | 145 ± 5 | 7.6 ± 0.9 | 7.6–9.9 | 136 ± 4 |
| Range | — | 0.094–0.112 | 138–156 | 5.6–9.2 | — | 129–146 |
| Peripheral blood T cells§ | ||||||
| Mean | 280 | 0.109 ± 0.004 | 78 ± 7 | 5.4 ± 1.0 | 5.4–7.0 | 29 ± 3 |
| Range | — | 0.082–0.121 | 44–100 | 2.5–10.2 | — | 17–38 |
Mean cell surface area was calculated using the expression, Scell = 4πr2 × 1.8, where r was the mean measured radius of the cells and 1.8 was a correction factor for the roughness of the T cell (29).
Mean Kd,ideal was calculated using a ∂max value of 0.23 in Eq. 31 for both Jurkat T cells and peripheral blood T cells.
Nt was calculated using Eq. 12. Calculation of Nt using Eq. 35 yielded values within 3–4% of those using Eq. 12.
Results represent mean, mean ± SE, and range of values from three independent determinations on Jurkat T cells.
Results represent mean, mean ± SE, and range of values from determinations on peripheral blood T cell samples from eight different individuals.
Our analysis also yields an estimate for the total number of CD2 molecules per cell (Nt value). Nt was calculated using Eq. 12 and the x-intercepts of the B/F vs. B × p binding data summarized in Table 2. According to this analysis, the calculated number of CD2 sites was 136 ± 4 × 103 molecules/cell for Jurkat T cells and 29 ± 3 × 103 molecules/cell for peripheral blood T cells (Table 2). These values were 20–60% larger than the Nt values determined using radiolabeled antibody assays. This finding may reflect the increased tendency of cells with a greater number of CD2 receptors to adhere to CD58-containing bilayers, which would bias the Nt determination to higher values (25).
Finally, we considered the effect of immobile CD2 molecules on the 2D Kd and Nt measurements. Based on this analysis (see Appendix B), the range of Kd,ideal values was found to be 7.6–9.9 molecules/μm2 for Jurkat T cells and 5.4–7.0 molecules/μm2 for peripheral blood T cells. The overlap of these ranges with the 2D Kd determined using Eq. 10 indicated that the small fraction of immobile CD2 molecules could be ignored without introducing significant error. Equation 35 was used to calculate Nt taking account of the binding of immobile CD2 to CD58 in the contact area. The Nt values determined using Eq. 35 were within 3–4% of those calculated using Eq. 12, indicating that immobile CD2 receptors did not contribute significantly to the Nt calculation.
DISCUSSION
The dynamic control of adhesive receptor-ligand interactions is essential for a variety of important biological processes, including immune synapse formation, lymphocyte migration, and cancer cell metastasis. We present here a method to quantify three mechanisms that regulate the strength of cell-cell adhesion: the number of adhesion receptors (Nt), the fraction of laterally mobile receptors (f), and the receptor affinity for ligand (2D Kd). All of these mechanisms are known to affect adhesive interactions; for example, the surface density of both LFA-1 and ICAM-1 is increased upon cytokine stimulation of cells (30–32), and changes in receptor affinity and lateral mobility control adhesion mediated by LFA-1 or CD2 (32–38). Although these parameters are often important features of quantitative models of cell adhesion (39), experimental determination of their contributions to adhesion has been difficult (30–36,38). Using quantitative fluorescence imaging techniques and artificial bilayers of defined composition, we have developed a methodology to quantify the roles of receptor-ligand affinity, receptor mobility, and receptor expression level in cell-cell adhesion.
We have found that the initial density of CD58 in the planar bilayer is an important parameter for the development of adhesion. Production of a stable cell-bilayer adhesion requires a minimum CD58 density of 20 molecules/μm2 (Fig. 1), suggesting that a minimum number of initial bonds is required to stabilize the adhesive interaction. In the physiological setting, CD2-CD58 interactions are subjected to shear forces that could shift this threshold. At CD58 densities greater than this threshold value, both CD2 and CD58 accumulate at the cell-bilayer interface; the contact area reaches a steady-state size over a time course of 30–60 min (Fig. 2). The initial density of CD58 in the bilayer determines both the contact area size and the level of steady-state accumulation of CD2-CD58 complexes in the contact area (Figs. 3–5).
