Abstract
Tlr elements are a novel family of ∼30 putative mobile genetic elements that are confined to the germ line micronuclear genome in Tetrahymena thermophila. Thousands of diverse germ line-limited sequences, including the Tlr elements, are specifically eliminated from the differentiating somatic macronucleus. Macronucleus-retained sequences flanking deleted regions are known to contain cis-acting signals that delineate elimination boundaries. It is unclear whether sequences within deleted DNA also play a regulatory role in the elimination process. In the current study, an in vivo DNA rearrangement assay was used to identify internal sequences required in cis for the elimination of Tlr elements. Multiple, nonoverlapping regions from the ∼23-kb Tlr elements were independently sufficient to stimulate developmentally regulated DNA elimination when placed within the context of flanking sequences from the most thoroughly characterized family member, Tlr1. Replacement of element DNA with macronuclear or foreign DNA abolished elimination activity. Thus, diverse sequences dispersed throughout Tlr DNA contain cis-acting signals that target these elements for programmed elimination. Surprisingly, Tlr DNA was also efficiently deleted when Tlr1 flanking sequences were replaced with DNA from a region of the genome that is not normally associated with rearrangement, suggesting that specific flanking sequences are not required for the elimination of Tlr element DNA.
Ciliates are unusual among eukaryotes in that each cell contains two structurally and functionally distinct nuclei. This phenomenon of nuclear dualism has apparently evolved to separate the two primary functions of the genome: transmission and expression of genetic information (14). The micronucleus, which is transcriptionally silent in vegetatively growing cells, is the germ line nucleus in ciliates. It contains an intact copy of the complete genome of the organism and participates primarily in the reciprocal exchange of gametic nuclei that occurs during conjugation, the sexual phase of the ciliate life cycle. In contrast, the somatic macronucleus contains a highly processed subset of the micronuclear genome and is responsible for essentially all nuclear transcription in vegetatively growing cells. In each sexual generation, the macronucleus is degraded, and a replacement is generated from a mitotic copy of the zygotic micronucleus formed during conjugation. As the new macronucleus differentiates, it undergoes dramatic genome reorganization involving chromosomal fragmentation and extensive DNA elimination (14).
The ciliate Tetrahymena thermophila provides a particularly useful system for the molecular analysis of programmed DNA elimination events (33). In the developing macronucleus, the five germ line-derived T. thermophila chromosomes are fragmented at 50 to 250 distinct chromosome breakage sequences, and telomeres are added de novo to the resulting subchromosomal molecules. In addition, an estimated 6,000 stretches of micronucleus-limited DNA are removed through programmed deletion events involving the excision of specific chromosomal segments and the ligation of macronucleus-retained flanking sequences. Rapid degradation of the internally eliminated sequences (IESs) ultimately results in a loss of ∼15% of the germ line genome from the mature macronucleus (6, 66). Tetrahymena IESs range from ∼0.6 to >20 kb in length, have highly diverse nucleotide sequences, and appear to be dispersed throughout the genome.
Although the biological function of programmed DNA elimination in Tetrahymena is currently unclear, the molecular events involved in this remarkable process exhibit many of the hallmarks of heterochromatin formation (reviewed in reference 31). Nucleosomes across IES regions are underacetylated during the early stages of macronuclear differentiation, and treatment of conjugating cells with the histone deacetylase inhibitor trichostatin A severely impairs elimination (17). In addition, IES regions show dramatic enrichment of methylated lysine-9 on histone H3 (K9-MeH3) compared to macronucleus-retained sequences during developmental stages preceding programmed DNA elimination (52), and K9-MeH3 is required for elimination to occur (Y. Liu and M. A. Gorovsky, personal communication). Two chromodomain-containing proteins, Pdd1p and Pdd3p, are expressed exclusively during macronuclear development and specifically localize to IES regions prior to elimination (41, 45). Like chromodomain-containing proteins from other systems, Pdd1p has been demonstrated to bind K9-MeH3 (3, 30, 36, 46, 52). The progeny of PDD1 somatic knockout cells fail to undergo programmed DNA elimination (15), and artificially tethering Pdd1p to an otherwise inactive IES is sufficient to restore deletion activity to that sequence (52). The mechanism by which heterochromatin factors initially target IES regions is unknown. It seems likely that this localization process involves some level of nucleotide specificity, since (i) specific sequences are consistently deleted from large populations of developing macronuclei and (ii) construct-based IESs transformed into developing macronuclei as “naked” DNA undergo accurate and reproducible elimination (33). Nevertheless, few distinguishable nucleotide or structural similarities have been observed among different IESs.
