Abstract
Circulating CD4+ CD8+ T lymphocytes have been described in the peripheral blood of humans and several animal species. However, the origin and functional properties of these cells remain poorly understood. In the present study, we evaluated the frequency, phenotype and function of peripheral CD4+ CD8+ T cells in rhesus macaques. Two distinct populations of CD4+ CD8+ T cells were identified: the dominant one was CD4hi CD8lo and expressed the CD8αα homodimer, while the minor population was CD4lo CD8hi and expressed the CD8αβ heterodimer. The majority of CD4hi CD8αlo T cells exhibited an activated effector/memory phenotype (CCR5lo CD7– CD28– HLA-DR+) and expressed relatively high levels of granzyme B. Intracellular cytokine staining assays demonstrated that the frequency of cytomegalovirus-specific T cells was enriched five-fold in CD4hi CD8αlo T cells compared to single-positive CD4+ T cells, whereas no consistent enrichment was observed for simian immunodeficiency virus (SIV)-specific T cells. Cross-sectional studies of SIV-infected animals demonstrated that the frequency of CD4hi CD8αlo T cells was lower in wild-type SIV-infected animals compared to uninfected controls, although prospective studies of SIV-infected animals demonstrated depletion of CD4hi CD8αlo lymphocytes only in a subset of animals. Taken together, these data suggest that CD4+ T cells expressing CD8α represent an effector/memory subset of CD4+ T cells and that this cell population can be depleted during the course of SIV infection.
Keywords: animal models/studies; primates; helper T cells (Th cells, Th0, Th1, Th2, Th3); memory cells; simian immunodeficiency virus
Introduction
CD4+ CD8+ T lymphocytes represent a minor subset of circulating T cells coexpressing both CD4 and CD8 molecules on their surface. In human peripheral blood, CD4+ CD8+ T cells are relatively infrequent, constituting on average 1–2% of CD3+ lymphocytes,1–3 but they have been observed at increased frequencies in some normal subjects4–7 and in individuals with a wide range of diseases such as T- and B-cell leukaemia,8,9 autoimmune diseases10 and bacterial or viral infections.11–13 CD4+ CD8+ T cells have also been described in other species, including mice,14 rats,15 swine16 and cynomolgus macaques17 and in some species they represent up to 20–60% of circulating T lymphocytes.16,18
The phenotype, origin and functional properties of CD4+ CD8+ T cells are still incompletely understood. Previous studies in rats and mice initially suggested that these cells might represent developing CD4+ CD8+ thymocytes prematurely released into the circulation.15,19,20 Other authors have proposed that CD4+ CD8+ T cells may originate extrathymically from CD4 single-positive (SP) T cells that acquired the ability to express CD8.8,12,21,22 Conversely, CD4+ CD8+ cells have also been proposed to arise from activated CD8 SP T cells that have up-regulated the surface expression of CD4.13,23,24
In the present study, we analysed the CD4+ CD8+ population in rhesus macaques, either uninfected or infected with wild-type or attenuated strains of simian immunodeficiency virus (SIV). We observed two distinct subpopulations of CD4+ CD8+ T cells: a dominant one, represented by CD4hi CD8lo T cells, expressing primarily the CD8αα homodimer, and a minor population, represented by CD4loCD8hi cells, expressing the CD8αβ heterodimer. CD4hi CD8αlo T cells exhibited an activated effector memory phenotype and expressed relatively low levels of CD7. In addition, as compared with CD4 SP T cells, CD4hi CD8αlo T cells had increased expression of Ki67 and granzyme B and lower levels of T-cell receptor excision circles (TRECs). Intracellular cytokine staining analysis revealed that the frequency of cytomegalovirus (CMV)-specific, but not SIV-specific cells was enriched in the CD4hi CD8αlo population compared with CD4hi CD8– cells. A significant fraction of CD4hi CD8αlo cells expressed the SIV co-receptor CCR5, and cross-sectional studies demonstrated a significantly lower level of CD4hi CD8αlo T cells in wild-type SIV-infected animals.
Materials and methods
Animals
Rhesus macaques (Macaca mulatta) analysed for this study included SIV-negative (naive) monkeys, animals inoculated with the live attenuated strains SIVmac239Δnef,25 or SIVmac239Δ326 and animals infected with the pathogenic strains SIVmac239 or SIVmac251. A subset of animals infected with attenuated SIV strains had been subsequently challenged with pathogenic SIV strains but maintained undetectable levels of plasma viraemia at the time of study. CMV-seropositive animals with naturally acquired CMV infection were identified by enzyme-linked immunosorbent assay as described previously.27 All animals were housed at the New England Primate Research Center and maintained in accordance with the guidelines of the Committee on Animals of the Harvard Medical School and the Guide for the Care and use of Laboratory Animals.28
Flow cytometric analysis
Surface phenotyping was performed directly on freshly drawn whole blood using standard procedures. In a few instances, phenotyping was performed on peripheral blood mononuclear cells (PBMCs) isolated from fresh heparinized blood by Ficoll density gradient centrifugation (Ficoll-Paque Pharmacia, San Diego, CA) and suspended in phosphate-buffered saline (PBS) containing 2% fetal bovine serum (FBS). Monoclonal antibodies (mAbs) were conjugated with fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophyll protein (PerCP) or allophycocyanin (APC) as indicated. Except where noted, all antibodies were obtained from BD Biosciences (San Diego, CA). The mAbs used for these studies included CD3-FITC (clone SP34), CD4-APC (clone SK3), CD7-PE (clone M-T701), CD8α-PerCP (clone SK1) and CD8β-PE (clone 2ST8.5H7 from Beckman/Coulter (Miami, FL); CD25-PE (clone 2A3), CD28-FITC or PE (clone CD28.2), CD95-PE (clone DX2) from Caltag, Burlingame, CA; CD45RA-FITC (clone 5H9), CD62L-PE (clone SK11), integrin-β7-PE (clone F1B504), and HLA-DR-PE (clone L243). The CD8β mAb 2ST8.5H7 recognizes an epitope that is either specific for CD8β or a conformational determinant of the CD8αβ heterodimer.29,30 Surface staining was carried out by standard procedures. Briefly, fresh whole blood (100 μl) was incubated with the antibodies for 30 min at 4°, protected from light. After staining, red blood cells were lysed with 1× fluorescence-acitvated cell sorter (FACS) Lysing Solution (BD Biosciences), remaining cells were then washed in wash buffer (PBS, 2% FBS) and fixed in 2% paraformaldehyde. Flow cytometric analysis was performed on a FACSCalibur flow cytometer (BD Biosciences), and data were acquired and analysed using CellQuest (BD Biosciences) or FlowJo (Tree Star, Inc. Ashland, OR). Six-colour flow cytometric analysis was performed using CD95-FITC, CCR5-PE, CD4-PerCP-Cy5·5, CD28 biotin with streptavidin APC and CD3 APC-Cy7 (all BD Biosciences) analysed on a FACS Vantage (BD Biosciences) using FlowJo. In selected instances (e.g. evaluation of expression of CCR5), a fluorescence minus one (FMO) control was performed to establish gates. The FMO control consisted of the omission of the antibody of interest from a typical four-colour tube, thus allowing background fluorescence in the empty channel to be assessed on the subpopulation defined by the three remaining colours.31
For analysis of granzyme B expression, surface staining with CD3-FITC, CD8-PerCP and CD4-APC was followed by fixation and permeabilization using CALTAG Fix-Perm, according to the manufacturer's instructions. Fixed and washed cells were then stained with anti-granzyme B-PE (clone GB12; Caltag Laboratories). For Ki67, Ki67-FITC (clone MIB-1; Coulter, Miami, FL) or an FITC-conjugated mouse isotype control were used after surface staining and subsequent permeabilization as described above.