Receptor accumulation in the contact area requires migration of the receptors to the site of adhesion, and therefore depends on the lateral mobility of both CD58 in the bilayer and CD2 on the cell surface. The lateral diffusion coefficients of CD58 and CD2 determine the kinetics of receptor migration and of contact area development. Because the diffusion coefficient of CD2 in the cell membrane (6–8 × 10−10 cm2 s−1) is approximately an order-of-magnitude less than that of CD58 in the bilayer (20,25,40–42), it is likely that the lateral diffusion of CD2 into the contact area is the rate-limiting step for adhesion development.
The lateral diffusion of cell surface receptors is a critical parameter for stable adhesion (40). Here, CD2 migration to the contact area results in a significant (up to fourfold) increase in CD2 density at the site of adhesion (Fig. 7). This dynamic property of the adhesion zone violates one of the assumptions used in analyzing receptor-ligand interactions by traditional (Scatchard) methods. We have therefore developed a new analysis to accommodate the dynamic nature of the interaction involving laterally mobile cell surface receptors and ligands. Our analysis makes only one fundamental assumption: we assume that, at steady-state, the free receptor density in the contact area is identical to the free receptor density outside the contact area. Quantitative fluorescence microscopy and fluorescence photobleaching recovery techniques are used to calculate the 2D Kd, total receptor number (Nt), receptor lateral mobility (f), cell surface area (Scell), and cell-bilayer contact area (Sb). Our model can be used to analyze the three fundamental regulatory mechanisms of cellular avidity modulation:
Altering the 2D Kd through a conformational change in the receptor;
Changing the mobile fraction of the receptor (f) to provide for increased receptor accumulation in the contact area; and
Altering the total receptor number (Nt) and thus the maximum number of potential bonds.
Moreover, the model reduces to the Scatchard form for the special case where p equals 1 (see Eqs. 10 and 15), i.e., where the receptors over the entire surface of the cell are able to participate in the interaction. Although p = 1 in a case such as the binding of soluble CD58 to cell surface CD2, for any cell-cell adhesion event the receptor-ligand binding interactions can occur only in the two-dimensional contact area, for which p becomes ≪1. Therefore, our model is a general analysis that is applicable to both two-dimensional and three-dimensional interactions, while the Scatchard analysis is a special case of this model.
The factor p is the ratio of the contact area to the total cell surface area. As shown in Fig. 3, p is also a function of CD58 density. Because the contact area size varies over a narrow range for productive cell-bilayer adhesions (65–78 μm2/cell), p is also confined to a small range (0.094–0.112) and it reaches a plateau at CD58 densities >40 molecules/μm2 (Fig. 3). Although our analysis implicitly treats the adhesion zone as a uniform distribution of receptors, it is possible that nonuniform patterns may contribute to the two-dimensional affinity in certain systems (43). The parameter f is the mobile fraction of free receptor molecules on the cell. The confocal FPR results (Table 1) indicate that 75% of CD2 molecules are laterally mobile in resting T cells, and that ligation of CD2 by CD58 in the contact area does not alter the f value. From our results and analysis, we find that f is the primary determinant of the rate and the maximum extent of receptor accumulation in the contact area.
Because it is experimentally difficult to measure separately the binding of mobile and immobile receptors, the determination of 2D Kd,ideal is challenging. Therefore, we have used an approximation to determine the 2D Kd. The general equations (Eqs. 16–18) assume that the mobile and immobile receptors have the same affinity for ligand. As we demonstrate in Appendix B, the deviation caused by this assumption is quite small. Equations 32 and 33 show that the range of Kd,ideal values is a function of f; when f = 50%, Kd,app is well within this range. Moreover, f has a minimal effect on the calculation of Nt, and this effect can be calculated using Eq. 35.