Analyses of three relatively short Tetrahymena IESs, the M, R, and mse2.9 elements, suggested that macronucleus-retained sequences flanking each element contain cis-acting regulatory signals that specify the boundaries of DNA elimination. In all cases, the sequences necessary for the accurate placement of rearrangement boundaries are located ∼50 to 70 bp outside the IES (9, 18, 22, 23, 39). Notably, no obvious nucleotide similarities have been detected among these cis-acting signals. Further deletion analyses of the M element suggested that, in addition to flanking regulatory sequences, cis-acting signals within micronucleus-limited DNA are required for programmed elimination (14, 65). However, the identity of these essential internal signals and their distribution within M element DNA are currently unclear. In light of these observations, it has been proposed that two distinct classes of cis-acting regulatory signals control the programmed deletion of Tetrahymena IESs: (i) internal sequences that serve to target an element for elimination (presumably through direct or indirect recruitment of heterochromatin complexes) and (ii) flanking regulatory signals within macronucleus-retained sequences that limit the spread of IES-associated heterochromatin and determine the location of rearrangement junctions (14, 65).
The M, R, and mse2.9 elements are relatively small (0.6 to 2.9 kb) and appear to have little protein-coding potential. However, the majority of Tetrahymena IESs belong to families of repetitive elements, at least some of which are transposon-like (12). Of these, the Tlr elements have been the most thoroughly characterized (21, 62). Approximately 30 Tlr family members are present in the micronuclear genome; all of them are eliminated from the developing macronucleus. Tlr elements consist of an ∼22-kb internal region flanked by long, complex terminal inverted repeats. The Tlr internal region is 90 to 97% conserved among family members at the nucleotide level and contains 15 major open reading frames. The conceptual translation products from several of the open reading frames resemble proteins encoded by transposable elements and viruses. In contrast to the high degree of nucleotide conservation observed within the Tlr internal region, Tlr terminal inverted repeats appear to vary in length and sequence from element to element. Although transposition of the Tlr elements has not been observed experimentally, based on their structure and copy number, it seems likely that Tlr elements represent a family of novel mobile genetic elements.
The most extensively analyzed Tlr family member is Tlr1. During macronuclear development, Tlr1 is eliminated as an IES with variable excision boundaries (47, 60). The 825-bp terminal inverted repeat of Tlr1 is located near, but not at, the rearrangement junctions. Obliteration of the macronucleus-destined DNA flanking the right side of Tlr1 results in aberrant processing of the element (48). Thus, it seems likely that this region contains cis-acting DNA signals that control the position of the adjacent elimination junction, analogous to the regulatory sequences located outside the M, R, and mse2.9 elements. The sequences involved in delimiting the left Tlr1 elimination boundary have not yet been identified.
In this report, we further analyze the mechanism of Tlr1 elimination by examining the internal sequences required in cis for the programmed deletion of this putative mobile genetic element. Our results demonstrate that multiple, nonoverlapping regions of Tlr1 contain cis-acting DNA signals that independently target robust programmed elimination activity in vivo when the DNA fragments are placed within the context of normal Tlr1 flanking sequences. In addition, we show that at least three of these Tlr DNA fragments are capable of inducing elimination when inserted within a sequence of the Tetrahymena genome that is not normally associated with programmed rearrangement. Thus, specific flanking sequences are not required for the programmed deletion of Tlr DNA.
MATERIALS AND METHODS
DNA rearrangement constructs.