Intracellular cytokine staining
Intracellular cytokine staining was performed as initially described in humans with modifications as previously described for optimal performance using macaque PBMC.27,32 PBMC were stimulated for 6 hr with superantigen or specific viral antigens. For optimal costimulation, cells were stimulated in the presence of mAbs specific for the costimulatory molecules, CD28 (clone 28·2) and CD49d (clone 9F10) at 10 μg/ml each, cross-linked with affinity-purified F(ab′)2 fragments of goat anti-mouse immunoglobulin G (Kirkegaard & Perry, Gaithersburg, MD), at 2·5 μg/ml in 50 mm Tris–HCl (pH 8·6). Superantigen stimulation was carried out using a combination of staphylococcal enterotoxin A (SEA, Sigma, St Louis, MO) and staphylococcal enterotoxin B (SEB, Sigma) each at a final concentration of 100 ng/ml (Sigma). For antigen-specific stimulation we used either a pool of overlapping SIV Gag peptides32 or rhesus CMV antigen (rhCMV).27 A cell lysate from uninfected cells was used as a negative control for rhCMV, as previously described.27 Brefeldin A (Sigma) or Golgi Stop was added at 10 μg/ml for the final 5 hr of stimulation. During the stimulation period cells were kept in RPMI-1640 (BioWhittaker, Walkersville, MD) supplemented with glutamine (2 mm) (Biofluids, Rockville, MD), penicillin-streptomycin (Biofluids), HEPES (15 mm) (Biofluids), and 10% FBS. At the end of the stimulation, cells were surface stained for 30 min at 4° with conjugated anti-CD4 and anti-CD8 mAbs and subsequently fixed and permeabilized by successive incubations in 1× FACS Lysing Solution and FACS permeabilizing solution (BD Biosciences). Permeabilized cells were finally incubated for 30 min at 4° with anti-CD69-PE (clone FN50) and with anti-cytokine mAbs: interferon-γ-FITC or -APC (clone B27) or tumour necrosis factor-α (TNF-α)-FITC or -APC (clone MAB11), washed, and kept overnight at 4° in 2% paraformaldehyde. At least 50 000 events were collected on a FACS Calibur, and the data were analysed using CellQuest or FlowJo. Antigen-specific cells were defined as T lymphocytes that coexpressed CD69 and the cytokine of interest after antigen-specific stimulation.
Cell sorting
CD4hi CD8αlo, CD4 SP and CD8 SP T lymphocytes were sorted from fresh PBMCs using a FACS Vantage. Briefly, cells were stained for 30 min at 4° with CD3-FITC, CD4-APC and CD8α-PerCP. After staining, cells were washed and fixed for 1 hr at 4° with 2% paraformaldehyde. After fixation, cells were washed again and resuspended at 10 × 106/ml in wash medium for subsequent sorting (performed in glass tubes). To isolate CD4 SP memory and CD4 SP naive T cells, cells were stained with CD4-APC, CD28-FITC and CD95-PE mAbs and then sorted. CD4 SP memory cells were defined as CD4+ CD95+ CD28+/–, while CD4 SP naive cells were defined as CD4+ CD28+ CD95–.33 The purity of the sorted populations was analysed using a FACS Calibur flow cytometer and found to be >90%. Sorted cells were used for DNA extraction and subsequent TREC analysis.
TREC assay
DNA was extracted from sorted T-cell populations using a PUREGENE DNA Isolation Kit (Gentra Systems, Inc., Minneapolis, MN) following incubation of the cell lysate overnight at 55° with proteinase K (Sigma) according to the manufacturer's instructions. DNA concentration was determined by spectrophotometry and 200 ng of DNA from each sample were used for TREC analysis. Signal joint TREC levels were determined using a quantitative real-time polymerase chain reaction (PCR) assay, as previously described.34,35 Real-time PCR analysis was performed on DNA extracted from total PBMCs or sorted populations of CD4+ CD8+, CD8 SP, CD4 SP memory (CD95+) and CD4 SP naive (CD95–) T cells. The DNA standard curve was generated from serial dilutions of a plasmid encoding the sequence of a M. mulatta TREC sequence that was kindly provided by Linqi Zhang and David Ho36 and diluted into genomic DNA from a macaque herpespapio-transformed B-cell line. Each 50-μl PCR mixture contained 200 ng DNA, the signal joint primers 1F (sense) (5′-CAGTGTGACATGGAGGGCTGAA-3′) (500 nm) and 1R (antisense) (5′GTGTCTCTGTCAACAAAGTTGATGC-3′) (500 nm)36 TREC probe and TaqMan universal PCR MasterMix (Applied Biosystems). PCR conditions consisted of one cycle of denaturation (95° for 5 min), followed by 50 cycles of amplification (94° for 30 seconds, 60° for 30 seconds, 72° for 30 seconds) using an ABI PRISM 7700 Sequence Detection System, Perkin Elmer (Wellesley, MA).
Statistical analysis
Statistical analysis was carried out using StatView (SAS Institute, Cary, NC). Analysis of variance (anova) and Fisher's protected least-significant-difference (PLSD) post hoc test were used for comparisons involving more than two groups.
Results
Identification of two distinct populations of CD4+ CD8+ T cells in the peripheral blood of rhesus macaques
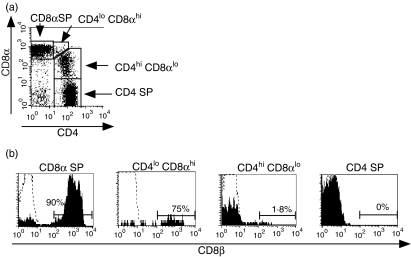
Surface staining of peripheral blood from multiple rhesus macaques revealed the presence of two distinct populations of CD4+ CD8+ T cells: a major population represented by CD4hi CD8lo lymphocytes and a minor population of CD4lo CD8hi cells (Fig. 1a). As an alternative approach to validate the separation of CD4+ CD8+ T cells into these subpopulations, we examined the expression of CD8β. In humans, CD4hi CD8lo T lymphocytes have been shown to express only the CD8αα isoform, whereas CD4lo CD8hi lymphocytes express the CD8αβ isoform.7,37 In uninfected macaques, we observed that CD8β was expressed on the majority of CD8 SP T cells (mean ±SD, 91·2 ± 5·7%) but was essentially absent on CD4 SP cells (Fig. 1b). As expected, a low percentage of CD4hi CD8lo cells expressed CD8β (mean ±SD, 2·7 ± 3·1%) compared to CD8 SP T cells, confirming that CD8 expression on CD4hiCD8lo cells consisted primarily of the CD8αα homodimer, as previously described in humans.7,37 In contrast, CD8αhi was expressed on a high percentage (mean ±SD, 83·1 ± 10·9%) of CD4lo CD8hi cells, suggesting that this minor population of cells is likely to be derived from CD8 SP T cells. The frequency of CD4hi CD8αlo T cells in uninfected macaques was variable, ranging from 1·8% to 18·5% in naive animals (mean ± SD, 6·1 ± 4·6%n = 14). However, the frequency of CD4hi CD8αlo T cells in individual animals was generally stable, with an average coefficient of variation of 20% (data obtained from between two and four determinations from eight animals over up to 8 months). In contrast to a previous report describing an increasing frequency of CD4hi CD8αlo cells in older cynomolgus macaques,17 cross-sectional analysis of the frequency of CD4hi CD8αlo T cells in normal animals ranging from 4 to 16 years old did not reveal any correlation with age (data not shown). As a result of the low and variable numbers of CD4lo CD8hi cells, we focused our subsequent analysis on CD4hi CD8αlo T cells.