The analysis used here allows determination of the physiologically relevant 2D Kd. For the CD2-CD58 interaction, the 2D Kd is 5.4–7.6 molecules/μm2. Importantly, this 2D Kd value is similar between Jurkat T cells and peripheral blood T cells, even though the total number of CD2 molecules on the Jurkat cell line is ∼4–5-fold greater than that on peripheral blood T cells (Table 2). The surface density of CD2 is similar on the two cell types (i.e., Nt/Scell = 100–190 molecules/μm2). The measured 2D Kd suggests that the formation of CD2-CD58 bonds between T cells and antigen-presenting cells (APCs) is highly favored at steady state. Rearrangement of Eq. 2 allows estimation of a lower limit for the ratio of bound to unbound CD2 receptors (RL/R) at the T cell-APC contact, based on the 2D Kd for the interaction and the initial CD58 site density on APCs. The initial CD58 site density, which ranges between 60 and 90 molecules/μm2 (44), predicts that the ratio of bound to unbound CD2 is ≥8–12 in Jurkat cells and ≥11–16 in peripheral blood T cells. These values would suggest that at least 88–94% of CD2 molecules at the contact area are engaged in adhesive interactions at steady state.
Utilizing the same Zhu-Golan analysis method, we have previously used the adhesion of Jurkat cells to supported planar bilayers to obtain a value of 1.1 molecules/μm2 for the 2D Kd of the CD2-CD58 interaction (25). Several experimental differences are likely to have contributed to the disparity between this value and the value obtained in this study. These differences include the method used to measure contact area size; the choice of blocking conditions; and the Jurkat clone used in the study. Here, we used the fluorescence of accumulated FITC-CD58 to define the contact area, whereas previous determinations used interference reflection microscopy (IRM). The fluorescence intensity threshold method requires that the membranes are within 15 nm for the CD2-CD58 interaction, whereas the IRM method requires that the membranes are less than a quarter wavelength apart (∼130 nm for green light). The two methods may therefore report different contact area sizes, although both methods are acceptable for this determination. Since the contact area sizes measured here are ∼1.5-fold larger than those measured by IRM, a similar magnitude of increase would be expected in the 2D Kd according to Eq. 10. Our earlier study also utilized nonfat dry milk rather than BSA to block nonspecific binding of cells to the bilayer. In other studies, we have found that use of nonfat dry milk decreases nonspecific lymphocyte-bilayer adhesion that emerges after specific interactions induce formation of a contact area, and thereby facilitates processes such as rapid cell migration (45). Interestingly, reduction of bilayer-bilayer interactions after specific adhesion would allow the dimensions of the CD2-CD58 interaction to determine the intermembrane spacing, resulting in a higher two-dimensional affinity due to greater confinement. We have used BSA here because it is better defined than nonfat dry milk, and our adhesion data suggest that Jurkat cell interactions with the bilayer are both specific and dependent on CD58 density (Fig. 1). The Jurkat clone used here is also different from that used in our previous experiments, and variability in Jurkat clones could contribute to the discrepancy in the 2D Kd measurements. Hahn et al. have suggested that cell activation influences the avidity of adhesion mediated by the CD2-CD58 interaction (34), and we have recently found that activation of the Jurkat clone used here induces a 2.5-fold enhancement in the two-dimensional affinity of CD2-CD58 (46). It is possible that the Jurkat clone used in our earlier study manifested a more activated phenotype than the clone used here. Comparing our earlier result with the findings presented here, we favor the lower affinity (7.6 molecules/μm2) measured here as the 2D Kd for the CD2-CD58 interaction in resting Jurkat cells, as it is supported by a direct comparison to the analogous measurement in peripheral blood T cells (Table 2).
To achieve a quantitative understanding of cell surface avidity regulation, new methods are required that measure the parameters affecting cell-cell adhesion. We have designed a system for quantitative monitoring of receptor-ligand interactions at two-dimensional interfaces, and have employed a new model to analyze the data in terms of a 2D Kd. This analysis should be useful for dissecting the specific mechanisms of avidity regulation used by a number of different immune cell adhesion molecules. In this work, we find that a threshold density of CD2-CD58 binding interactions is required for the stable adhesion of resting T cells. We show that the 2D Kd of the CD2-CD58 interaction is 5.4–7.6 molecules/μm2 in both Jurkat T cells and peripheral blood T cells. Our analysis provides a general method for discerning the contribution of two-dimensional receptor affinity to adhesive interactions, and could be particularly useful in characterizing systems that undergo conformational changes resulting in affinity modulation. It is well known, for example, that cell stimulation modulates the avidity of receptors including CD2 and LFA-1 (33,34,42). By using our method to determine the 2D Kd, the roles of receptor affinity and mobility (clustering) in these interactions could be separated. Explicit incorporation of these quantitative measures of two-dimensional affinity should also be of interest for new models that include the contributions of receptor flexibility and conformational changes to adhesive interactions (47). As alluded to above, we have recently reported the results of studies examining the effect of cell activation on the parameters that regulate adhesion mediated by the CD2-CD58 interaction (46).
