Abstract
The ability to measure the abundance of many proteins precisely and simultaneously in experimental samples is an important, recent advance for static and dynamic, as well as descriptive and predictive, biological research. The value of multiplexed protein measurement is being established in applications such as comprehensive proteomic surveys, studies of protein networks and pathways, validation of genomic discoveries and clinical biomarker development. As standards do not yet exist that bridge all of these applications, the current recommended best practice for validation of results is to approach study design in an iterative process and to integrate data from several measurement technologies. This review describes current and emerging multiplexed protein measurement technologies and their applications, and discusses the remaining challenges in this field.
Longitudinal and cross-sectional studies to measure the levels of individual proteins in cells, tissues, extracts and biological fluids have been a long-standing and crucial component of progress in life-science research and clinical practice. Such measurements have three key purposes. First, they are surrogates (or biological markers) for organ activity, disease processes or drug action — examples include serum cardiac troponin I measurement for diagnosis of myocardial infarction1 and B-type natriuretic peptide measurement for diagnosis of congestive heart failure2. Second, they are proxies for activity when the measurement of organ, cell, pathway or protein activity is onerous — for example, using the titre of influenza-antigen-specific immunoglobulin rather than virus-neutralizing activity for the determination of immunity. Third, they are an end measure — for example, measuring the mass of protein in a timed urine sample when assessing glomerular function.
Longitudinal study.
A study in which measurements are made over a time course within an individual or individuals.
Cross-sectional study.
A study in which measurements are made at a single time point in many individuals.
Single nucleotide polymorphisms.
A specific location in a DNA sequence at which different people can have a different DNA base. Differences in a single base could change the protein sequence, leading to disease, or have no known consequences.
The accepted ‘gold standard’ for single-protein measurement is immunoassay, which exploits the diversity and specificity of antigen binding by immunoglobulins. Commonly used affinity ligands are monoclonal immunoglobulins or their antigen-binding domains, or polyclonal antisera. These assays use either single antigen-specific antibodies (as in immunohistochemistry) or, more commonly, two antibodies (as in ‘sandwich’ or competitive immunoassays). A wide range of labelling and signal-enhancement strategies have been developed that allow ligand binding to be detected through association of the ligand with a ‘read-out’ antibody that has particular fluorescent, colorimetric, histochemical or radioactive properties, or through changes in density or mass. Assay formats exist to measure proteins in solutions (enzyme-linked immunosorbent assays (ELISA) and immunospot assays), on the surface of cells (flow cytometry), within cells (immunohistochemical and immunofluorescent microscopy) and in organs (in vivo imaging with labelled antibodies). The most widely used format is ELISA, which has a well-established typical specification for measurement of single proteins in a solution; a lower limit of reliable quantitation of ∼1 pg ml−1; a dynamic range of 3 logs; coefficients of variation between replicate measurements of 5–20%; undetectable non-specific binding in the dynamic range; time to first-result of 1–2 h; and 96- or 384-well formats.
Recently, there has been increasing interest in the simultaneous measurement of many proteins in experimental samples. Predominantly, this interest has been prompted by the impact that highly multiplexed assays have had on genomics. During the past ten years, comprehensive surveys of genes, transcripts and single nucleotide polymorphisms have become commonplace for genetics and molecular biology discoveries. Furthermore, it is now recognized that single nucleic-acid measurements can lead to spurious conclusions in, for example, studies of transcriptional regulation or heritability of complex traits3,4.
Multiplexed measurement is logical for biological discovery with proteins because they constitutively function within networks, pathways, complexes and families5-7. The activity of an individual protein is dependent not only on its abundance, but also on the effects of interacting, modifying, antagonistic and synergistic proteins. Cytokine biology provides an example of the complexity of protein networks and inadequacy of monoplex measurement. In living systems, effects on target-cell activities are a dynamic aggregate of multiple agonist and antagonist cytokines, associated modifier proteins, ligands, receptors and receptor antagonists. Measurement of the level of a single cytokine in vivo is therefore a poor surrogate for activity, and integration of results from multiplexed measurement of component cytokines in a network is much more likely to be descriptive of biological processes (see later for further discussion).
The same caveat applies to the measurement of single proteins as surrogates (or biological markers) that are predictive of organ activity, disease process or drug action. In diagnostic parlance, the lack of concordance between individual protein abundance and clinical status or outcome can be referred to as a false-positive or false-negative result. The usefulness of diagnostic tests can be measured by positive predictive value (PPV) and negative predictive value (NPV). A logical advance in biomarker-based and activity-based protein measurement is surrogate protein selection predicated by a comprehensive understanding of protein networks, pathways and dynamics.
Until recently, specific, sensitive multiplexed protein measurements have not been generally undertaken because of technical challenges (BOX 1). These challenges include the diverse physico-chemical properties of proteins, their lability and the interferences introduced as a by-product of multiplexed measurement reagents. Solution-phase immunoassays, for example, have proven difficult to multiplex for these reasons. The diversity in dynamic range of protein concentrations in living systems is also challenging. For example, detectable proteins in human serum range in abundance from grams to tenths of picograms per ml. Furthermore, serum protein abundance can change by as much as 10,000-fold on stimulation. This is particularly true of many of the proteins that are most frequently measured, such as acute-phase reactant pentraxins or chemokines. A practical impediment to multiplexed protein measurement has been the expectation of a scientific community reared with the specifications of ELISAs that has been reluctant to deploy ratiometric or semi-quantitative multiplexing technologies. Nevertheless, reports of discoveries made using such technologies are starting to drive adoption of these approaches. This article briefly reviews technologies for multiplexed protein measurement, emerging standards, applications and remaining challenges.
Box 1 Protein array substrates and attachment chemistries
Five types of protein arrays are in common use today:
Planar glass or silicon chips.
These were the first surfaces to be used in protein arrays. Advantages include low cost, low lot-to-lot variability, low fluorescent background and extreme ‘flatness’ (important for accurate array scanning). However, glass or silicon protein chips require the onerous application of a surface chemistry for protein attachment; many attachment chemistries have been described6,58. Given a practical limit of multiplexing of sandwich immunoassays of ∼50, the parallel processing advantage of planar arrays is typically achieved by assaying many samples in partitioned sub-arrays on a single chip. The disadvantages of glass or silicon chips are the difficulties and lot-to-lot variability associated with attachment chemistry, and denaturation of most of the attached capture agent. Recent innovations have been silicon chips, either in a conventional array132 or in an etched, ‘compact disc’ format133.
Flow cytometric microbead assays.
‘Suspension’ arrays involve attachment of the capture agent for protein assays to microspheres, and their enumeration by flow cytometry129,134. Microspheres (and assays) can be multiplexed by using beads that differ in size or fluorescence intensity (with a fluorescent dye non-overlapping in emission spectrum to that used for the specific protein assay). Attractive aspects of suspension arrays are fluid-phase kinetics (which are faster than the solid-phase kinetics of planar arrays), custom multiplexing and greater precision (due to measurements of hundreds of beads for each analyte). Disadvantages of multiplexing of sandwich immunoassays are similar to those associated with bead and chip assays. The major limitation of cytometric bead assays is sample throughput. Microbead assays are available from Becton, Dickinson and Company (Cytometric Bead Array) and the partners of Luminex Corp. (xMAP).
Arrays in plastic microwells.
Enzyme-linked immunosorbent assays (ELISAs) in 96- and 384-well plastic plates have dominated monoplex protein measurement for many years. Genometrix pioneered the printing of arrays in 96-well plate format135. Multiplexed, microplate immunoassays are available in a 384-well format that features up to 16 assays per well from Pierce Biotechnology Inc. (SearchLight). Sample consumption is greater than on chips and sensitivity is lower.
Nitrocellulose on chips.
Nitrocellulose binds proteins in a non-covalent, irreversible manner. Modified nitrocellulose membranes have been used for many years for the detection of specific proteins that have been resolved by gel electrophoresis and western blotting. Attachment of nitrocellulose membranes to glass slides combines the advantages of microarrays with the protein binding capacity, long-term stability of printed proteins and assay methods of traditional blots. Nitrocellulose arrays are available from Whatman PLC (FAST Slides).
Three-dimensional gels.
Coating glass slides with a porous, hydrophilic, three-dimensional surface chemistry comprising a crosslinked polymer containing amine-reactive groups was initially described by Khrapko et al136. The advantages of three-dimensional gels are reaction kinetics that are more similar to fluid-phase than solid-phase kinetics (due to holding attached biomolecules, such as capture antibodies, away from the surface of the slide), low fluorescent background and reduced protein denaturation56,59. These benefits translate into approximately tenfold higher sensitivity than planar glass. The disadvantages of three-dimensional gel microarrays are cost and novelty (and as such have an unproven track record). Several types of three-dimensional microarray are available, for example, CodeLink from GE HealthCare Co. or HydroGel from PerkinElmer Inc.