Figure 1.
Differential expression of CD8β on different subsets of CD4+ CD8+ T cells. Peripheral whole blood from an uninfected rhesus macaque was analysed by four-colour flow cytometry for expression of CD8β on the indicated populations of lymphocytes. (a) Expression of CD4 and CD8α on CD3+ T lymphocytes. (b) Expression of CD8β on different subsets of T lymphocytes. The percentage of CD8β+ lymphocytes is indicated for each histogram. Dotted lines represent the isotype control. Results are representative of a total of 36 separate experiments.
Phenotypic characterization of CD4hi CD8αlo T cells
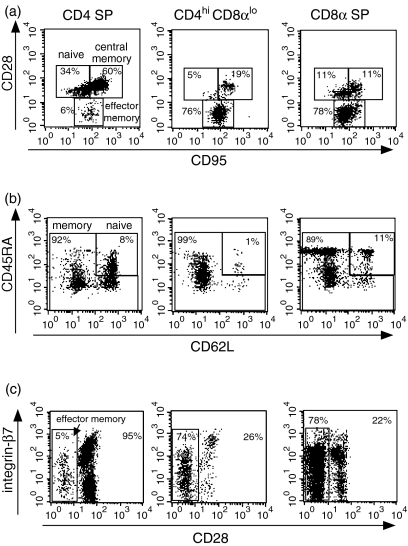
To better characterize CD4hi CD8αlo T cells in rhesus macaques, we examined the expression of a panel of naive/memory and activation markers. Previous work in rhesus macaques has demonstrated that subpopulations of naive, effector and central memory CD4+ T cells can be identified based on the expression of CD28 and CD95 in combination with other markers such as integrin-β7.33 Specifically, naive macaque CD4+ T cells can be delineated by their CD95lo CD28hi integrin-β7 phenotype, while naive CD8+ T cells are defined by their CD95lo CD28int CD11alo phenotype. In contrast, memory cells are more heterogeneous. However, both CD4+ and CD8+ memory subsets are CD95hi, and effector memory cells can be defined by their lack of CD28 expression.33
In this study, we adopted these phenotypic criteria used for CD4 and CD8 SP cells to characterize CD4hi CD8αlo cells. Based on analysis of CD28 and CD95 expression, the majority (∼80%) of CD4hi CD8αlo lymphocytes displayed an effector memory phenotype (CD28– CD95+), while only minor fractions of central memory and naive phenotype cells were observed in the CD4hi CD8αlo population (Fig. 2a, Table 1). Analysis of expression of CD45RA, CD62L, integrin-β7 and CD28 in combination confirmed the significant enrichment of effector memory phenotype cells in the CD4hi CD8αlo population, as evidenced by a predominant CD45RA–/lo CD62L– integrin-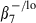 phenotype (Fig. 2b,c).
phenotype (Fig. 2b,c).
Figure 2.
Phenotypic analysis of CD4hi CD8αlo T cells. CD4 SP, CD4hi CD8αlo and CD8 SP lymphocytes were analysed for the coexpression of (a) CD28 and CD95; (b) CD45RA and CD62L; (c) CD28 and β7. Gates used for the identification of central/memory, effector/memory and naive lymphocytes are shown, along with the percentage of cells in each gated population. Data are representative of 12 separate animals.
Table 1.
Expression of naive/memory markers in different subsets of T lymphocytes
| % of T lymphocytes | |||
|---|---|---|---|
| Subset | Central memory (CD28+/CD95+) | Effector memory (CD28–/CD95+) | Naive (CD28+/CD95–) |
| CD4 SP | 41·6 ± 12·3 | 8·0 ± 8·4 | 51·2 ± 14·8 |
| CD4hi CD8αlo | 14·1 ± 8·6 | 78·2 ± 11·3 | 5·8 ± 5·3 |
| CD8 SP | 13·1 ± 4·4 | 63·3 ± 13·9 | 23·1 ± 11·3 |
Results are expressed as the percentage of T cells coexpressing the indicated memory/naive markers. Data (mean ± SD) were obtained from 12 uninfected macaques.
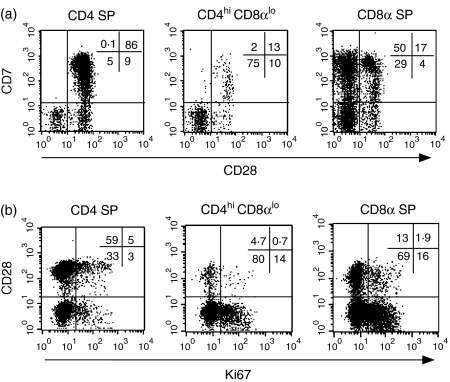
We also evaluated the expression of CD7, which is expressed on T cells early in ontogeny, including on CD4+ CD8+ thymocytes as well as on most mature T cells in peripheral blood.38 Expression of CD7 and CD28 differed markedly in CD4 SP and CD4hi CD8αlo cells: CD4hi CD8αlo cells were predominantly CD7– CD28– (mean ±SD, 73% ± 14, n = 12), while CD4 SP T cells were largely CD7+ CD28+ (79 ± 13%) (Fig. 3a). Based on previous reports suggesting that CD4+ CD8+ cells in humans and macaques represented an activated T-cell population with lytic activity,11,39–41 we also examined the expression of the activation markers CD25 and HLA-DR, as well as the serine protease granzyme B. As compared with CD4 SP lymphocytes, CD4hi CD8αlo cells had significantly increased expression of HLA-DR but not CD25 (Table 2). There was also a marked increase in the percentage of CD4hi CD8αlo cells expressing granzyme B compared with CD4 SP cells. Overall, the percentage of granzyme B in the CD4hi CD8αlo subset was comparable to the CD8 SP subset (Table 2).
Figure 3.
Expression of CD7, Ki67 and CD28 on CD4 SP, CD4hi CD8αlo and CD8 SP lymphocytes. (a) Analysis of CD7 and CD28 expression. The percentages of cells corresponding to each quadrant are indicated on the top right side of the plot. Data are representative of five independent experiments from different animals. (b) Analysis of Ki67 and CD28 expression. The indicated gates for expression of Ki67 were determined based on the analysis of lymphocytes stained with an isotype control anti-mouse-FITC after fixation and permeabilization of surface-stained cells. Plots are representative of eight separate animals.
Table 2.
Expression of activation markers, granzyme B and Ki67 in CD4hi CD8αlo and single positive (SP) T lymphocytes
| T lymphocyte subpopulation | ||||
|---|---|---|---|---|
| Marker | No. | CD4 SP | CD4hi CD8αlo | CD8 SP |
| CD25 | 16 | 16·6 ± 6·0 | 7·8 ± 7·0 | 3·4 ± 1·8 |
| HLA-DR | 10 | 17·6 ± 10·7 | 53·1 ± 14·0 | 41·4 ± 13·2 |
| Granzyme B | 9 | 4·0 ± 4·9 | 48·6 ± 19·7 | 47·5 ± 20·6 |
| Ki67 | 8 | 8·1 ± 1·9 | 13·6 ± 2·9 | 14 ± 4·5 |
Results represent the mean ± SD of the percentages of the specific subpopulation expressing the indicated molecule. All data are from uninfected macaques. The difference in Ki67 expression between CD4 SP cells and CD4hi CD8αlo cells is significant (P = 0·01, Wilcoxon signed rank test).