Acknowledgments
We thank James Miller for FITC-CD58 preparation and for site-density determinations on Jurkat T cells, Naishadh Desai for reagents, Hemant Thatte for preparation of human peripheral blood mononuclear cells, and Timothy Springer for helpful discussions.
This work was supported by National Institutes of Health grants No. HL32854 and No. HL70819 (to D.E.G.), and AI43542 (to M.L.D.). C.W.C. and D.E.G. are grateful for support from the Alexander and Margaret Stewart Trust.
APPENDIX A: NOMENCLATURE
See Table 3 below.
TABLE 3.
Glossary of symbols
| Symbol | Definition | Units | Measurement* |
|---|---|---|---|
| B | Bound ligand density in contact area | [molecules/μm2] | (FLcontact − FLbilayer − FLautofluor) ÷ specific activity [FL/molecule]. |
| Bmax | Maximum bound ligand density in contact area | [molecules/μm2] | |
| F | Free ligand density (assumed to be equal in and out of contact area) | [molecules/μm2] | FLbilayer ÷ specific activity [FL/molecule]. |
| [R] | Density of free receptor in contact area | [molecules/μm2] | |
| [L] | Density of free ligand in contact area | [molecules/μm2] | |
| [RL] | Density of receptor-ligand complex in contact area | [molecules/μm2] | |
| [R]t | Density of total receptors | [molecules/μm2] | |
| [R]b | Density of bound receptors | [molecules/μm2] | |
| [RM] | Density of free mobile receptors | [molecules/μm2] | |
| [RI] | Density of free immobile receptors | [molecules/μm2] | |
| BM | Density of bound mobile receptors | [molecules/μm2] | |
| BI | Density of bound immobile receptors | [molecules/μm2] | |
| Kd,ideal | Ideal dissociation constant | [molecules/μm2] | |
| Kd,app | Apparent dissociation constant | [molecules/μm2] | Negative reciprocal slope of B/F vs. B × p plot. |
| Nt | Total number of CD2 molecules per cell | [molecules/cell] | Iodinated IgG or Fab binding; (X × Scell)/f. |
| Nb | Total number of bound ligand molecules | [molecules/contact] | B × Sb. |
| Scell | Surface area of cell | [μm2] | 4πr2 × 1.8, where r is the measured radius of the cell (see Results). |
| Sb | Size of contact area | [μm2] | Interference reflection microscopy (IRM); fluorescence intensity threshold method (see Experimental Procedures). |
| Sb,max | Maximal size of contact area | [μm2] | |
| p | Sb ÷ Scell | ||
| f | Fractional mobility | [%] | Fluorescence photobleaching recovery (FPR). |
| δmax | Maximal fractional deviation of B × p | ||
| X | x-intercept | ||
| Y | y-intercept |
APPENDIX B: MODEL VALIDATION
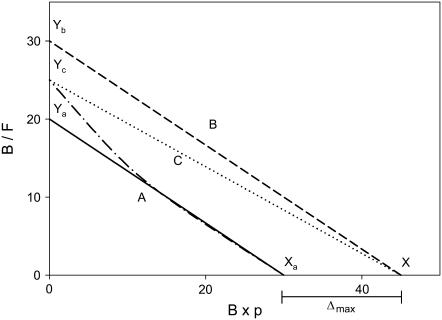
At a given B/F, the difference (Δ) between the values of (BM × p + BI) and (BM + BI) × p is
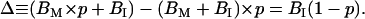 |
(20) |
The maximum values of BM (BM,max) and BI (BI,max) are defined by
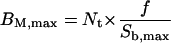 |
(21) |
and
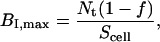 |
(22) |
respectively, where Sb,max is the maximum contact area at Bmax. Therefore, the maximum Δ (Δmax), i.e., the difference between the x-intercepts of the two plots at B/F = 0, is defined by Eqs. 20–22:
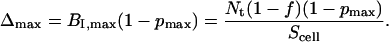 |