Multiplexed protein measurement
Although the specific needs of biologists for multiplexed protein measurement are heterogeneous, they can be grouped into four main categories that follow a logical research and development succession: surveys of changes in protein abundance; modelling networks, pathways, and physiological and disease states; biomarker validation; and clinical diagnostics (TABLE 1). The optimal technology for multiplexed protein measurement varies among the four main types of application, together with the associated degree of multiplexing, desired specification and type of experimental design. The two principal types of technologies in use today in these applications are mass spectrometry and protein arrays. Mass spectrometry is typically used for comprehensive proteomic surveys and has previously been reviewed in this journal8, and so will be covered only briefly in this article, which focuses primarily on protein arrays.
Table 1.
Applications and technologies for multiplexed protein measurement
| Application category | Technology | Approximate multiplexing | Technology characteristics | Desired specification | Experimental design | Refs |
|---|---|---|---|---|---|---|
| Surveys of most, large changes in protein abundance | ‘Shotgun’ mass spectrometry | 2,500 | Universal, open architecture | Semi-quantitative; CV 30% | Iterative, cross-sectional; 50 samples | 137 |
| Modelling networks, pathways, physiological and disease states | Bead and chip protein arrays; indirect immunoassays | 250 | Multiple sets of canonical optimized assays | Quantitative; CV 10% | Longitudinal or dose-response; 500 samples | 7 |
| Biomarker validation; outcome surrogates in clinical trials | Bead and chip protein arrays; sandwich immunoassays | 25 | Flexible platform with rapid assay development | Quantitative; CV 5% | Cross-sectional or longitudinal; 500 samples | 115 |
| Panel-based clinical diagnostics | Multiplexed immunodiagnostic platform for rapid, point-of-care testing or automated central laboratory testing | 5 | Specific, highly optimized sets of assays | Quantitative; CV 3%; rapid format | Single sample comparison with reference range; >10,000 samples | 127 |
CV, coefficients of variation.
Cytokine.
Cytokines comprise several hundred small, soluble proteins that are powerful mediators of target cell activities such as migration, activation, phagocytosis, proliferation and apoptosis. Cytokines are secreted by producer cells into extracellular fluids, and act on target cells through binding to specific cell-surface receptors.
Positive predictive value (PPV). The probability that the patient has the disease when restricted to those who test positive.
Negative predictive value (NPV). An assessment of reliability of a negative test. The probability that the patient does not have the disease when restricted to those who test negative.
Proteomic surveys by mass spectrometry
The typical purpose of comprehensive proteomic survey experiments is to be initial exploratory tools for hypothesis generation. Usually, this involves the identification of a set of proteins with different levels of expression between two states. The technology specifications for comprehensive surveys of protein levels are: applicability to many sample types, including tissue lysates; ability to measure simultaneously most (thousands) of the proteins/peptides in a small sample volume in up to 100 samples; and moderate price.
The technology of choice for proteomic surveys today is ‘shotgun’ mass spectrometry. Many technology variations exist that differ in specificity, sensitivity, dynamic range, ease of use and cost. However, all are based on conversion of intact proteins in a sample into peptide fragments, followed by their volatilization, measurement of their mass-to-charge ratio (m/z ratio) and intensity, and then off-line protein identification by comparison with a database of peptides and their masses. Mass spectrometry has an open architecture, and is therefore not constrained by hypotheses regarding which groups of proteins are differentially expressed. Experiments typically involve the comparison of results from two, small (∼10–50), matched groups of samples, with identification of many intensity peaks that represent peptide expression differences. Database analysis translates these m/z ratio intensity differences into high-likelihood protein-abundance differences. Experiments are typically repeated, with protein abundance differences ‘discovered’ in a first study, and the differences ‘validated’ in a subsequent study. Mass spectrometry is ideal for rapid, cost-effective, initial proteomic surveys of experimental systems. However, it has limitations. First, analytical sensitivity for multiplexed protein measurement is much lower than ELISA, and limits the usefulness of the technology with small or ‘rare’ proteins, such as cytokines in blood samples. Sensitivity can, however, be enhanced by sample preparation to remove high-abundance proteins9,10 (for example, albumin and immunoglobulins in plasma samples). Second, extensive sample preparation is frequently necessary, especially for intracellular or membrane-associated proteins, which can introduce systematic bias. Furthermore, many proteins are labile, and degradation during sample preparation or storage greatly impairs protein identification. Third, automated protein identification by database lookup is frequently plagued by false positives (due to imprecision in m/z ratio measurement, non-specific peptide cleavage, or incomplete or inaccurate peptide database entries) and false negatives (due to orphan peptide fragments from small proteins). Bioinformatic algorithms are typically used to select expression differences in proteins with high likelihood identities. The development of empirical databases of expressed peptides in specific tissues, using methods such as multi-dimensional protein identification technology11-13, promises to assist in the correct identification of differentially expressed proteins. Similarly, use of statistical approaches, such as False Discovery Rate or Significance Analysis of Microarrays14-18, that incorporate appropriate adjustments for multiple observations will assist in the identification of true protein abundance differences. Fourth, mass spectrometry is semi-quantitative and best used for identification of gross differences in expression (≥fivefold). Specialized techniques do exist to make results more quantitative, but these require the use of radiolabelling steps beyond the scope of many experiments. Fifth, day-to-day, run-to-run, operator-to-operator and machine-to-machine variation continue to constrain use of mass spectrometry to experiments in which all the samples are run in a single batch. However, coefficients of variation are improving, and efforts are underway to ‘normalize’ or standardize spectra, thereby enabling benchmarking19. A recent advance — multiplexed reaction monitoring — provides significant improvement in sensitivity and coefficients of variation, and has the potential to extend the applicability of mass spectrometry from biomarker discovery to multiplexed protein measurement20.
For these reasons, multiplexed protein measurement by mass spectrometry is currently best used for hypothesis-generating, rather than hypothesis-testing, experiments. In experimental systems for which there is little knowledge of relevant protein networks or pathways, mass spectrometry can rapidly provide lists of candidate proteins and pathways7. An example of this is the discovery of serum protein biomarker candidates that are predictive of drug efficacy. Rigorous attention to experimental technique and bioinformatic analysis is crucial for success21,22.
Arrays for multiplexed protein measurement
Conventional immunoassay platforms have limited multiplexing capacity and high sample volume requirement. Since the original description by Ekins23,24 and early advances by Ward25,26, Schreiber27, Snyder28 and Brown29, high-throughput multiplex immunoassays that measure hundreds of proteins in complex biological matrices have become significant tools for quantitative proteomics studies, diagnostic discovery and biomarker-assisted drug development5,6,30-39.
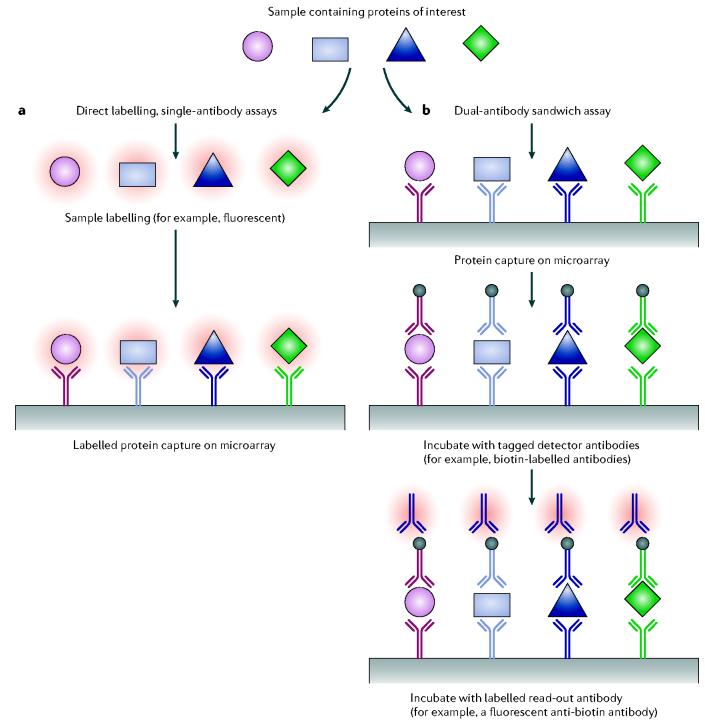
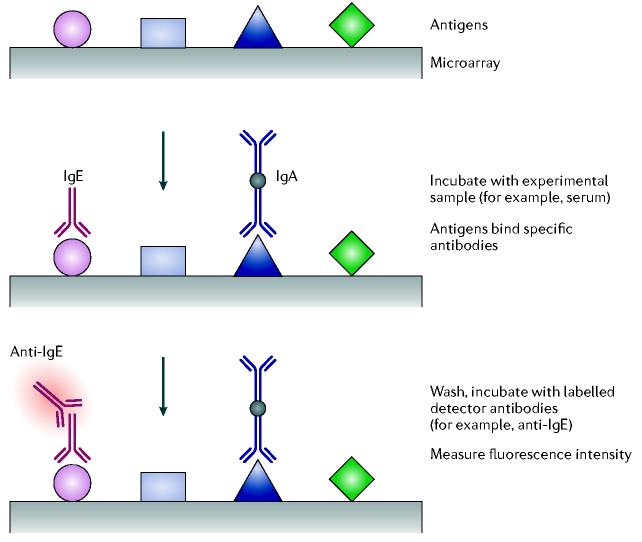
Three broad categories of antibody microarray experimental formats have been described (FIGS 1 and 2): first, direct labelling, single-capture antibody experiments29,40,41; second, dual antibody (capture and read-out antibody), sandwich immunoassays25,26,42-46; and third, antigen- or peptide-capture arrays, with single read-out antibodies47-51. In the direct labelling method, all proteins in a sample are tagged, thereby providing a means for detecting bound proteins following incubation on an antibody microarray (FIG. 1a). In the sandwich immunoassay format, proteins captured on an antibody micro-array are detected using a cocktail of labelled detection antibodies, with each antibody being matched to one of the spotted antibodies (FIG. 1b). In the antigen-capture arrays, the antigens are printed on the array and ligands in an experimental sample are detected on the basis of binding a labelled read-out antibody (FIG. 2). In each case, the spatial location of the specific capture reagent on the microarray defines the identity of analyte measured at that location: advantages and disadvantages of these formats are compared in TABLE 2.