Based on expression of Ki67, a marker for proliferating lymphocytes,42 CD4hi CD8αlo T cells proliferated at a significantly higher rate than CD4 SP T cells (Table 2). To further define the central and effector/memory profile of proliferating CD4hi CD8αlo T cells, we performed a four-colour flow cytometric analysis, combining analysis of expression of CD4, CD8 and CD28 following intracellular staining for Ki67. The majority of proliferating (Ki67+) CD4hi CD8αlo T cells were CD28–, consistent with the fact that most CD4hi CD8αlo cells lacked expression of CD28 (Fig. 3b).
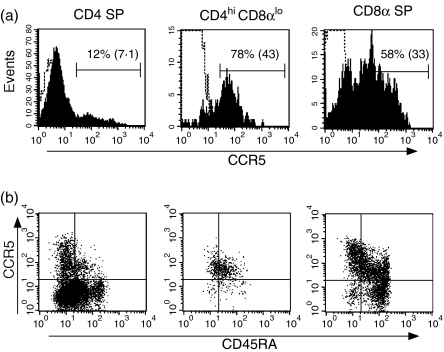
CCR5 is the dominant coreceptor for most SIV strains43 and is selectively expressed on a population of CD4+ CD45RA– T lymphocytes in peripheral blood that is rapidly depleted early in the course of SIV infection.44 Analysis of CCR5 expression on macaque CD4hi CD8lo cells revealed low but significant levels of CCR5 (Fig. 4a). This pattern of expression was in contrast to the bimodal distribution of CCR5 observed on CD4 SP cells, for which most CD4 SP cells were CCR5–/dim but a subpopulation of cells were CCR5hi (Fig. 4a). Analysis of CCR5 in combination with CD45RA demonstrated that CD4hi CD8αlo cells largely constituted a CD45RAlo CCR5lo population, in contrast to the more heterogeneous patterns of expression of these molecules on CD4 and CD8 SP cells (Fig. 4b). Six-colour flow cytometric analysis demonstrated that the distinct pattern of CCR5lo expression on CD4hi CD8αlo cells did not simply reflect the increased fraction of CD28– effector memory cells but also was true of the smaller fraction of naive and central memory phenotype CD4hi CD8αlo cells as well (data not shown).
Figure 4.
Expression of CCR5 on CD4hi CD8αlo lymphocytes. (a) Expression of CCR5 on CD4 SP, CD4hi CD8αlo and CD8 SP lymphocytes. The dotted line indicated staining obtained with an isotype control. The percentage of cells expressing CCR5 is shown, along with the geometric mean fluorescent intensity of the total population of CD4 SP, CD4hi CD8αlo and CD8 SP lymphocytes in parentheses. Results are representative of nine animals.
Examination of the relationship between expression of specific phenotypic markers and the frequency of CD4hi CD8lo T lymphocytes in our study animals did not reveal any statistically significant correlation between any of the phenotypic markers and the frequency of CD4hi CD8αlo cells.
Cytokine production in CD4hi CD8αlo lymphocytes following stimulation with viral antigens
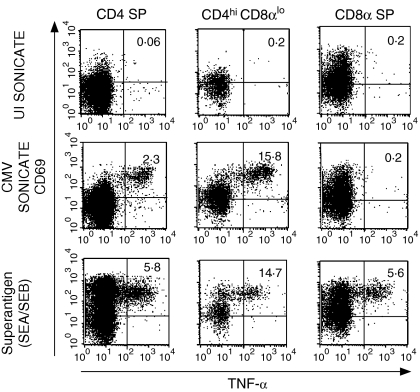
Based in part on the phenotypic evidence that CD4hi CD8αlo cells represented a predominant effector memory population, we investigated the ability of this population to respond to viral antigens. Initial studies were conducted in rhCMV-seropositive macaques infected with attenuated strains of SIV (SIVΔnef and SIVmac239Δ3) that have previously been shown to have strong CD4+ T-cell responses to rhCMV and SIV.27,45 Antigen-specific T lymphocytes were identified based on up-regulation of the early T-lymphocyte activation molecule CD69 and secretion of TNF-α. Previous work from our laboratory has demonstrated that analysis of TNF-α identifies a higher frequency of macaque CD4+ T lymphocytes responding to CMV and SIV than revealed by analysis of interferon-γ alone.32 A significant fraction (11·7 ± 5·4%) of CD4hi CD8αlo cells secreted TNF-α following stimulation with CMV antigen, representing on average a fivefold enrichment compared with CD4 SP T cells (Fig. 5). A comparable degree of enrichment was observed when interferon-γ secretion was examined in PBMC from SIV-uninfected animals (data not shown). However, analysis of the frequency of SIV-specific T cells in CD4 SP and CD4hi CD8αlo lymphocytes from macaques infected with attenuated strains of SIV did not reveal any significant enrichment of SIV-specific T cells in the CD4hi CD8αlo population. Of six animals examined, five exhibited an average 67% decrease in the frequency of SIV Gag-specific T cells in CD4hi CD8αlo T cells compared with CD4 SP T cells, while the sixth animal had a fourfold enrichment (data not shown).
Figure 5.
Detection of CMV-specific responses by CD4hi CD8αlo lymphocytes using intracellular cytokine staining. PBMC from an SIVΔnef-vaccinated, CMV-seropositive monkey were stimulated with control antigen from uninfected (UI) cells, whole virus-rhCMV antigen, or superantigen (SEA/SEB). The percentages of cells expressing CD69 and TNF-α are indicated in the right upper quadrant of each plot. Results are representative of nine animals.
Analysis of TRECs in CD4hi CD8αlo T cells
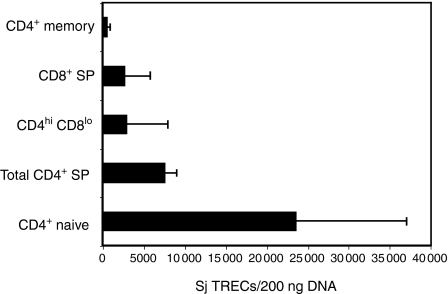
As an alternative approach to characterizing macaque CD4hi CD8αlo T cells, we determined the number of TRECs in different T-cell subsets. Signal joint TRECs are stable episomal molecules that are generated during T-cell receptor (TCR) rearrangement in the thymus and progressively diluted out during subsequent rounds of cell division.34,46 We quantified the number of signal joint TRECs in sorted populations of lymphocytes using real-time PCR primers and probes specific for macaque TRECs.36 The level of TRECs in CD4hi CD8αlo cells was sevenfold lower than observed in naive CD4+ T cells and twofold lower that the total CD4+ T-cell population (Fig. 6). Levels of TRECs in CD4hi CD8αlo T cells were not significantly different from those observed in CD4+ memory T cells. The relatively low levels of TRECs observed in the CD4hi CD8αlo T-cell population are consistent with the phenotypic evidence that these cells represent an effector memory T-cell population and do not represent recent thymic emigrants.
Figure 6.
Analysis of TRECs in CD4hi CD8αlo T cells. Purified populations of CD4hi CD8αlo, CD4 SP and CD8 SP T cells were obtained by cell sorting from five rhesus macaques. CD4 SP T cells were further sorted into memory and naive subsets based on their CD28 and CD95 expression. TREC analysis was performed by real-time PCR on 200 ng of DNA extracted from the indicated subpopulations of T cells. The difference in the level of TRECs between CD4hi CD8αlo and CD4 SP naive cells is significant (P = 0·001, Fisher's PLSD). Sj: signal joint.