(23) |
Let
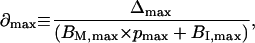 |
(24) |
i.e., ∂max represents the maximum fractional deviation of B × p from (BM × p + BI). Combining Eqs. 20–24, we have
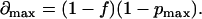 |
(25) |
This analysis is described graphically in Fig. 9, where Line A represents B/F vs. B × p, with an x-intercept of Xa. (Note that the actual curve, as shown by the dotted line, should deviate from the linear approximation, particularly in the region of small B × p values.) Line B is an imaginary line parallel to Line A and separated from Line A by Δmax in the x direction. Line C, the imaginary ideal plot of B/F vs. (BM × p + BI), lies between Line A and Line B. X is the x-intercept of both Line B and Line C. Ya, Yb, and Yc are the y-intercepts of Line A, Line B, and Line C, respectively, in the order
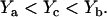 |
(26) |
Let
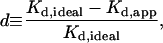 |
(27) |
where Kd,ideal represents the ideal Kd corresponding to Line C, Kd,app represents the approximate Kd obtained from Line A, and d is the fractional deviation of Kd,app from Kd,ideal. From Eqs. 24, 25, and 27,
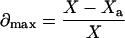 |
(28) |
and
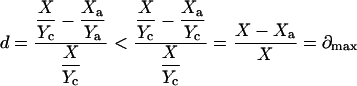 |
(29) |
or
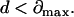 |
(30) |
Therefore,
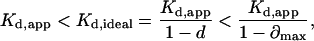 |
(31) |
or
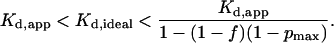 |
(32) |
FIGURE 9.
Range of deviation in the B/F vs. B × p plot caused by using (B × p) to approximate (BM × p + BI). Line A is the linearized approximate plot of B/F vs. B × p, with an x-intercept of Xa and a y-intercept of Ya. (The dotted line shows the plot before linearization.) Line B is an imaginary line parallel to Line A, separated from Line A by Δmax in the x direction. Line C is the imaginary ideal plot of B/F vs. (BM × p + BI). Lines B and C share the same x-intercept, X. Yb and Yc are the y-intercepts of Lines B and C, respectively.
According to Eq. 32, the range of Kd,app is defined by the experimental values of f and pmax. For example, when f = 0.5 and pmax = 0.1, Kd,app < Kd,ideal < 1.8 × Kd,app.
In general, pmax ≪ 1 and Eq. 32 can be simplified to
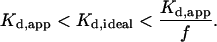 |
(33) |
Finally, we consider the contribution of immobile receptors to the estimate of total receptor number described by Eq. 12. From Eqs. 18, 23, and 25, we have
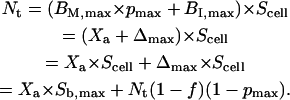 |
(34) |
Therefore,
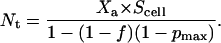 |
(35) |
When pmax ≪ 1, Eq. 35 is nearly identical to Eq. 12. Therefore, we conclude that the effect of an immobile receptor population is minor in our experimental system. Models that include an explicit treatment of the immobile receptor fraction have reached similar conclusions, finding no major deviations from the analysis reported here (48).
De-Min Zhu's present address is Merck Research Laboratories, Merck & Co., WP78-302, West Point, PA 19486.
Christopher Cairo's present address is Dept. of Chemistry, University of Alberta, Edmonton, Alberta T6G 2G2, Canada.