Tflank-series rearrangement constructs were derived from pHWT.cam, which consists of the previously described WT.cam construct (48) cloned between two NotI sites in the polylinker region of bacterial plasmid vector pHSS6 (49). Tflank.IRL, Tflank.IRR, and Tflank were generated by inverse PCR with pHWT.cam as a template and primer sets Tlr1L:1961-1938- Tlr1R:891-911, Tlr1L:894-872- Tlr1R:35-56, and Tlr1L:894-872- Tlr1R:891-911, respectively. The nucleotide sequences of these and other primers used in this study are shown in Table 1. The resulting fragments containing deletions of the left half, the right half, or all of the Tlr1 inverted repeat sequence were incubated with the Klenow enzyme, phosphorylated with T4 polynucleotide kinase, gel purified, and circularized with T4 DNA ligase to generate plasmids pHTflank.IRL, pHTflank.IRR, and pHTflank, respectively.
TABLE 1.
PCR primer sequences
| Primer name | Primer sequence (5′ to 3′) |
|---|---|
| Tlr1L:25-47 | GGAATTCGAAAAATGATAAATACAACCTCC |
| Tlr1L:894-872 | CACGAGTAGCAAATGAAATTATA |
| Tlr1L:895-914 | AGAGAATTTACAATCGGAGC |
| Tlr1L:1961-1938 | ATAGCTGTTTAGAAGATTTGATGA |
| Tlr1R:35-56 | AGACTAACAGGAATGAATGAAG |
| Tlr1R:891-911 | TCTCGTGTATCAGAAAGAAATG |
| pMBR4C1:3554-3576 | AATATGTGGCAGTTCGCAATGTG |
| pMBR4C1:4218-4196 | TATGACAAGGATTAGGATAAGAG |
| pMBR2:1864-1885 | TAAGATGTCTGAAGCAATCAAC |
| pMBR2:2657-2635 | TGATTTACTTCTACTGGCTATTG |
| pMBR2:6359-6380 | GTTGGTGGATAGGAAATTAGTG |
| pMBR2:7698-7676 | GGTCTGGAATCAATGTACGAATG |
| IntB:3194-3213 | AGGATATGCAGCAACTTTAC |
| IntB:3937-3914 | TAAATCTGTTATTTGTCCCTGTTG |
| Tlint:2563-2582 | CTACAGTATTCTCCTTACCA |
| T1int:3384-3365 | TGGATTTATTCAGGAGGTAG |
| Cyd1:168-192 | ATACAGTTTTGATGACTTGGTAATG |
| Cyd1:585-607 | CAGAAAAACTGCCTTACTTTACT |
| Cyd1:1103-1084 | TTAGCACAAAGCAAAACTGG |
| αTub:1428-1449 | GTATCCAAGTCGGTAACGCCTG |
| αTub:2526-2447 | TCAGCGATGGCAGTAGAGTTG |
| Cat:257-277 | TCACTGGATATACCACCGTTG |
| Cat:1058-1038 | ACCAGCAATAGACATAAGCGG |
| C3 | TAGCAATTTAACTGTGATAAACTACCGCA |
| C4 | GATAAGCTGTCAAACATGAGAATTCCGG |
| R2 | ACTATGATTCCTCGTAAGCTTTCACTTACA |
| R3 | AAACATCTCATTGATAACTAACTGT |
Constructs Tflank.IN1 to Tflank.IN5 were generated by ligating DNA fragments from previously cloned internal segments of Tlr elements to the empty Tlr1 site in pHTflank. Fragment IN1 was amplified from Tlr clone pMBR4C1 (GenBank accession number AF451860) with primers pMBR4C1:3554-3576 and pMBR4C1:4218-4196. Fragments IN2 and IN3 were amplified from Tlr clone pMBR2 (GenBank accession number AF451863) with primer sets pMBR2:1864-1885- pMBR2:2657-2635 and pMBR2:6359-6380- pMBR2:7698-7676, respectively. Fragment IN4 was amplified from Tlr clone IntB (GenBank accession number AF232243) with primers IntB:2563-2582 and IntB:3937-3914. Fragment IN5 was PCR amplified from Tlr1 clone Tlr1 Int (GenBank accession number AF232246) with primers T1int:2563-2582 and T1int:3384-3365. The resulting circularized plasmids were designated pHTflank.IN1 to pHTflank.IN5.