Figure 1.
Schematic representation of the two antibody microarray experimental formats. a | Direct labelling, single-capture antibody experiments. All proteins in a sample are labelled (red haloes), thereby providing a means for detecting bound proteins following incubation on an antibody microarray. b | Dual-antibody (capture and read-out antibody) sandwich immunoassays. Proteins captured on an antibody microarray are detected by a cocktail of tagged detection antibodies, which are matched to the spotted antibodies. The detector antibody tag is then measured by binding of a labelled (red halo) read-out antibody.
Figure 2.
Schematic representation of antigen or peptide capture arrays. Antigens are printed on the array, which is then incubated with an experimental sample containing antibodies. Binding of the antibodies to the antigens can then be detected by binding a secondary, anti-immunoglobulin (a read-out antibody) that has been fluorescently labelled (red halo). Ig, immunoglobulin.
Table 2.
Pros and cons of array formats for multiplexed protein measurement
| Format | Advantages | Disadvantages | Refs |
|---|---|---|---|
| Single antibody | Capacity to multiplex hundreds of analytes | Currently semi-quantitative (ratiometric) | 29 |
| Require single capture antibodies | Low specificity | ||
| Low sensitivity | |||
| Dual antibody | High specificity | Limited to ∼50 analytes per array | 25,26 |
| High sensitivity | Requires matched antibody pairs | ||
| Quantitative | |||
| Antigen array | Capacity to multiplex hundreds of analytes | Generally limited to assays of antibodies, for example, autoantibody detection, infectious disease serodiagnosis | 47,49 |
| Require single read-out antibody | |||
| Capacity to measure certain activities | Variability in specificity, sensitivity and quantitativeness |
Various microarray substrates have been described, including nylon membranes, plastic microwells, planar glass slides, gel-based arrays and beads in suspension arrays42,45,52-59. Numerous studies have been carried out to optimize antibody attachment to the microarray substrate60-62. In addition, various signal generation and signal enhancement strategies have been used in antibody arrays, including colorimetry, radioactivity, fluorescence, chemiluminescence, quantum dots, other nanoparticles, enzyme-linked assays, resonance light scattering, tyramide signal amplification, rolling circle amplification and eTag protein proximity assays6,25,63-65. Although most arrays use recombinant or monoclonal antibodies, alternative high-affinity ligands have recently been used in microarrays66-70.
Each of these formats and procedures has distinct advantages and disadvantages, relating broadly to sensitivity, specificity, dynamic range, multiplexing capability, precision, throughput and ease of use. In general, multiplexed microarray immunoassays are ambient analyte assays71, involving picolitre antibody ‘spots’ and microlitre sample volumes. Besides low requirements for reagent and sample, miniaturization of reactions confers the benefits of reduced reaction times (as a consequence of short diffusion distances) and improved signal-to-noise ratios72.
Despite the heterogeneity of formats and procedures available for multiplexed protein measurement, the ‘gold standard’ for specification remains the ELISA, albeit with additional specifications for a multiplexed assay environment. Unique considerations in assembling multiplexed immunoassays include the elimination of assay cross-reactivity, configuration of multianalyte sensitivities, achievement of dynamic ranges appropriate for biological relevance when carried out in diverse matrices and biological states, and optimization of reagent manufacturing and chip production to achieve acceptable reproducibility. Specifications and standards for antibody microarrays are being developed73-75. Specifically, requirements have been described for comprehensive validation programmes to identify and minimize antibody cross-reaction under highly multiplexed conditions, controls to normalize sample replicate measurements, and quality control testing of reagents, antibody microarrays, sensitivity, dynamic range and platform precision. A key remaining challenge for many analytes is the availability of high-affinity reagents with specificity in multiplexed assays, because almost all monoclonal antibody pairs were selected for performance in a monoplex assay format. Another challenge, in common with other high-throughput technologies, is the development of statistical approaches to integrate data from replicated assays and to reveal the significance of biomarker findings.
Arrays for modelling networks and pathways
Modelling gene or protein networks or pathways in disease states is a recent field of endeavour. Proteins usually function in interdependent networks. The goal of model development is to understand inter-relationships among the members of a protein network or family in a disease state. Precise models enable prediction of the biological responses to a perturbation, and detection of novel associations among members76.
Model development is a mathematical exercise. Two types of mathematical approach are being implemented in static and dynamic biological systems. The standard approach in biology has been reductionist analysis — division of the system into component variables and ‘solving’ a differential equation for each with empirical data77. Although biological systems are generally complex and nonlinear, much of our current biological knowledge has been derived from this type of deterministic, reduction-ist analysis78. Furthermore, disease states are frequently associated with linear dynamics79 (or, more accurately, with the breakdown of multi-scale fractal complexity). For these reasons, reductionist methods are likely to remain useful for the foreseeable future for quantitative prediction of responses to perturbation of networks. Notable recent examples of progress are the development of dynamic models of yeast responses to hyperosmolar shock80 and treatment of chronic myeloid leukaemia with imatinib81. The second, newer approach to proteomic modelling is finite-state modelling, which uses the changes in gene3,4 or protein expression76 that follow perturbation of a particular node to identify the topology of complex networks. This approach, although relatively recently introduced to systems biology76,82, has proven useful in other nonlinear systems and enables detection of novel associations among elements76 or assessment of network robustness.
Current biological experimentation, particularly for multiplexed protein measurement, mandates that these modelling approaches must have the flexibility to use noisy, incomplete data. When modelling networks, pathways and disease states, the technological specifications should include the following: assays for many of the major elements in that network or pathway (typically 25–200 proteins); measurement of those within the biological dynamic range with reasonable precision (coefficients of variation of ≤15%); and the ability to measure many samples (≥500) without confounding run-to-run imprecision. The technologies of choice for this application are immunofluorescent bead or planar arrays. Experiments typically involve following the time course or dose response in multiple individuals or following multiple treatments or interventions. As with ELISAs, fluorescent intensities are converted back to mass units using standard curves, and several replicate observations are made for each protein and sample to calculate standard errors of values73.
One field of research that is being dramatically affected by this use of protein arrays is the characterization of humoral immunity in common allergies34,47,83-87, autoimmune disorders31,32,49,88-92, cancers37,93,94 and infectious diseases33,38,95-99. Comprehensive characterization of changes in humoral immunity is starting to become possible through the development of protein chips containing hundreds or thousands of potential allergens, autoantigens or epitopes arrayed as microscopic spots on planar glass slides (FIG. 2). Arrays are incubated with serum, then washed, ‘developed’ by incubation with a fluorescently tagged anti-immunoglobulin (for example, anti-IgE for allergy detection) or anti-immunoglobulin subtype, and quantified in a fluorescence scanner. Antigen arrays are simple to develop and calibrate. Hundreds of antibody specificities can be screened simultaneously by class, subclass and titre.
Clinical uses of such protein arrays include disease classification on the basis of reactive or autoreactive epitopes, monitoring disease progression by measuring the change in epitope dynamics, immunoglobulin class or sub-class switching, and monitoring disease activity by analysis of the antibody titre. In limited comparisons with traditional ‘monoplex’ in vitro diagnostics, multiplexed antigen arrays have shown similar or improved diagnostic sensitivity and specificity83,85. Although studies so far have been limited to descriptions of states or differences between states, antigen arrays have the power to enable development of quantitative mathematical models of the dynamics of humoral immune systems in health and disease100,101. Furthermore, the development of high-throughput peptide expression systems has the potential to greatly expand the repertoire of such studies by allowing a broad array of epitopes to be surveyed simultaneously for humoral immune responses.
A related, highly innovative development is the construction of microarrays of substrates for protein activities or protein modifications28,36,50,51,102-110. Rather than measuring multiplexed protein abundance changes between states or in disease processes, such arrays measure changes in both specific protein activity and in global patterns of activity.