References
- 1.Dustin, M. L., S. K. Bromley, M. M. Davis, and C. Zhu. 2001. Identification of self through two-dimensional chemistry and synapses. Annu. Rev. Cell Dev. Biol. 17:133–157. [DOI] [PubMed] [Google Scholar]
- 2.Davis, S. J., S. Ikemizu, M. K. Wild, and P. A. van der Merwe. 1998. CD2 and the nature of protein interactions mediating cell-cell recognition. Immunol. Rev. 163:217–236. [DOI] [PubMed] [Google Scholar]
- 3.Tangye, S. G., J. H. Phillips, and L. L. Lanier. 2000. The CD2-subset of the Ig superfamily of cell surface molecules: receptor-ligand pairs expressed by NK cells and other immune cells. Semin. Immunol. 12:149–157. [DOI] [PubMed] [Google Scholar]
- 4.Doyle, C., and J. L. Strominger. 1987. Interaction between CD4 and class-II MHC molecules mediates cell-adhesion. Nature. 330:256–259. [DOI] [PubMed] [Google Scholar]
- 5.Luescher, I. F., E. Vivier, A. Layer, J. Mahiou, F. Godeau, B. Malissen, and P. Romero. 1995. CD8 modulation of T-cell antigen receptor-ligand interactions on living cytotoxic T-lymphocytes. Nature. 373:353–356. [DOI] [PubMed] [Google Scholar]
- 6.Matsui, K., J. J. Boniface, P. A. Reay, H. Schild, B. F. Destgroth, and M. M. Davis. 1991. Low affinity interaction of peptide-MHC complexes with T-cell receptors. Science. 254:1788–1791. [DOI] [PubMed] [Google Scholar]
- 7.O'Rourke, A. M., J. R. Apgar, K. P. Kane, E. Martz, and M. F. Mescher. 1991. Cytoskeletal function in CD8-cell and T-cell receptor-mediated interaction of cytotoxic T-lymphocytes with class-I protein. J. Exp. Med. 173:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber, S., A. Traunecker, F. Oliveri, W. Gerhard, and K. Karjalainen. 1992. Specific low-affinity recognition of major histocompatibility complex plus peptide by soluble T-cell receptor. Nature. 356:793–796. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, W. C., E. Menu, B. E. Bierer, and Y. Shimizu. 1993. CD2: A multifunctional coreceptor involved in T-cell adhesion and activation. In Lymphocyte Adhesion Molecules. R. G. Landes, Austin, TX. 105–134.
- 10.Harvey, J. E., N. Hogg, R. C. Landis, and Y. Shimizu. 1993. LFA-1 and the ICAMs. In Lymphocyte Adhesion Molecules. R. G. Landes, Austin, TX. 26–53.
- 11.Springer, T. A. 1990. Adhesion receptors of the immune-system. Nature. 346:425–434. [DOI] [PubMed] [Google Scholar]
- 12.Thompson, C. B. 1995. Distinct roles for the co-stimulatory ligands B7-1 and B7-2 in T-helper cell-differentiation. Cell. 81:979–982. [DOI] [PubMed] [Google Scholar]
- 13.Dustin, M. L., P. Selvaraj, R. J. Mattaliano, and T. A. Springer. 1987. Anchoring mechanisms for LFA-3 cell-adhesion glycoprotein at membrane-surface. Nature. 329:846–848. [DOI] [PubMed] [Google Scholar]
- 14.van der Merwe, P. A., S. J. Davis, A. S. Shaw, and M. L. Dustin. 2000. Cytoskeletal polarization and redistribution of cell-surface molecules during T cell antigen recognition. Semin. Immunol. 12:5–21. [DOI] [PubMed] [Google Scholar]
- 15.Irvine, D. J., M. A. Purbhoo, M. Krogsgaard, and M. M. Davis. 2002. Direct observation of ligand recognition by T cells. Nature. 419:845–849. [DOI] [PubMed] [Google Scholar]
- 16.Bockenstedt, L. K., M. A. Goldsmith, M. Dustin, D. Olive, T. A. Springer, and A. Weiss. 1988. The CD2 ligand LFA-3 activates T-cells but depends on the expression and function of the antigen receptor. J. Immunol. 141:1904–1911. [PubMed] [Google Scholar]
- 17.Davis, S. J., E. A. Davies, A. N. Barclay, S. Daenke, D. L. Bodian, E. Y. Jones, D. I. Stuart, T. D. Butters, R. A. Dwek, and P. A. van der Merwe. 1995. Ligand-binding by the immunoglobulin superfamily recognition molecule CD2 is glycosylation-independent. J. Biol. Chem. 270:369–375. [DOI] [PubMed] [Google Scholar]