Effect of SIV infection on the frequency of CD4hi CD8αlo lymphocytes
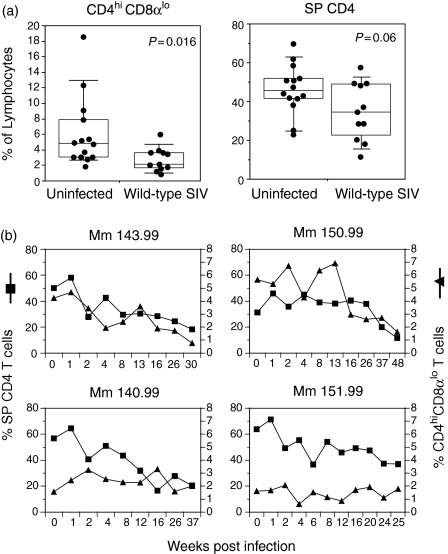
In the light of evidence that CD4hi CD8αlo lymphocytes represented an activated memory population that expressed CCR5, we examined whether CD4hi CD8αlo cells were depleted during the course of SIV infection. We first examined the frequency of CD4hi CD8αlo lymphocytes in cross-sectional studies of animals infected with pathogenic strains of SIV as compared with SIV-controls. Animals were analysed at the time of necropsy for advanced acquired immunodeficiency syndrome (geometric mean viral load 3·9 × 107 copy equivalents/ml). The frequency of CD4hiCD8αlo T lymphocytes was 56% lower in animals infected with pathogenic strains of SIV as compared with normal animals (P = 0·016, Mann–Whitney U-test) (Fig. 7a). The magnitude of depletion of CD4hi CD8αlo lymphocytes in SIV-infected animals exceeded that of CD4 SP lymphocytes, which were reduced by only 26% (P = 0·06, Mann–Whitney).
Figure 7.
Effect of SIVmac239 infection on the frequency of CD4 SP and CD4hi CD8αlo lymphocytes. (a) Cross-sectional analysis of the frequency of CD4hi CD8αlo and CD4 SP T cells in uninfected and wild-type SIV-infected macaques. P-values calculated by the Mann–Whitney U-test are shown. (b) Longitudinal analysis of the frequency of CD4+ and CD4hi CD8αlo lymphocytes at the indicated times after infection in rhesus macaques inoculated with pathogenic SIVmac239.
We also examined the effect of SIV infection on CD4hi CD8αlo lymphocytes in longitudinal studies in seven rhesus macaques inoculated with pathogenic SIVmac239. In four of these animals we observed a significant decline of CD4+ T cells during the observation period. In two of these four animals (Mm 143.99 and Mm 150.99), the decrease of CD4 SP T cells was accompanied by a decline of CD4hi CD8αlo T cells (Fig. 7b, top panels), suggesting that CD4hi CD8αlo T cells are depleted in a subset of animals during SIV infection, in concert with SP CD4 T cells. However, no significant changes were observed in the percentage of CD4hi CD8αlo T cells during the period of observation of the other five SIV-infected animals. There was no significant correlation between the preinfection frequency of CD4hi CD8αlo cells and the rate of CD4+ T-cell depletion.
Discussion
Mature T lymphocytes characteristically express either CD4 or CD8 alone, while CD4+ CD8+ lymphocytes have been classically considered to represent immature thymocytes. However, there has been increasing recognition of distinct subsets of mature T lymphocytes that express both CD4 and CD8. Our present study was designed to assess the phenotype and function of CD4+ CD8+ T lymphocytes in rhesus macaques and to investigate the effects of SIV infection on this distinct subset of cells. We identified two subsets of CD4+ CD8+ T lymphocytes in the peripheral blood of rhesus macaques. The dominant subset, CD4hi CD8lo, expressed the CD8αα homodimer and the minor subset, CD4lo CD8hi, expressed the CD8αβ heterodimer. CD4hi CD8αlo T cells exhibited an effector memory phenotype, as previously reported in humans and several animal species.23 Interestingly, the frequency of CMV-specific cells was enriched in CD4hi CD8αlo cells compared with CD4 SP cells, further reinforcing the classification of these cells as differentiated effector memory T cells. Finally, CD4hi CD8αlo cells expressed intermediate levels of CCR5 and were depleted in SIV-infected animals.
Efforts to understand the origin of CD4+ CD8+ T lymphocytes have been confounded by the failure of some investigators to distinguish the heterogeneous populations of CD4+ CD8+ T lymphocytes. CD4+ CD8+ T lymphocytes may include CD4hi CD8lo, CD4lo CD8hi and CD4hi CD8hi subsets. In many species, including humans, monkeys and swine, most peripheral CD4+ CD8+ T cells in adults are CD4hi CD8lo, whereas the other subsets of CD4+ CD8+ T cells are generally less frequent.23,47 The CD4hi CD8lo population in other species has also been observed to express the CD8αα homodimer, while the CD4lo CD8hi subset expresses CD8β, as we observed in rhesus macaques.7,17,47 CD4hi CD8αlo cells can comprise a significant fraction of T lymphocytes in normal rhesus macaques (up to 18%, mean 6%), whereas the frequency of CD4hi CD8αlo T cells in humans appears to be lower (mean 1·5–2·0%).40,48 Several previous publications have provided evidence that CD4hi CD8αlo T lymphocytes represent a differentiated memory T-cell population derived from CD4 SP T lymphocytes. Phenotypic analysis of CD4hi CD8αlo T cells has demonstrated that they exhibit a characteristic effector memory phenotype (CD28– CD45RA–), while lacking markers characteristic of naive or immature lymphocytes (CD1a,b, CD7, CD62L).3,12,17,23,40 The phenotypic characterization of CD4hi CD8αlo T cells as a differentiated memory T-cell population is also supported by the finding of a lower TREC content and shorter telomere length in these cells.40 As discussed in more detail below, CD4hi CD8αlo T lymphocytes also express multiple effector molecules such as perforin and are able to mediate antiviral responses against CMV and human immunodeficiency virus type 1 (HIV-1).12,40 Further evidence that CD4hi CD8αlo T cells represent a differentiated CD4+ T-cell population comes from the finding in humans and macaques that TCR sequences of CD4hi CD8αlo cells can be found in CD4 SP T cells but not CD8 SP T cells.4,39
Our data support the concept of rhesus macaque CD4hi CD8αlo T cells as an effector memory T-cell population derived from CD4 SP T cells. This conclusion is strongly supported by the phenotypic characterization of CD4hi CD8αlo cells as a CD28– CD95+ population that lacks the expression of markers characteristic of naive cells (i.e. CD7, CD45RA, CD62L).33 The lower TREC content of macaque CD4hi CD8αlo T cells than naive CD4+ T cells reinforces the phenotypic evidence that CD4hi CD8αlo cells represent an effector memory population. Finally, the increased frequency of CD4hi CD8αlo T cells responding to stimulation with whole CMV lysate also supports their origin in the CD4 SP T-cell population, because responses of SP CD8α T cells to rhCMV lysate are negligible, and this response has been previously shown to be major histocompatibility complex (MHC) class II-restricted.27 These findings are similar to those reported for CD4hi CD8αlo lymphocytes in cynomolgus macaques, which have previously been reported to have an effector memory phenotype and to increase with age.17 Notably, cynomolgus CD4hi CD8αlo T lymphocytes have been shown to express perforin and granzyme mRNA and to be able to mediate direct cytolysis of target cells.39
While CD4+ T lymphocytes have been classically viewed primarily as T helper cells that modulate effector function by other immune cells, there is mounting evidence that subpopulations of CD4+ T lymphocytes can mediate direct effector function. In mice, evidence for direct effector function of CD4+ T cells has been observed in vivo for control of influenza,49 the γ-herpesvirus MHV-6850 and Friend retrovirus infection.51 The mechanisms by which murine CD4+ T lymphocytes mediate direct effector functions against virus-infected cells are still incompletely defined but appear to involve both secretion of interferon-γ and direct cytolysis by CD4+ T cells.50,51 In humans, subpopulations of CD4+ T cells that are able to mediate in vitro cytolysis of cells expressing CMV and HIV antigens have been described.12,52,53 Increased frequencies of peripheral blood CD4+ T cells expressing either perforin54 or granzymes A and B53 have been described in HIV-seropositive and CMV-seropositive subjects, respectively. The fact that rhesus CD4hi CD8αlo T lymphocytes expressed levels of granzyme B at a frequency similar to that of CD8 SP T lymphocytes strongly suggests that CD4hi CD8αlo T cells represent a subset of CD4+ T cells that is able to mediate direct antiviral activity. Taken together, these data suggest that CD4hi CD8αlo cells may represent a CD4+ effector cell population able to mediate the control of chronic viral infections such as CMV.