As noted above, a second area in which protein arrays are enabling new understanding of networks, pathways and disease states is in cytokine biology43,45,111-114. Multiplexed, immunoassay arrays allow quantitative, comprehensive measurement of cytokine families and networks. Cytokine arrays require less multiplexing than antigen arrays, but are technically more demanding, having the following requirements: they must be sandwich immunoassays (with antibodies capable of detecting two epitopes); pg ml−1 sensitivity; and a dynamic range of 3 logs (without detectable cross reactivity at the lower quantitation limits). Formats available for multiplexed cytokine measurement include bead arrays, planar glass arrays and nitrocellulose membrane arrays. Such arrays enable the multiplexed measurement of time course and dose response of cytokine production by specific leukocyte lineages following treatment with various lectins or ionophores or in disease states. These studies are essential for the validation of findings from gene expression or shotgun mass spectrometry studies, and are crucial for the development of mathematical models of cytokine networks. Such models permit prediction of responses to various stimuli and downstream changes in bioactivities. One such study examined cytokine network responses of dendritic cells to in vitro treatment with defensins114. RNAse, which was included in the study as a control treatment, was, unexpectedly, found to elicit dose-dependent secretion of pro-inflammatory cytokines. Subsequent studies confirmed that RNAse A superfamily proteins activate dendritic cells in a manner similar to that of tumour-necrosis factor — a hitherto unknown property.
21 CFR compliant.
In compliance with Title 21 of the Code of Federal Regulations. This is the principal law enforced by the US FDA.
Protein arrays for biomarker development
A biomarker is a measurable ‘biological marker’ that correlates with a specific outcome or state: this subject has been reviewed recently in this journal115 and is only briefly discussed here. As applied to drug development, the biomarker hypothesis states that changes in levels of blood or tissue proteins, individually or multiplexed, are highly and specifically characteristic of disease states and therapeutic outcomes. The basis of this hypothesis is that biological systems are adaptive and that challenges to homeostasis effect characteristic topological perturbations of protein networks.
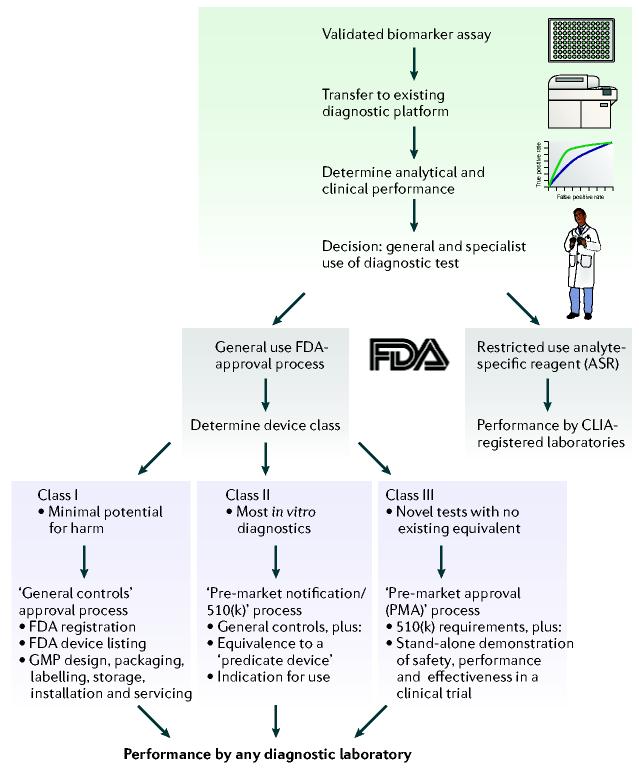
Biomarker development occurs in a four-step process of discovery, followed by replication in independent cohorts, validation of diagnostic sensitivity and specificity, and, finally, translation into a clinical diagnostic test or surrogate endpoint in a clinical study (FIG. 3). Candidate biomarkers are triaged at each stage of development. A biomarker can be a single protein (such as prostate-specific antigen), a panel of proteins or a combination of one or more proteins and other clinical measures, together with an algorithm that integrates individual values. In cases in which individual biomarkers lack adequate PPV or NPV, combinations of 3–10 protein profiles can be effective, providing a panel and algorithm with high PPV and NPV.
Figure 3.
Steps for the development of validated biomarkers into a clinical diagnostic test. Optimized, multiplexed assays are developed for validated biomarkers and transferred to an immunodiagnostic platform. Then specifications are determined regarding analytical and clinical performance. Finally, in vitro diagnostic development is driven by regulatory, manufacturing and marketing considerations. CLIA, Clinical Laboratory Improvement Amendment (the US law to ensure quality laboratory testing); GMP, good manufacturing practice.
Given the magnitude of the current monoplex immunodiagnostic market (US$12 billion annually), interest is burgeoning in protein biomarker development for: first, early diagnosis, differential diagnosis, disease staging and determination of disease prognosis; second, patient selection and as surrogate endpoints during clinical trials of investigational drugs; and third, companion diagnostics for guidance in drug prescribing.
Although mass spectrometry is an excellent technology for the identification of biomarker candidates, it is currently inadequate for the later stages of biomarker development, or use in 21 CFR-compliant clinical trials or regulated in vitro diagnostics116. The technology specifications for multiplexed protein measurement for biomarker development in clinical studies are onerous: multiplexed assays must be available for most biomarker candidate proteins (typically 5–25 analytes) and be of sufficient sensitivity to measure analytes within the biological dynamic range; multiplexed assays must have similar precision to an ELISA (coefficients of variation of ≤10%); assays should have high-throughput capability; and have sample volume requirements and good laboratory practice (GLP) or good manufacturing practice (GMP) development times that concur with ‘go/no-go’ decision timelines. The technologies of choice for clinical studies are fluorescent bead or chip immunoassay arrays or ELISAs. Experiments typically involve analysis of samples from multiple time points or dose intensities in all patients in a clinical study, and comparison of biomarker value means or medians in groups of patients that differ in primary or secondary endpoints.
Most protein biomarker publications so far have involved the early diagnosis, differential diagnosis, disease staging or determination of disease prognosis37,41,117-122. Serum and plasma are the most widely examined specimen types until now, although interesting studies in saliva, tears, breath condensates, cerebrospinal fluid and tissue lysates have also been reported123-125. Studies have either used mass spectrometry for protein biomarker discovery, followed by array-based replication, or arrays both for biomarker discovery and replication123. No studies have been published that have taken biomarkers through the entire development process, and it is anticipated that many of the candidate biomarkers identified so far will lack sufficient predictive value for diagnostic development.
In particular, biomarkers are of great current interest in clinical trials of investigational drugs, given their potential ability to reduce trial duration and cohort size, or detect drug efficacy with greater sensitivity. Although only a few publications have substantiated this premise, most pharmaceutical companies are actively exploring the use of biomarkers in clinical trials. Two specific types of biomarkers that are highly prized are ‘bridging biomarkers’ that are useful both in preclinical and clinical development, and ‘surrogate markers’ that are sufficiently validated to substitute for a primary or secondary clinical endpoint or outcome in a clinical trial. In this author's experience, multiplexed protein measurement in Phase I and II clinical trials for several therapeutic areas can yield useful, objective measures of drug mechanism-of-action, effect and efficacy. However, there has yet to be an example of a drug for which approval was expedited by the use of multiplexed protein measurement. Furthermore, claims for the use of multiplexed protein measurement in drug safety in clinical studies are, in this author's opinion, premature.
Arrays for panel-based clinical diagnostics
Multiplexed protein measurement is not new to clinical chemistry. However, until now, multichannel analysers (both discrete and continuous flow) have generally used large amounts of sample and reagents, carried out multiplexing by serial performance of single assays, and have not incorporated immunoassay or other ligand-binding assay techniques for low-concentration analytes. Furthermore, physicians have integrated the results of several single protein analyte measurements for the evaluation of organ function in hepatic, renal, myocardial, endocrine, immunological or infectious diseases. One example of a multiprotein diagnostic panel in common use is a combination of elevated levels of cardiac isoenzymes of protein markers, such as creatinine phosphokinase, troponin T, aldolase and lactose dehydrogenase, in serum in the diagnosis of acute myocardial infarction.
Two significant, recent advances in immuno-diagnostics are the development of miniaturized, multiplexed assays and panel algorithms that translate multiple analyte concentrations into a specific diagnostic or prognostic index23,72,117. Advantages of miniaturized, multiplexed assays are reduced time to first result, reduced cost of goods and smaller sample volume requirements (which is particularly important for paediatric assays). Early publications have engendered considerable excitement about biomarker-panel-based diagnostics or prognostics, especially for early diagnosis of cancer. It must be emphasized, however, that replication in independent cohorts, analytical validation by transfer to a qualified platform and biological validation by incrimination of a network or pathway in a particular disease process are essential for biomarker panels to have analytical and statistical rigour. With few exceptions, biomarker-based diagnostic candidates have not yet been examined for adequate PPV, NPV or area-under-the-curve of receiver–operator-characteristics126. Serum or plasma protein biomarker panels in development include those for differentiation of acute thrombotic and haemorrhagic stroke117, and as objective outcome measures in sepsis and community acquired pneumonia.