- 18.van der Merwe, P. A., A. N. Barclay, D. W. Mason, E. A. Davies, B. P. Morgan, M. Tone, A. K. C. Krishnam, C. Ianelli, and S. J. Davis. 1994. Human cell-adhesion molecule CD2 binds CD58 (LFA-3) with a very-low affinity and an extremely fast dissociation rate but does not bind CD48 or CD59. Biochemistry. 33:10149–10160. [DOI] [PubMed] [Google Scholar]
- 19.van der Merwe, P. A., M. H. Brown, S. J. Davis, and A. N. Barclay. 1993. Affinity and kinetic-analysis of the interaction of the cell-adhesion molecules rat CD2 and CD48. EMBO J. 12:4945–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dustin, M. L., L. M. Ferguson, P. Y. Chan, T. A. Springer, and D. E. Golan. 1996. Visualization of CD2 interaction with LFA-3 and determination of the two-dimensional dissociation constant for adhesion receptors in a contact area. J. Cell Biol. 132:465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scatchard, G. 1949. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 51:660–672. [Google Scholar]
- 22.Liu, S. Q. J., W. C. Hahn, B. E. Bierer, and D. E. Golan. 1995. Intracellular mediators regulate CD2 lateral diffusion and cytoplasmic Ca2+ mobilization upon CD2-mediated T-cell activation. Biophys. J. 68:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vollger, L. W., D. T. Tuck, T. A. Springer, B. F. Haynes, and K. H. Singer. 1987. Thymocyte binding to human thymic epithelial cells is inhibited by monoclonal-antibodies to CD-2 and LFA-3 antigens. J. Immunol. 138:358–363. [PubMed] [Google Scholar]
- 24.Dustin, M. L., D. Olive, and T. A. Springer. 1989. Correlation of CD2 binding and functional-properties of multimeric and monomeric lymphocyte function-associated antigen-3. J. Exp. Med. 169:503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dustin, M. L., D. E. Golan, D. M. Zhu, J. M. Miller, W. Meier, E. A. Davies, and P. A. van der Merwe. 1997. Low affinity interaction of human or rat T cell adhesion molecule CD2 with its ligand aligns adhering membranes to achieve high physiological affinity. J. Biol. Chem. 272:30889–30898. [DOI] [PubMed] [Google Scholar]
- 26.Bromley, S. K., A. Iaboni, S. J. Davis, A. Whitty, J. M. Green, A. S. Shaw, A. Weiss, and M. L. Dustin. 2001. The immunological synapse and CD28–CD80 interactions. Nat. Immunol. 2:1159–1166. [DOI] [PubMed] [Google Scholar]
- 27.Fraker, P. J., and J. C. Speck. 1978. Protein and cell-membrane iodinations with a sparingly soluble chloramide, 1,3,4,6-tetrachloro-3A,6A-diphenylglycoluril. Biochem. Biophys. Res. Commun. 80:849–857. [DOI] [PubMed] [Google Scholar]
- 28.Sayre, P. H., R. E. Hussey, H. C. Chang, T. L. Ciardelli, and E. L. Reinherz. 1989. Structural and binding analysis of a two-domain extracellular CD2 molecule. J. Exp. Med. 169:995–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mege, J. L., C. Capo, A. M. Benoliel, C. Foa, R. Galindo, and P. Bongrand. 1986. Quantification of cell-surface roughness—a method for studying cell mechanical and adhesive properties. J. Theor. Biol. 119:147–160. [DOI] [PubMed] [Google Scholar]
- 30.Dustin, M. L., R. Rothlein, A. K. Bhan, C. A. Dinarello, and T. A. Springer. 1986. Induction by IL-1 and interferon-γ—tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1). J. Immunol. 137:245–254. [PubMed] [Google Scholar]
- 31.Pober, J. S., M. P. Bevilacqua, D. L. Mendrick, L. A. Lapierre, W. Fiers, and M. A. Gimbrone. 1986. Two distinct monokines, interleukin-1 and tumor-necrosis-factor, each independently induce biosynthesis and transient expression of the same antigen on the surface of cultured human vascular endothelial cells. J. Immunol. 136:1680–1687. [PubMed] [Google Scholar]
- 32.Shimizu, Y., G. A. Vanseventer, K. J. Horgan, and S. Shaw. 1990. Regulated expression and binding of 3 VLA (β-1) integrin receptors on T-cells. Nature. 345:250–253. [DOI] [PubMed] [Google Scholar]
- 33.Dustin, M. L., and T. A. Springer. 1989. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature. 341:619–624. [DOI] [PubMed] [Google Scholar]