Our finding of depletion of CD4hi CD8αlo T lymphocytes in a subset of SIV-infected macaques highlights the fact that SIV and HIV strains preferentially infect and deplete distinct subsets of memory CD4+ T lymphocytes. This preferential infection occurs as a result of the fact that most primary SIV and HIV strains utilize CCR5 as a co-receptor and the preferential expression of CCR5 on a subset of memory CD4+ T lymphocytes.43,55 We observed that the majority of CD4hi CD8αlo T lymphocytes expressed intermediate levels of CCR5 and were CD45RAlo. Based on earlier reports that CCR5+ memory CD4+ T lymphocytes are preferentially depleted by SIV during primary infection,55,56 this suggests that the CD4hi CD8αlo effector memory CD4+ T-cell population is likely to be infected during the course of the SIV infection. Our data are consistent with observations in SIV-infected cynomolgus macaques, in which CD4hi CD8αlo T cells were depleted as early as 1 month after infection.57 The ability of SIV and HIV58 to preferentially deplete subsets of memory CD4+ T lymphocytes may lead to immunodeficiency that is incompletely reflected by the analysis of total CD4+ T-cell counts, as shown by the fact that SIV-infected macaques often develop opportunistic infections at CD4+ T-cell counts above 200 per mm3.59
Identification of a significant subpopulation of CD4+ T cells expressing CD8α, which can represent over 50% of all CD4+ T cells in some animals, adds a cautionary note to the interpretation of studies in which administration of mAbs specific for CD8α are used to deplete CD8α+ T lymphocytes.60,61 Although results from these studies have often been interpreted to reflect the selective depletion of CD8α SP T lymphocytes, CD8α is expressed on several other lymphocyte populations in macaques, including natural killer cells and B cells.62 While the increases in SIV replication that have been observed following administration of CD8α-specific antibodies may most likely be the result of depletion of CD8+ T lymphocytes, the potential contributions of other CD8α-expressing effector populations such as natural killer cells and CD4hi CD8αlo T cells need to be better addressed.
In addition to serving as a phenotypic marker of CD4+ effector memory T cells, expression of CD8αα on these cells may serve a functional role. Several important functional differences between CD8αα and CD8αβ have been described.63 The extracytoplasmic domain of CD8α predominantly mediates interactions with the conserved α3 domain of the MHC class I heavy chain64 and the cytoplasmic tail of CD8α interacts with the p56 lck tyrosine kinase.65 However, the CD8β chain plays a key role in the ability of CD8 to function as a TCR coreceptor66 a feature that may be related to the fact while the CD8αβ heterodimer is localized with lipid rafts, the CD8αα homodimer is excluded.67 Thus, the interaction of CD8αα with MHC class I might serve to increase the binding affinity of CD4hi CD8αlo cells to their cognate target cells, without significantly affecting TCR-mediated signalling. The factors that regulate up-regulation of CD8αα on a subpopulation of CD4+ T lymphocytes are not well understood. Up-regulation of expression of CD8αα on CD4 SP T cells has been described following lymphocyte activation or incubation with IL-4.22,68
In conclusion, our data indicate that rhesus macaque CD4hi CD8αlo cells represent a distinct subset of memory CD4+ T cells that express several effector molecules and are selectively enriched for CMV-specific cells. Moreover, CD4hi CD8αlo cells express the SIV co-receptor CCR5 and are depleted in a subset of macaques during the course of SIV infection. However, further studies are necessary to better understand the function of CD4hi CD8αlo T cells and their role in controlling viral infections in vivo.
Acknowledgments
We thank Michelle Connole for cell sorting, Amy Barabasz for flow cytometric analysis, Jackie Gillis and Stephen Braun for helpful discussions, Angela Carville and members of the New England Primate Research Center Primate Medicine Division for expert animal care, Linqi Zhang and David Ho for providing the cloned M. mulatta TREC sequence, and Carolyn O'Toole and Noel Bane for manuscript preparation. This work was supported by National Institutes of Health grants RR00168, AI43890, AI45314 and AI62412.
Abbreviations
- APC
allophycocyanin
- BD
Becton Dickinson
- CMV
cytomegalovirus
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- FMO
fluorescence minus one
- HIV
human immunodeficiency virus
- mAb
monoclonal antibody
- PBMC
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PE
phycoerythrin
- PerCP
Peridinin chlorophyll protein
- PLSD
protected least-significant difference
- rhCMV
rhesus cytomegalovirus
- SIV
simian immunodeficiency virus
- SP
single positive
- TREC
T-cell receptor excision circle
References
- 1.Blue ML, Daley JF, Levine H, Schlossman SF. Co-expression of T4 and T8 on peripheral blood T cells demonstrated by two-color fluorescence flow cytometry. J Immunol. 1985;134:228–34. [PubMed] [Google Scholar]
- 2.Lanier LL, Phillips JH. Evidence for three types of human cytotoxic lymphocytes. Immunol Today. 1986;7:132–4. doi: 10.1016/0167-5699(86)90076-9. [DOI] [PubMed] [Google Scholar]
- 3.Ortolani C, Forti E, Radin E, Cibin R, Cossarizza A. Cytofluorimetric identification of two populations of double positive (CD4+ CD8+) T lymphocytes in human peripheral blood. Biochem Biophys Res Commun. 1993;191:601–9. doi: 10.1006/bbrc.1993.1260. [DOI] [PubMed] [Google Scholar]
- 4.Colombatti A, Doliana R, Schiappacassi M, Argentini C, Tonutti E, Feruglio C, Sala P. Age-related persistent clonal expansions of CD28(–) cells: phenotypic and molecular TCR analysis reveals both CD4(+) and CD4(+) CD8(+) cells with identical CDR3 sequences. Clin Immunol Immunopathol. 1998;89:61–70. doi: 10.1006/clin.1998.4580. [DOI] [PubMed] [Google Scholar]
- 5.Prince H, Golding J, York J. Characterization of circulating CD4+ CD8+ lymphocytes in healthy individuals prompted by identification of a blood donor with a markedly elevated level of CD4+ CD8+ lymphocytes. Clin Diagn Lab Immunol. 1994;1:597–605. doi: 10.1128/cdli.1.5.597-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sala P, Tonutti E, Feruglio C, Florian F, Colombatti A. Persistent expansions of CD4+ CD8+ peripheral blood T cells. Blood. 1993;1:1546–52. [PubMed] [Google Scholar]
- 7.Tonutti E, Sala P, Feruglio C, Yin Z, Colombatti A. Phenotypic heterogeneity of persistent expansions of CD4+ CD8+ T cells. Clin Immunol Immunopathol. 1994;73:312–20. doi: 10.1006/clin.1994.1204. [DOI] [PubMed] [Google Scholar]
- 8.Bagot M, Echchakir H, Mami-Chouaib F, et al. Isolation of tumor-specific cytotoxic CD4+ and CD4+ CD8dim+ T-cell clones infiltrating a cutaneous T-cell lymphoma. Blood. 1998;91:4331–41. [PubMed] [Google Scholar]
- 9.Mizuki M, Tagawa S, Machii T, et al. Phenotypical heterogeneity of CD4+ CD8+ double-positive chronic T lymphoid leukemia. Leukemia. 1998;12:499–504. doi: 10.1038/sj.leu.2400978. [DOI] [PubMed] [Google Scholar]