Development of fully validated biomarkers into a clinical diagnostic test involves several steps (FIG. 3). The first phase involves multiplexed immunoassay development, in which optimized, multiplexed assays are developed for validated biomarkers and transferred to an immunodiagnostic platform. This stage is process delimited and guided by strict specifications and quality controls. The result is a panel of 2–10 dual-antibody sandwich, direct or indirect, immunoassays with documented specifications regarding multiplexed lower limit of reliable quantitation, precision, interferences, component stability, dynamic range, ease-of-use and turn-around time. The next stage is final in vitro diagnostic development, which is driven by regulatory, manufacturing and marketing considerations. Few established platforms exist for multiplexed in vitro diagnostics, and these capture the analytes of interest on chromatographic devices127,128, on multiple labelled particles129-131 or a single surface132.
Conclusions
Multiplexed protein measurement is a rapidly advancing field (see TABLE 3 for primary, published manuscripts illustrating examples of uses of multiplexed protein measurement technologies) that has the broadest potential of any existing ‘-omics’ technology to transform drug discovery and development in the next 10 years. During this period, multiplexed protein abundance, activation state and activity measurement will become broadly applied for target validation and, with adaptation, will be applied to miniaturized, multiplexed high-throughput screens. In particular, quantitative, multiplexed protein abundance measurements will become obligatory for the validation of the biological significance of results of functional genomics studies. Guided by FDA prompting, tort concerns and pipeline needs, biomarker data from multiplexed protein measurement technologies will become increasingly the norm, both in Investigational New Drug (IND) approval submissions and Phase IV (post-marketing) studies. Inexorably, we are entering an era of personalized medicine, in which inclusion criteria for clinical studies will be based increasingly on a patient's genetic or proteomic profile, surrogate endpoints will become more prevalent and approved indications will more routinely include biomarker-based diagnostics. In tandem, multiplexed protein measurement for biomarker-based diagnostic and prognostic testing will become the largest growth segment of the immunodiagnostics industry. Finally, multiplexed protein measurement has the capacity to identify surrogates that will be integrated into clinical indices, treatment algorithms, and, ultimately, into dynamic disease models that permit real-time, data-driven patient management.
Table 3.
Proof-of-concept applications of array-based multiplexed protein measurement
| Application | Technology | Disease or biological process | Refs |
|---|---|---|---|
| Autoantibody profiling | Antigen arrays | Allergy | 47,83,84 |
| Autoimmunity | 31,32,49 | ||
| Cancer | 138 | ||
| Antibody response profiling | Antigen arrays | Immunization, exposure to infectious agents | 33,139 |
| Diagnosis of parasite differential | 140 | ||
| Determination of response to infection | 141 | ||
| Cytokine network profiling, enzyme substrate profiling | Antibody arrays, peptide arrays | Determination of antibody specificity | 102 |
| Discovery of new biological activities of substances | 28,114,103-109 | ||
| Clinical biomarker development | Antibody arrays | Inflammatory bowel disease | 119 |
| Sepsis | Personal communication | ||
| Lung and ovarian cancer | 121,122 | ||
| IND studies | 142 | ||
| Cytokine network profiling | Antibody arrays | Development of network/pathway models | 5,45,114 |
| Understanding disease process or mechanism of action | Antibody arrays | Severe combined immunodeficiency | Unpublished observations |
| Cerebral palsy | 118,143 | ||
| Systemic lupus erythematosus | Unpublished observations | ||
| IND studies | 142 | ||
| Detecting adverse events | Antibody arrays | Immunization | Unpublished observations |
| Clinical diagnoses | Antibody arrays | Point-of-care stroke | 117 |
| Diagnosis of myocardial infarction | 126,128 | ||
| Screening of drugs of abuse | 127 | ||
| Diagnosis of infection differential |
IND, Investigational New Drug application.
Acknowledgements
A Deo lumen, ab amicis auxiliam (to God for illumination, to friends for help). This work was partially supported by a National Institutes of Health grant. Thanks to J. Huntley, D. Gessler and the referees for critically reviewing this manuscript.
Footnotes
Competing interests statement
The author declares competing financial interests: see web version for details.
FURTHER INFORMATION
PRoteomics IDEntifications (PRIDE): http://www.ebi.ac.uk/pride
Expert Protein Analysis System: http://www.expasy.org
Plasma Proteome Database: http://www.plasmaproteomedatabase.org
Global Proteome Machine: http://www.thegpm.org
Human Protein Reference Database: http://www.hprd.org
PhysioNet: http://www.physionet.org
Access to this interactive links box is free online.
References
- 1.Katus HA, et al. Enzyme linked immuno assay of cardiac troponin T for the detection of acute myocardial infarction in patients. J. Mol. Cell. Cardiol. 1989;21:1349–1353. doi: 10.1016/0022-2828(89)90680-9. [DOI] [PubMed] [Google Scholar]
- 2.Mukoyama M, et al. Increased human brain natriuretic peptide in congestive heart failure. N. Engl. J. Med. 1990;323:757–758. doi: 10.1056/NEJM199009133231114. [DOI] [PubMed] [Google Scholar]
- 3.Laubenbacher R, Stigler B. A computational algebra approach to the reverse engineering of gene regulatory networks. J. Theor. Biol. 2004;229:523–537. doi: 10.1016/j.jtbi.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Luscombe NM, et al. Genomic analysis of regulatory network dynamics reveals large topological changes. Nature. 2004;431:308–312. doi: 10.1038/nature02782. [DOI] [PubMed] [Google Scholar]
- 5.Kingsmore SF, Patel DD. Multiplexed protein profiling on antibody-based microarrays by rolling circle amplification. Curr. Opin. Biotechnol. 2003;14:74–81. doi: 10.1016/s0958-1669(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 6.Schweitzer B, Kingsmore SF. Measuring proteins on microarrays. Curr. Opin. Biotechnol. 2002;13:14–19. doi: 10.1016/s0958-1669(02)00278-1. [DOI] [PubMed] [Google Scholar]
- 7.Petricoin EF, et al. Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J. Clin. Oncol. 2005;23:3614–3621. doi: 10.1200/JCO.2005.02.509. [DOI] [PubMed] [Google Scholar]
- 8.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics, translating benchside promise into bedside reality. Nature Rev. Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 9.Tang HY, et al. A novel four-dimensional strategy combining protein and peptide separation methods enables detection of low-abundance proteins in human plasma and serum proteomes. Proteomics. 2005;5:3329–3342. doi: 10.1002/pmic.200401275. [DOI] [PubMed] [Google Scholar]
- 10.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 11.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nature Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 12.Kislinger T, Gramolini AO, Maclennan DH, Emili A. Multidimensional Protein Identification Technology (MudPIT), technical overview of a profiling method optimized for the comprehensive proteomic investigation of normal and diseased heart tissue. J. Am. Soc. Mass Spectrom. 2005;16:1207–1220. doi: 10.1016/j.jasms.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Washburn MP. Utilisation of proteomics datasets generated via multidimensional protein identification technology (MudPIT) Brief. Funct. Genomic. Proteomic. 2004;3:280–286. doi: 10.1093/bfgp/3.3.280. [DOI] [PubMed] [Google Scholar]
- 14.Storey JD, Tibshirani R. Statistical significance for genome wide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao LP, Prentice R, Shen F, Hsu L. On the assessment of statistical significance in disease-gene discovery. Am. J. Hum. Genet. 1999;64:1739–1753. doi: 10.1086/512072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weller JI, Song JZ, Heyen DW, Lewin HA, Ron M. A new approach to the problem of multiple comparisons in the genetic dissection of complex traits. Genetics. 1998;150:1699–1706. doi: 10.1093/genetics/150.4.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacayo NJ, et al. Gene expression profiles at diagnosis in de novo childhood AML patients identify FLT3 mutations with good clinical outcomes. Blood. 2004;104:2646–2654. doi: 10.1182/blood-2003-12-4449. [DOI] [PubMed] [Google Scholar]
- 19.Haab BB, et al. Immunoassay and antibody microarray analysis of the HUPO Plasma Proteome Project reference specimens: systematic variation between sample types and calibration of mass spectrometry data. Proteomics. 2005;5:3278–3291. doi: 10.1002/pmic.200401276. [DOI] [PubMed] [Google Scholar]