- 34.Hahn, W. C., S. J. Burakoff, and B. E. Bierer. 1993. Signal transduction pathways involved in T-cell receptor-induced regulation of CD2 avidity for CD58. J. Immunol. 150:2607–2619. [PubMed] [Google Scholar]
- 35.Haverstick, D. M., and L. S. Gray. 1992. Lymphocyte adhesion mediated by lymphocyte function-associated antigen-1. 2. Interaction between phorbol ester-sensitive and cAMP-sensitive pathways. J. Immunol. 149:397–402. [PubMed] [Google Scholar]
- 36.Kim, M., C. V. Carman, W. Yang, A. Salas, and T. A. Springer. 2004. The primacy of affinity over clustering in regulation of adhesiveness of the integrin α(L)β 2. J. Cell Biol. 167:1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kucik, D. F., M. L. Dustin, J. M. Miller, and E. J. Brown. 1996. Adhesion-activating phorbol ester increases the mobility of leukocyte integrin LFA-1 in cultured lymphocytes. J. Clin. Invest. 97:2139–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardi, R., L. Inverardi, C. Rugarli, and J. R. Bender. 1992. Antigen-receptor complex stimulation triggers protein kinase-C-dependent CD11a/CD18-cytoskeleton association in T-lymphocytes. J. Cell Biol. 116:1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qi, S. Y., J. T. Groves, and A. K. Chakraborty. 2001. Synaptic pattern formation during cellular recognition. Proc. Natl. Acad. Sci. USA. 98:6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan, P. Y., M. B. Lawrence, M. L. Dustin, L. M. Ferguson, D. E. Golan, and T. A. Springer. 1991. Influence of receptor lateral mobility on adhesion strengthening between membranes containing LFA-3 and CD2. J. Cell Biol. 115:245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, S. Q. J., and D. E. Golan. 1999. T-cell stimulation through the T-cell receptor CD3 complex regulates CD2 lateral mobility by a calcium/calmodulin-dependent mechanism. Biophys. J. 76:1679–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shamri, R., V. Grabovsky, J. M. Gauguet, S. Feigelson, E. Manevich, W. Kolanus, M. K. Robinson, D. E. Staunton, U. H. von Andrian, and R. Alon. 2005. Lymphocyte arrest requires instantaneous induction of an extended LFA-1 conformation mediated by endothelium-bound chemokines. Nat. Immunol. 6:497–506. [DOI] [PubMed] [Google Scholar]
- 43.Hategan, A., K. Sengupta, S. Kahn, E. Sackmann, and D. E. Discher. 2004. Topographical pattern dynamics in passive adhesion of cell membranes. Biophys. J. 87:3547–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergwelt-Baildon, M. S., R. H. Vonderheide, B. Maecker, N. Hirano, K. S. Anderson, M. O. Butler, Z. N. Xia, W. Y. Zeng, K. W. Wucherpfennig, L. M. Nadler, and J. L. Schultze. 2002. Human primary and memory cytotoxic T lymphocyte responses are efficiently induced by means of CD40-activated B cells as antigen-presenting cells: potential for clinical application. Blood. 99:3319–3325. [DOI] [PubMed] [Google Scholar]
- 45.Dustin, M. L., S. K. Bromley, Z. Y. Kan, D. A. Peterson, and E. R. Unanue. 1997. Antigen receptor engagement delivers a stop signal to migrating T lymphocytes. Proc. Natl. Acad. Sci. USA. 94:3909–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, D. M., M. L. Dustin, C. W. Cairo, H. S. Thatte, and D. E. Golan. 2006. Mechanisms of cellular avidity regulation in CD2–CD58 mediated T cell adhesion. ACS Chem. Biol. 1:649–658. [DOI] [PubMed] [Google Scholar]
- 47.Qi, S. Y., M. Krogsgaard, M. M. Davis, and A. K. Chakraborty. 2006. Molecular flexibility can influence the stimulatory ability of receptor-ligand interactions at cell-cell junctions. Proc. Natl. Acad. Sci. USA. 103:4416–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao, J. Y., Y. Yu, and M. L. Dustin. 2005. A model for CD2/CD58-mediated adhesion strengthening. Ann. Biomed. Eng. 33:483–493. [DOI] [PubMed] [Google Scholar]