- 10.Matsui M, Fukuyama H, Akiguchi I, Kameyama M. Circulating CD4+ CD8+ cells in myasthenia gravis: supplementary immunological parameter for long-term prognosis. J Neurol. 1989;236:329–35. doi: 10.1007/BF00314374. [DOI] [PubMed] [Google Scholar]
- 11.Weiss L, Roux A, Garcia S, Demouchy C, Haeffner-Cavaillon N, Kazatchkine MD, Gougeon ML. Persistent expansion, in human immunodeficiency virus-infected person, of V beta-restricted CD4+ CD8+ T lymphocytes that express cytotoxicity-associated molecules and are committed to produce interferon-gamma and tumor necrosis factor-alpha. J Infect Dis. 1998;178:1158–62. doi: 10.1086/515674. [DOI] [PubMed] [Google Scholar]
- 12.Suni MA, Ghanekar SA, Houck DW, Maecker HT, Wormsley SB, Picker LJ, Moss RB, Maino VC. CD4(+) CD8(dim) T lymphocytes exhibit enhanced cytokine expression, proliferation and cytotoxic activity in response to HCMV and HIV-1 antigens. Eur J Immunol. 2001;31:2512–20. doi: 10.1002/1521-4141(200108)31:8<2512::aid-immu2512>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 13.Sullivan YB, Landay AL, Zack JA, Kitchen SG, Al-Harthi L. Upregulation of CD4 on CD8+ T cells: CD4dim CD8bright T cells constitute an activated phenotype of CD8+ T cells. Immunology. 2001;103:270–80. doi: 10.1046/j.1365-2567.2001.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasahara T, Tamauchi H, Ikewaki N, Kubota K. Unique properties of a cytotoxic CD4+ CD8+ intraepithelial T-cell line established from the mouse intestinal epithelium. Microbiol Immunol. 1994;38:191–9. doi: 10.1111/j.1348-0421.1994.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 15.Kenny E, Mason D, Pombo A, Ramirez F. Phenotypic analysis of peripheral CD4+ CD8+ T cells in the rat. Immunology. 2000;101:178–84. doi: 10.1046/j.1365-2567.2000.00071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuckermann FA, Husmann RJ. Functional and phenotypic analysis of porcine peripheral blood CD4/CD8 double-positive T cells. Immunology. 1996;87:500–12. [PMC free article] [PubMed] [Google Scholar]
- 17.Akari H, Terao K, Murayama Y, Nam KH, Yoshikawa Y. Peripheral blood CD4+ CD8+ lymphocytes in cynomolgus monkeys are of resting memory T lineage. Int Immunol. 1997;9:591–7. doi: 10.1093/intimm/9.4.591. [DOI] [PubMed] [Google Scholar]
- 18.Pescovitz MD, Sakopoulos AG, Gaddy JA, Husmann RJ, Zuckermann FA. Porcine peripheral blood CD4+ CD8+ dual expressing T-cells. Vet Immunol Immunopathol. 1994;43:53–62. doi: 10.1016/0165-2427(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 19.Bonomo A, Kehn PJ, Shevach EM. Premature escape of double-positive thymocytes to the periphery of young mice. Possible role in autoimmunity. J Immunol. 1994;152:1509–14. [PubMed] [Google Scholar]
- 20.Res P, Blom B, Hori T, Weijer K, Spits H. Downregulation of CD1 marks acquisition of functional maturation of human thymocytes and defines a control point in late stages of human T cell development. J Exp Med. 1997;185:141–51. doi: 10.1084/jem.185.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe N, DeRosa SC, Cmelak A, Hoppe R, Herzenberg LA, Herzenberg LA, Roederer M. Long-term depletion of naive T cells in patients treated for Hodgkin's disease. Blood. 1997;90:3662–72. [PubMed] [Google Scholar]
- 22.Paliard X, Malefijt RW, de Vries JE, Spits H. Interleukin-4 mediates CD8 induction on human CD4+ T-cell clones. Nature. 1988;335:642–4. doi: 10.1038/335642a0. [DOI] [PubMed] [Google Scholar]
- 23.Zuckermann FA. Extrathymic CD4/CD8 double positive T cells. Vet Immunol Immunopathol. 1999;72:55–66. doi: 10.1016/s0165-2427(99)00118-x. [DOI] [PubMed] [Google Scholar]
- 24.Flamand L, Crowley RW, Lusso P, Columbini-Hatch S, Margolis DM, Gallo RC. Activation of CD8+ T lymphocytes through the T cell receptor turns on CD4 gene expression: implications for HIV pathogenesis. Proc Natl Acad Sci USA. 1998;95:3111–16. doi: 10.1073/pnas.95.6.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kestler HW, III, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, Desrosiers RC. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–62. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 26.Wyand MS, Manson KH, Garcia-Moll M, Montefiori D, Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70:3724–33. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaur A, Hale CL, Noren B, Kassis N, Simon MA, Johnson RP. Decreased frequency of cytomegalovirus (CMV) -specific CD4+ T lymphocytes in simian immunodeficiency virus-infected rhesus macaques: inverse relationship with CMV viremia. J Virol. 2002;76:3646–58. doi: 10.1128/JVI.76.8.3646-3658.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anonymous. The Institute of Laboratory Animal Resources, National. Research Council, Guide for the Care and Use of Laboratory Animals. Washington, DC: National Institutes of Health; 1996. pp. 86–123. [Google Scholar]
- 29.DiSanto JP, Knowles RW, Flomenberg N. The human Lyt-3 molecule requires CD8 for cell surface expression. Embo J. 1988;7:3465–70. doi: 10.1002/j.1460-2075.1988.tb03221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norment AM, Littman DR. A second subunit of CD8 is expressed in human T cells. EMBO J. 1988;7:3433–9. doi: 10.1002/j.1460-2075.1988.tb03217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roederer M. Spectral compensation for flow cytometry. Visualization artifacts, limitations, and caveats. Cytometry. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 32.Gauduin M-C, Kaur A, Ahmad S, Yilma T, Lifson JD, Johnson RP. Optimization of intracellular cytokine staining for the quantitation of antigen-specific CD4+ T cell responses in rhesus macaques. J Immunol Meth. 2004;288:61–79. doi: 10.1016/j.jim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 33.Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ. Development and homeostasis of T cell memory in rhesus macaque. J Immunol. 2002;168:29–43. doi: 10.4049/jimmunol.168.1.29. [DOI] [PubMed] [Google Scholar]
- 34.Douek DC, McFarland RD, Keiser PH, et al. Changes in thymic function with age and during the treatment of HIV infection. Nature. 1998;396:690–5. doi: 10.1038/25374. [DOI] [PubMed] [Google Scholar]
- 35.Sodora DL, Douek DC, Silvestri G, et al. Quantification of thymic function by measuring T cell receptor excision circles within peripheral blood and lymphoid tissues in monkeys. Eur J Immunol. 2000;30:1145–53. doi: 10.1002/(SICI)1521-4141(200004)30:4<1145::AID-IMMU1145>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Chakrabarti LA, Lewin SR, Zhang L, et al. Normal T-cell turnover in sooty mangabeys harboring active simian immunodeficiency virus infection. J Virol. 2000;74:1209–23. doi: 10.1128/jvi.74.3.1209-1223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macchi B, Graziani G, Zhang J, Mastino A. Emergence of double-positive CD4/CD8 cells from adult peripheral blood mononuclear cells infected with human T cell leukemia virus type I (HTLV-1) Cell Immunol. 1993;149:376–89. doi: 10.1006/cimm.1993.1163. [DOI] [PubMed] [Google Scholar]