- 20.Anderson NL, Hunter CL. Quantitative mass spectrometric MRM assays for major plasma proteins. Mol. Cell. Proteomics. 2005 Dec 6; doi: 10.1074/mcp.M500331-MCP200. Initial description of multiplexed plasma protein measurement by modified mass spectrometry with sufficient precision for biomarker validation. [DOI] [PubMed] [Google Scholar]
- 21.Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinformatics. 2003;4:24. doi: 10.1186/1471-2105-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J, Coombes KR, Morris JS, Baggerly KA. The importance of experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief. Funct. Genomic. Proteomic. 2005;3:322–331. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- 23.Ekins RP, Chu FW. Multianalyte microspot immunoassay — microanalytical ‘compact disk’ of the future. Clin. Chem. 1991;37:1955–1967. [PubMed] [Google Scholar]
- 24.Ekins RP. Multi-analyte immunoassay. J. Pharm. Biomed. Anal. 1989;7:155–168. doi: 10.1016/0731-7085(89)80079-2. A pioneering publication of multiplexed protein measurement. [DOI] [PubMed] [Google Scholar]
- 25.Lizardi PM, et al. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nature Genet. 1998;19:225–232. doi: 10.1038/898. The first description of signal amplification to permit high-sensitivity protein arrays. [DOI] [PubMed] [Google Scholar]
- 26.Schweitzer B, et al. Inaugural article: immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc. Natl Acad. Sci. USA. 2000;97:10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, et al. Analysis of yeast protein kinases using protein chips. Nature Genet. 2000;26:283–289. doi: 10.1038/81576. The first description of whole-proteome arrays. [DOI] [PubMed] [Google Scholar]
- 29.Haab BB, Dunham MJ, Brown PO. Protein microarrays for highly parallel detection and quantitation of specific proteins and antibodies in complex solutions. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-2-research0004. RESEARCH0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walter G, Bussow K, Cahill D, Lueking A, Lehrach H. Protein arrays for gene expression and molecular interaction screening. Curr. Opin. Microbiol. 2000;3:298–302. doi: 10.1016/s1369-5274(00)00093-x. [DOI] [PubMed] [Google Scholar]
- 31.Hueber W, Utz PJ, Steinman L, Robinson WH. Autoantibody profiling for the study and treatment of autoimmune disease. Arthritis Res. 2002;4:290–295. doi: 10.1186/ar426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson WH, Steinman L, Utz PJ. Protein arrays for autoantibody profiling and fine-specificity mapping. Proteomics. 2003;3:2077–2084. doi: 10.1002/pmic.200300583. [DOI] [PubMed] [Google Scholar]
- 33.Bacarese-Hamilton T, Gray J, Crisanti A. Protein microarray technology for unraveling the antibody specificity repertoire against microbial proteomes. Curr. Opin. Mol. Ther. 2003;5:278–284. [PubMed] [Google Scholar]
- 34.Deinhofer K, et al. Microarrayed allergens for IgE profiling. Methods. 2004;32:249–254. doi: 10.1016/j.ymeth.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 35.Utz PJ. Protein arrays for studying blood cells and their secreted products. Immunol. Rev. 2005;204:264–282. doi: 10.1111/j.0105-2896.2005.00251.x. [DOI] [PubMed] [Google Scholar]
- 36.Hirabayashi J. Oligosaccharide microarrays for glycomics. Trends Biotechnol. 2003;21:141–143. doi: 10.1016/S0167-7799(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 37.Haab BB. Antibody arrays in cancer research. Mol. Cell. Proteomics. 2005;4:377–383. doi: 10.1074/mcp.M500010-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Bacarese-Hamilton T, Ardizzoni A, Gray J, Crisanti A. Protein arrays for serodiagnosis of disease. Methods Mol. Biol. 2004;264:271–283. doi: 10.1385/1-59259-759-9:271. [DOI] [PubMed] [Google Scholar]
- 39.Predki PF. Functional protein microarrays: ripe for discovery. Curr. Opin. Chem. Biol. 2004;8:8–13. doi: 10.1016/j.cbpa.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Zhou H, et al. Two-color, rolling-circle amplification on antibody microarrays for sensitive, multiplexed serum-protein measurements. Genome Biol. 2004;5:R28. doi: 10.1186/gb-2004-5-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JC, et al. Antibody microarray profiling of human prostate cancer sera: antibody screening and identification of potential biomarkers. Proteomics. 2003;3:56–63. doi: 10.1002/pmic.200390009. [DOI] [PubMed] [Google Scholar]
- 42.Huang RP. Detection of multiple proteins in an antibody-based protein microarray system. J. Immunol. Methods. 2001;255:1–13. doi: 10.1016/s0022-1759(01)00394-5. [DOI] [PubMed] [Google Scholar]
- 43.Huang RP, Huang R, Fan Y, Lin Y. Simultaneous detection of multiple cytokines from conditioned media and patient's sera by an antibody-based protein array system. Anal. Biochem. 2001;294:55–62. doi: 10.1006/abio.2001.5156. [DOI] [PubMed] [Google Scholar]
- 44.Wiese R, Belosludtsev Y, Powdrill T, Thompson P, Hogan M. Simultaneous multianalyte ELISA performed on a microarray platform. Clin. Chem. 2001;47:1451–1457. [PubMed] [Google Scholar]
- 45.Schweitzer B, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nature Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang CC, et al. Array-based multiplexed screening and quantitation of human cytokines and chemokines. J. Proteome Res. 2002;1:337–343. doi: 10.1021/pr0255203. [DOI] [PubMed] [Google Scholar]
- 47.Wiltshire S, et al. Detection of multiple allergen-specific IgEs on microarrays by immunoassay with rolling circle amplification. Clin. Chem. 2000;46:1990–1993. One of the first publications to describe antigen arrays for the measurement of specific immunoglobulins. [PubMed] [Google Scholar]
- 48.Holt LJ, Bussow K, Walter G, Tomlinson IM. By-passing selection: direct screening for antibody-antigen interactions using protein arrays. Nucleic Acids Res. 2000;28:E72. doi: 10.1093/nar/28.15.e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robinson WH, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nature Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 50.Lesaicherre ML, Uttamchandani M, Chen GY, Yao SQ. Antibody-based fluorescence detection of kinase activity on a peptide array. Bioorg. Med. Chem. Lett. 2002;12:2085–2088. doi: 10.1016/s0960-894x(02)00378-5. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc. Natl Acad. Sci. USA. 2003;100:9330–9335. doi: 10.1073/pnas.1633513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Jager W, te Velthuis H, Prakken BJ, Kuis W, Rijkers GT. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin. Diagn. Lab. Immunol. 2003;10:133–139. doi: 10.1128/CDLI.10.1.133-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moreno-Bondi MC, Alarie JP, Vo-Dinh T. Multi-analyte analysis system using an antibody-based biochip. Anal. Bioanal. Chem. 2003;375:120–124. doi: 10.1007/s00216-002-1626-y. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz-Taylor LA, et al. Monolayers of derivatized poly(l-lysine)-grafted poly(ethylene glycol) on metal oxides as a class of biomolecular interfaces. Proc. Natl Acad. Sci. USA. 2001;98:852–857. doi: 10.1073/pnas.98.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pawlak M, et al. Zeptosens' protein microarrays: a novel high performance microarray platform for low abundance protein analysis. Proteomics. 2002;2:383–393. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 56.Nallur G, et al. Protein and nucleic acid detection by rolling circle amplification on gel-based microarrays. J. Biomed. Microdevices. 2003;5:117–125. [Google Scholar]
- 57.Angenendt P, Glokler J, Konthur Z, Lehrach H, Cahill DJ. 3D protein microarrays: performing multiplex immunoassays on a single chip. Anal. Chem. 2003;75:4368–4372. doi: 10.1021/ac034260l. [DOI] [PubMed] [Google Scholar]
- 58.Angenendt P, Glokler J, Sobek J, Lehrach H, Cahill DJ. Next generation of protein microarray support materials: evaluation for protein and antibody microarray applications. J. Chromatogr. A. 2003;1009:97–104. doi: 10.1016/s0021-9673(03)00769-6. [DOI] [PubMed] [Google Scholar]
- 59.Rubina AY, et al. Hydrogel-based protein microchips: manufacturing, properties, and applications. Biotechniques. 2003;34:1008–1022. doi: 10.2144/03345rr01. [DOI] [PubMed] [Google Scholar]
- 60.Shao W, et al. Optimization of rolling circle amplified protein microarrays for multiplexed protein profiling. J. Biomed. Biotechnol. 2003;4:1–9. doi: 10.1155/S1110724303209268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peluso P, et al. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- 62.Angenendt P, Glokler J. Evaluation of antibodies and microarray coatings as a prerequisite for the generation of optimized antibody microarrays. Methods Mol. Biol. 2004;264:123–134. doi: 10.1385/1-59259-759-9:123. [DOI] [PubMed] [Google Scholar]