- 38.Aandahl EM, Sandberg JK, Beckerman KP, Tasken K, Moretto WJ, Nixon DF. CD7 is a differentiation marker that identifies multiple CD8 T cell effector subsets. J Immunol. 2003;170:2349–55. doi: 10.4049/jimmunol.170.5.2349. [DOI] [PubMed] [Google Scholar]
- 39.Nam KH, Akari H, Terao K, Shibata H, Kawamura S, Yoshikawa Y. Peripheral blood extrathymic CD4(+) CD8(+) T cells with high cytotoxic activity are from the same lineage as CD4(+) CD8(–) T cells in cynomolgus monkeys. Int Immunol. 2000;12:1095–103. doi: 10.1093/intimm/12.7.1095. [DOI] [PubMed] [Google Scholar]
- 40.Nascimbeni M, Shin E-C, Chiriboga L, Kleiner DE, Rehermann B. Peripheral CD4+ CD8+ T cells are differentiated effector memory cells with antiviral functions. Blood. 2004;104:478–86. doi: 10.1182/blood-2003-12-4395. [DOI] [PubMed] [Google Scholar]
- 41.Rentenaar RJ, Wever PC, van Diepen FN, Schellekens PT, Wertheim PM, ten Berge IJ. CD4dull CD8bright double-positive T-lymphocytes have a phenotype of granzyme B+ CD8+ memory T-lymphocytes. Nephrol Dial Transplant. 1999;14:1430–4. doi: 10.1093/ndt/14.6.1430. [DOI] [PubMed] [Google Scholar]
- 42.Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–15. [PubMed] [Google Scholar]
- 43.Chen Z, Zhou P, Ho DD, Landau NR, Marx PA. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–14. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veazey RS, Tham IC, Mansfield KG, et al. Identifying the target cell in primary simian immunodeficiency virus (SIV) infection: highly activated memory CD4+ T cells are rapidly eliminated in early SIV infection in vivo. J Virol. 2000;74:57–64. doi: 10.1128/jvi.74.1.57-64.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gauduin M-C, Glickman RL, Ahmad S, Yilma T, Johnson RP. Immunization with live attenuated simian immunodeficiency virus induces strong type 1 T helper responses and β-chemokine production. Proc Natl Acad Sci USA. 1999;96:14031–6. doi: 10.1073/pnas.96.24.14031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong F, Chen CH, Cooper MD. Thymic function can be accurately monitored by the level of recent T cell emigrants in the circulation. Immunity. 1998;8:97–104. doi: 10.1016/s1074-7613(00)80462-8. [DOI] [PubMed] [Google Scholar]
- 47.Parel Y, Chizzolini C. CD4+ CD8+ double positive (DP) T cells in health and disease. Autoimmun Rev. 2004;3:215–20. doi: 10.1016/j.autrev.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz JE, Forman MA, Lifton MA, et al. Expression of the CD8αβ-heterodimer on CD8(+) T lymphocytes in peripheral blood lymphocytes of human immunodeficiency virus– and human immunodeficiency virus+ individuals. Blood. 1998;92:198–206. [PubMed] [Google Scholar]
- 49.Graham MB, Braciale VL, Braciale TJ. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J Exp Med. 1994;180:1273–82. doi: 10.1084/jem.180.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Christensen JP, Cardin RD, Branum KC, Doherty PC. CD4(+) T cell-mediated control of a gamma-herpesvirus in B cell-deficient mice is mediated by IFN-gamma. Proc Natl Acad Sci USA. 1999;96:5135–40. doi: 10.1073/pnas.96.9.5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwashiro M, Peterson K, Messer RJ, Stromnes IM, Hasenkrug KJ. CD4(+) T cells and gamma interferon in the long-term control of persistent friend retrovirus infection. J Virol. 2001;75:52–60. doi: 10.1128/JVI.75.1.52-60.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norris PJ, Sumaroka M, Brander C, et al. Multiple effector functions mediated by human immunodeficiency virus-specific CD4(+) T-cell clones. J Virol. 2001;75:9771–9. doi: 10.1128/JVI.75.20.9771-9779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaunders JJ, Dyer WB, Wang B, et al. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 2004;103:2238–47. doi: 10.1182/blood-2003-08-2765. [DOI] [PubMed] [Google Scholar]
- 54.Appay V, Zaunders JJ, Papagno L, et al. Characterization of CD4+ CTLs ex vivo. J Immunol. 2002;168:5954–8. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 55.Veazey RS, Mansfield KG, Tham IC, Carville AC, Shvetz DE, Forand AE, Lackner AA. Dynamics of CCR5 expression by CD4+ T cells in lymphoid tissues during simian immunodeficiency virus infection. J Virol. 2000;74:11001–7. doi: 10.1128/jvi.74.23.11001-11007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattapallil JJ, Douek DC, Hill BJ, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–7. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- 57.Akari H, Nam KH, Mori K, Otani I, Shibata H, Adachi A, Terao K, Yoshikawa Y. Effects of SIVmac infection on peripheral blood CD4+ CD8+ T lymphocytes in cynomolgus macaques. Clin Immunol. 1999;91:321–9. doi: 10.1006/clim.1999.4700. [DOI] [PubMed] [Google Scholar]
- 58.Brenchley JM, Schacker TW, Ruff LE, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–59. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lackner A. Pathology of simian immunodeficiency virus induced disease. Curr Top Microbiol Immunol. 1994;188:35–59. doi: 10.1007/978-3-642-78536-8_3. [DOI] [PubMed] [Google Scholar]
- 60.Schmitz JE, Kuroda MJ, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 61.Jin X, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–8. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webster RL, Johnson RP. Delineation of multiple subpopulations of natural killer cells in rhesus macaques. Immunology. 2005;115:206–14. doi: 10.1111/j.1365-2567.2005.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gangadharan D, Cheroutre H. The CD8 isoform CD8αα is not a functional homologue of the TCR co-receptor CD8αβ. Curr Opin Immunol. 2004;16:264–70. doi: 10.1016/j.coi.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 64.Salter RD, Benjamin RJ, Wesley PK, et al. A binding site for the T-cell co-receptor CD8 on the alpha 3 domain of HLA-A2. Nature. 1990;345:41–6. doi: 10.1038/345041a0. [DOI] [PubMed] [Google Scholar]
- 65.Hoeveler A, Malissen B. The cysteine residues in the cytoplasmic tail of CD8 alpha are required for its coreceptor function. Mol Immunol. 1993;30:755–64. doi: 10.1016/0161-5890(93)90147-4. [DOI] [PubMed] [Google Scholar]
- 66.Wong JS, Wang X, Witte T, Nie L, Carvou N, Kern P, Chang HC. Stalk region of beta-chain enhances the coreceptor function of CD8. J Immunol. 2003;171:867–74. doi: 10.4049/jimmunol.171.2.867. [DOI] [PubMed] [Google Scholar]
- 67.Arcaro A, Gregoire C, Boucheron N, Stotz S, Palmer E, Malissen B, Luescher IF. Essential role of CD8 palmitoylation in CD8 coreceptor function. J Immunol. 2000;165:2068–76. doi: 10.4049/jimmunol.165.4.2068. [DOI] [PubMed] [Google Scholar]
- 68.Gao MH, Walz M, Kavathas PB. Post-transcriptional regulation associated with control of human CD8 alpha expression of CD4+ T cells. Immunogenetics. 1996;45:130–5. doi: 10.1007/s002510050180. [DOI] [PubMed] [Google Scholar]