- 63.Liang RQ, Tan CY, Ruan KC. Colorimetric detection of protein microarrays based on nanogold probe coupled with silver enhancement. J. Immunol. Methods. 2004;285:157–163. doi: 10.1016/j.jim.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 64.Huang R, et al. Enhanced protein profiling arrays with ELISA-based amplification for high-throughput molecular changes of tumor patients' plasma. Clin. Cancer Res. 2004;10:598–609. doi: 10.1158/1078-0432.ccr-0697-03. [DOI] [PubMed] [Google Scholar]
- 65.Chan-Hui PY, Stephens K, Warnock RA, Singh S. Applications of eTag trade mark assay platform to systems biology approaches in molecular oncology and toxicology studies. Clin. Immunol. 2004;111:162–174. doi: 10.1016/j.clim.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 66.Winssinger N, et al. PNA-encoded protease substrate microarrays. Chem. Biol. 2004;11:1351–1360. doi: 10.1016/j.chembiol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 67.Gilbert I, et al. Double chip protein arrays using recombinant single-chain Fv antibody fragments. Proteomics. 2004;4:1417–1420. doi: 10.1002/pmic.200300736. [DOI] [PubMed] [Google Scholar]
- 68.Naffin JL, et al. Immobilized peptides as high-affinity capture agents for self-associating proteins. Chem. Biol. 2003;10:251–259. doi: 10.1016/s1074-5521(03)00049-8. [DOI] [PubMed] [Google Scholar]
- 69.Stich N, et al. Phage display antibody-based proteomic device using resonance-enhanced detection. J. Nanosci. Nanotechnol. 2002;2:375–381. doi: 10.1166/jnn.2002.111. [DOI] [PubMed] [Google Scholar]
- 70.Bock C, et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics. 2004;4:609–618. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 71.Saviranta P, et al. Evaluating sandwich immunoassays in microarray format in terms of the ambient analyte regime. Clin. Chem. 2004;50:1907–1920. doi: 10.1373/clinchem.2004.037929. [DOI] [PubMed] [Google Scholar]
- 72.Ekins RP. Ligand assays: from electrophoresis to miniaturized microarrays. Clin. Chem. 1998;44:2015–2030. [PubMed] [Google Scholar]
- 73.Perlee L, et al. Development and standardization of multiplexed antibody microarrays for use in quantitative proteomics. Proteome Sci. 2004;2:9. doi: 10.1186/1477-5956-2-9. One of the first papers to suggest standards and quality controls for multiplexed immunoassays. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pang S, et al. A comparability study of the emerging protein array platforms with established ELISA procedures. J. Immunol. Methods. 2005;302:1–12. doi: 10.1016/j.jim.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 75.Poetz O, et al. Protein microarrays for antibody profiling: specificity and affinity determination on a chip. Proteomics. 2005;5:2402–2411. doi: 10.1002/pmic.200401299. [DOI] [PubMed] [Google Scholar]
- 76.Sachs K, Perez O, Pe'er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 77.Aderem A, Smith KD. A systems approach to dissecting immunity and inflammation. Semin. Immunol. 2004;16:55–67. doi: 10.1016/j.smim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 78.Sorger PK. A reductionist's systems biology: opinion. Curr. Opin. Cell Biol. 2005;17:9–11. doi: 10.1016/j.ceb.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 79.Goldberger AL, et al. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation. 2000;101:E215–E220. doi: 10.1161/01.cir.101.23.e215. [DOI] [PubMed] [Google Scholar]
- 80.Klipp E, Nordlander B, Kruger R, Gennemark P, Hohmann S. Integrative model of the response of yeast to osmotic shock. Nature Biotechnol. 2005;23:975–982. doi: 10.1038/nbt1114. [DOI] [PubMed] [Google Scholar]
- 81.Michor F, et al. Dynamics of chronic myeloid leukemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 82.Palumbo MC, Colosimo A, Giuliani A, Farina L. Functional essentiality from topology features in metabolic networks: a case study in yeast. FEBS Lett. 2005;579:4642–4646. doi: 10.1016/j.febslet.2005.07.033. [DOI] [PubMed] [Google Scholar]
- 83.Mullenix MC, Wiltshire S, Shao W, Kitos G, Schweitzer B. Allergen-specific IgE detection on microarrays using rolling circle amplification: correlation with in vitro assays for serum IgE. Clin. Chem. 2001;47:1926–1927. [Google Scholar]
- 84.Kim TE, et al. Quantitative measurement of serum allergen-specific IgE on protein chip. Exp. Mol. Med. 2002;34:152–158. doi: 10.1038/emm.2002.22. [DOI] [PubMed] [Google Scholar]
- 85.Jahn-Schmid B, et al. Allergen microarray: comparison of microarray using recombinant allergens with conventional diagnostic methods to detect allergen-specific serum immunoglobulin E. Clin. Exp. Allergy. 2003;33:1443–1449. doi: 10.1046/j.1365-2222.2003.01784.x. [DOI] [PubMed] [Google Scholar]
- 86.Fall BI, et al. Microarrays for the screening of allergen-specific IgE in human serum. Anal. Chem. 2003;75:556–562. doi: 10.1021/ac026016k. [DOI] [PubMed] [Google Scholar]
- 87.Lebrun SJ, Petchpud WN, Hui A, McLaughlin CS. Development of a sensitive, colorometric microarray assay for allergen-responsive human IgE. J. Immunol. Methods. 2005;300:24–31. doi: 10.1016/j.jim.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 88.Feng Y, et al. Parallel detection of autoantibodies with microarrays in rheumatoid diseases. Clin. Chem. 2004;50:416–422. doi: 10.1373/clinchem.2003.023994. [DOI] [PubMed] [Google Scholar]
- 89.Hueber W, et al. Antigen microarray profiling of autoantibodies in rheumatoid arthritis. Arthritis Rheum. 2005;52:2645–2655. doi: 10.1002/art.21269. [DOI] [PubMed] [Google Scholar]
- 90.Kamachi M, et al. Human autoimmune sera as molecular probes for the identification of an autoantigen kinase signaling pathway. J. Exp. Med. 2002;196:1213–1225. doi: 10.1084/jem.20021167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Robinson WH, et al. Protein microarrays guide tolerizing DNA vaccine treatment of autoimmune encephalomyelitis. Nature Biotechnol. 2003;21:1033–1039. doi: 10.1038/nbt859. [DOI] [PubMed] [Google Scholar]
- 92.Lueking A, et al. Profiling of alopecia areata autoantigens based on protein microarray technology. Mol. Cell. Proteomics. 2005;4:1382–1390. doi: 10.1074/mcp.T500004-MCP200. [DOI] [PubMed] [Google Scholar]
- 93.Qiu J, et al. Development of natural proteinmicroarrays for diagnosing cancer based on an antibody response to tumor antigens. J. Proteome. Res. 2004;3:261–267. doi: 10.1021/pr049971u. [DOI] [PubMed] [Google Scholar]
- 94.Bouwman K, et al. Microarrays of tumor cell derived proteins uncover a distinct pattern of prostate cancer serum immunoreactivity. Proteomics. 2003;3:2200–2207. doi: 10.1002/pmic.200300611. [DOI] [PubMed] [Google Scholar]
- 95.Tong M, et al. A multiplexed and miniaturized serological tuberculosis assay identifies antigens that discriminate maximally between TB and non-TB sera. J. Immunol. Methods. 2005;301:154–163. doi: 10.1016/j.jim.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 96.Steller S, et al. Bacterial protein microarrays for identification of new potential diagnostic markers for Neisseria meningitidis infections. Proteomics. 2005;5:2048–2055. doi: 10.1002/pmic.200401097. [DOI] [PubMed] [Google Scholar]
- 97.Cepok S, et al. Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. J. Clin. Invest. 2005;115:1352–1360. doi: 10.1172/JCI23661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bacarese-Hamilton T, Mezzasoma L, Ardizzoni A, Bistoni F, Crisanti A. Serodiagnosis of infectious diseases with antigen microarrays. J. Appl. Microbiol. 2004;96:10–17. doi: 10.1046/j.1365-2672.2003.02111.x. [DOI] [PubMed] [Google Scholar]
- 99.Li B, et al. Protein microarray for profiling antibody responses to Yersinia pestis live vaccine. Infect. Immun. 2005;73:3734–3739. doi: 10.1128/IAI.73.6.3734-3739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quintana FJ, et al. Functional immunomics: microarray analysis of IgG autoantibody repertoires predicts the future response of mice to induced diabetes. Proc. Natl Acad. Sci. USA. 2004;101(Suppl 2):14615–14621. doi: 10.1073/pnas.0404848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davies DH, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl Acad. Sci. USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Michaud GA, et al. Analyzing antibody specificity with whole proteome microarrays. Nature Biotechnol. 2003;21:1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 103.Schweitzer B, Predki P, Snyder M. Microarrays to characterize protein interactions on a whole-proteome scale. Proteomics. 2003;3:2190–2199. doi: 10.1002/pmic.200300610. [DOI] [PubMed] [Google Scholar]
- 104.Gembitsky DS, Lawlor K, Jacovina A, Yaneva M, Tempst P. A prototype antibody microarray platform to monitor changes in protein tyrosine phosphorylation. Mol. Cell. Proteomics. 2004;3:1102–1118. doi: 10.1074/mcp.M400075-MCP200. [DOI] [PubMed] [Google Scholar]
- 105.Diks SH, et al. Kinome profiling for studying lipopolysaccharide signal transduction in human peripheral blood mononuclear cells. J. Biol. Chem. 2004;279:49206–49213. doi: 10.1074/jbc.M405028200. [DOI] [PubMed] [Google Scholar]
- 106.Galanina OE, Mecklenburg M, Nifantiev NE, Pazynina GV, Bovin NV. GlycoChip: multiarray for the study of carbohydrate-binding proteins. Lab. Chip. 2003;3:260–265. doi: 10.1039/b305963d. [DOI] [PubMed] [Google Scholar]
- 107.Vanov SS, et al. Antibodies immobilized as arrays to profile protein post-translational modifications in mammalian cells. Mol. Cell. Proteomics. 2004;3:788–795. doi: 10.1074/mcp.M300130-MCP200. [DOI] [PubMed] [Google Scholar]
- 108.Ratner DM, et al. Probing protein-carbohydrate interactions with microarrays of synthetic oligosaccharides. Chembiochem. 2004;5:379–382. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 109.Uttamchandani M, Chan EW, Chen GY, Yao SQ. Combinatorial peptide microarrays for the rapid determination of kinase specificity. Bioorg. Med. Chem. Lett. 2003;13:2997–3000. doi: 10.1016/s0960-894x(03)00633-4. [DOI] [PubMed] [Google Scholar]
- 110.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl Acad. Sci. USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nagorsen D, Marincola FM, Panelli MC. Cytokine and chemokine expression profiles of maturing dendritic cells using multiprotein platform arrays. Cytokine. 2004;25:31–35. doi: 10.1016/j.cyto.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 112.Huang R, et al. Connexin 43 suppresses human glioblastoma cell growth by down-regulation of monocyte chemotactic protein 1, as discovered using protein array technology. Cancer Res. 2002;62:2806–2812. [PubMed] [Google Scholar]
- 113.Traicoff JL, et al. Profiling the expression of mitogen-induced T-cell proteins by using multi-membrane dot-blotting. Biochem. Biophys. Res. Commun. 2004;323:355–360. doi: 10.1016/j.bbrc.2004.08.093. [DOI] [PubMed] [Google Scholar]
- 114.Yang D, et al. Human ribonuclease A superfamily members, eosinophil-derived neurotoxin and pancreatic ribonuclease, induce dendritic cell maturation and activation. J. Immunol. 2004;173:6134–6142. doi: 10.4049/jimmunol.173.10.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nature Rev. Drug Discov. 2003;2:566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 116.Hortin GL. Can mass spectrometric protein profiling meet desired standards of clinical laboratory practice? Clin. Chem. 2005;51:3–5. doi: 10.1373/clinchem.2004.043281. [DOI] [PubMed] [Google Scholar]
- 117.Reynolds MA, et al. Early biomarkers of stroke. Clin. Chem. 2003;49:1733–1739. doi: 10.1373/49.10.1733. One of the best plasma protein biomarker development papers. [DOI] [PubMed] [Google Scholar]
- 118.Kaukola T, et al. Cerebral palsy is characterized by protein mediators in cord serum. Ann. Neurol. 2004;55:186–194. doi: 10.1002/ana.10809. [DOI] [PubMed] [Google Scholar]
- 119.Kader HA, et al. Protein microarray analysis of disease activity in pediatric inflammatory bowel disease demonstrates elevated serum PLGF, IL-7, TGF-β1 and IL-12p40 levels in Crohn's disease and ulcerative colitis patients in remission versus active disease. Am. J. Gastroenterol. 2004;99:1–10. doi: 10.1111/j.1572-0241.2004.40819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Szodoray P, Alex P, Brun JG, Centola M, Jonsson R. Circulating cytokines in primary Sjogren's syndrome determined by a multiplex cytokine array system. Scand. J. Immunol. 2004;59:592–599. doi: 10.1111/j.0300-9475.2004.01432.x. [DOI] [PubMed] [Google Scholar]
- 121.Gao WM, et al. Distinctive serum protein profiles involving abundant proteins in lung cancer patients based upon antibody microarray analysis. BMC Cancer. 2005;5:110. doi: 10.1186/1471-2407-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mor G, et al. Serum protein markers for early detection of ovarian cancer. Proc. Natl Acad. Sci. USA. 2005;102:7677–7682. doi: 10.1073/pnas.0502178102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kastenbauer S, Angele B, Sporer B, Pfister HW, Koedel U. Patterns of protein expression in infectious meningitis: a cerebrospinal fluid protein array analysis. J. Neuroimmunol. 2005;164:134–139. doi: 10.1016/j.jneuroim.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 124.Varnum SM, Woodbury RL, Zangar RC. A protein microarray ELISA for screening biological fluids. Methods Mol. Biol. 2004;264:161–172. doi: 10.1385/1-59259-759-9:161. [DOI] [PubMed] [Google Scholar]
- 125.Sack RA, et al. Membrane array characterization of 80 chemokines, cytokines, and growth factors in open- and closed-eye tears: angiogenin and other defense system constituents. Invest. Ophthalmol. Vis. Sci. 2005;46:1228–1238. doi: 10.1167/iovs.04-0760. [DOI] [PubMed] [Google Scholar]
- 126.Mitchell AM, Brown MD, Menown IB, Kline JA. Novel protein markers of acute coronary syndrome complications in low-risk outpatients: a systematic review of potential use in the emergency department. Clin. Chem. 2005;51:2005–2012. doi: 10.1373/clinchem.2005.052456. [DOI] [PubMed] [Google Scholar]
- 127.Buechler KF, et al. Simultaneous detection of seven drugs of abuse by the Triage panel for drugs of abuse. Clin. Chem. 1992;38:1678–1684. [PubMed] [Google Scholar]
- 128.Wu AH, Smith A, Christenson RH, Murakami MM, Apple FS. Evaluation of a point-of-care assay for cardiac markers for patients suspected of acute myocardial infarction. Clin. Chim. Acta. 2004;346:211–219. doi: 10.1016/j.cccn.2004.03.036. [DOI] [PubMed] [Google Scholar]
- 129.Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman JR. Advanced multiplexed analysis with the FlowMetrix™ system. Clin. Chem. 1997;43:1749–1756. [PubMed] [Google Scholar]
- 130.Klutts JS, Liao RS, Dunne WM, Jr, Gronowski AM. Evaluation of a multiplexed bead assay for assessment of Epstein–Barr virus immunologic status. J. Clin. Microbiol. 2004;42:4996–5000. doi: 10.1128/JCM.42.11.4996-5000.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shovman O, et al. Evaluation of the BioPlex™ 2200 ANA screen: analysis of 510 healthy subjects: incidence of natural/predictive autoantibodies. Ann. NY Acad. Sci. 2005;1050:380–388. doi: 10.1196/annals.1313.120. [DOI] [PubMed] [Google Scholar]
- 132.Jenison R, La H, Haeberli A, Ostroff R, Polisky B. Silicon-based biosensors for rapid detection of protein or nucleic acid targets. Clin. Chem. 2001;47:1894–1900. [PubMed] [Google Scholar]
- 133.Inganäs M, et al. Integrated microfluidic compact disc device with potential use in both centralized and point-of-care laboratory settings. Clin. Chem. 2005;51:1985–1987. doi: 10.1373/clinchem.2005.053181. [DOI] [PubMed] [Google Scholar]
- 134.Scillian JJ, et al. Early detection of antibodies against rDNA-produced HIV proteins with a flow cytometric assay. Blood. 1989;73:2041–2048. [PubMed] [Google Scholar]
- 135.Mendoza LG, et al. High-throughput microarray-based enzyme-linked immunosorbent assay (ELISA) Biotechniques. 1999;27:778–780. doi: 10.2144/99274rr01. 782–786, 788. [DOI] [PubMed] [Google Scholar]
- 136.Khrapko KR, et al. A method for DNA sequencing by hybridization with oligonucleotide matrix. DNA Seq. 1991;1:375–388. doi: 10.3109/10425179109020793. [DOI] [PubMed] [Google Scholar]
- 137.Shevchenko A, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl Acad. Sci. USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang X, et al. Autoantibody signatures in prostate cancer. N. Engl. J. Med. 2005;353:1224–1235. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 139.Pedersen SK, et al. An immunoproteomic approach for identification of clinical biomarkers for monitoring disease: application to cystic fibrosis. Mol. Cell. Proteomics. 2005;4:1052–1060. doi: 10.1074/mcp.M400175-MCP200. [DOI] [PubMed] [Google Scholar]
- 140.Sharp SE, Suarez CA, Duran Y, Poppiti RJ. Evaluation of the Triage micro parasite panel for detection of Giardia lamblia, Entamoeba histolytica/Entamoeba dispar, and Cryptosporidium parvum in patient stool specimens. J. Clin. Microbiol. 2001;39:332–334. doi: 10.1128/JCM.39.1.332-334.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Davies DH, et al. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl Acad. Sci. USA. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee JW, et al. Method validation and measurement of biomarkers in nonclinical and clinical samples in drug development: a conference report. Pharm. Res. 2005;22:499–511. doi: 10.1007/s11095-005-2495-9. [DOI] [PubMed] [Google Scholar]
- 143.Kaukola T, et al. Chorioamnionitis and cord serum proinflammatory cytokines: lack of association with brain damage and neurologic outcome in very preterm infants. Pediatr. Res. 2005 Sep 23; (doi: 10.1203/01. PDR.0000182596.66175.EE) [Google Scholar]