The Tflank.Cyd, Tflank.Tub, and Tflank.Cat rearrangement constructs were assembled by using a similar strategy. The 936-bp Cyd fragment was PCR amplified with primers Cyd1:168-192 and Cyd1:1103-1084 (Table 1) from clone pCyd1X-B, which contains a segment of the Cyd1 T. thermophila macronuclear DNA locus (GenBank accession number L34029). The 1,099-bp Tub fragment of the α-tubulin gene (GenBank accession number M86723) was amplified from macronuclear DNA with primers αTub:1428-1449 and αTub:2526-2447. An 802-bp fragment of the Tn9 chloramphenicol acetyltransferase gene (GenBank accession number V00622) was PCR amplified from pHWT.cam with primers Cat:257-277 and Cat:1058-1038 to yield the Cat fragment. The resulting products were ligated between the Tlr1 flanking sequences in pHTflank to yield plasmids pHTflank.Cyd, pHTflank.Tub, and pHTflank.Cat.
Cflank-series rearrangement constructs were derived from bacterial plasmid pBCydH. In order to generate pBCydH, the ∼1,150-kb HindIII fragment of Cyd1 was excised from pCyd1X-B and cloned into the HindIII site of pBluescript KSΔPstI; the latter is a modified form of the vector in which the PstI site was removed from pBluescript by linearization with PstI followed by Klenow treatment and recircularization with T4 DNA ligase. Cflank.IN2, Cflank.IN5, Cflank.Tub, and Cflank.Cat were assembled by ligating the IN2, IN5, Tub, and Cat PCR products, respectively, to the unique PstI site in the Cyd1X portion of pBCydH. In each case, PstI-digested pBCydH was blunt ended with the Klenow enzyme and dephosphorylated with calf intestinal phosphatase prior to ligation. In order to generate the Cflank.IRL construct, the left side of the Tlr1 inverted repeat was PCR amplified from pHTflank with primers Tlr1L:895-914 and Tlr1L:1961-1938 and ligated to PstI-linearized pBCydH. Cflank-series constructs were excised from pBluescript KSΔPstI with HindIII and ligated to the HindIII site in the polylinker region of pHSS6 to yield pHCflank-series plasmids.
Once assembled, pHTflank-series and pHCflank-series plasmid constructs were excised from pHSS6 with NotI and ligated to the unique NotI site in Tetrahymena ribosomal DNA (rDNA) vector pD5H8 (22). The structures of the resulting pDTflank-series and pDCflank-series plasmids, respectively, were verified by DNA sequencing.
Tetrahymena conjugation and transformation.
T. thermophila strains CU427 and CU428 (kindly provided by Peter Bruns, Cornell University) were grown in PPYS (2% Proteose Peptone, 0.1% yeast extract, 0.003% Sequestrine) at 30°C to a density of ∼3 × 105 cells/ml and then washed in 10 mM Tris-HCl (pH 7.4). Following 12 to 16 h of starvation, conjugation was initiated by mixing strains of complementary mating types. Approximately 2 h past the anlagen II stage (43) of Tetrahymena nuclear development (usually ∼10 h postmixing), mating pairs were washed and concentrated to approximately 2 × 107 cells/ml in 10 mM HEPES (pH 7.5), mixed with ∼10 μg of each pD-series plasmid or pD5H8 as a control, and transformed by electroporation as described by Gaertig and Gorovsky (20). Electroporated cells were diluted to approximately 2 × 104 cells/ml and pipetted into 96-well microtiter plates. At 18 to 20 h postelectroporation, paromomycin was added to each well to a final concentration of 130 μg/ml. Transformants were identified as saturated Tetrahymena cultures in microtiter wells 3 to 5 days after the addition of the drug. Eight or nine independent transformant cell lines from each transformation were picked at random for molecular analysis (see below).
In vivo DNA rearrangement assay.
The in vivo rearrangement assay for DNA elimination makes use of a T. thermophila transformation system (67). pD5H8 (Fig. 1A) (22), a pUC18-based bacterial vector containing Tetrahymena micronuclear rDNA, is transformed into developing macronuclei of conjugating T. thermophila cells. Vector rDNA is subsequently processed in a manner similar to that of endogenous rDNA. The pD5H8 rDNA locus is excised from the plasmid at chromosomal breakage sequences and converted to the extrachromosomal palindromic structure characteristic of macronuclear rDNA (Fig. 1A). Telomeric repeats are added de novo to the rDNA, and the bacterial vector sequence is lost. The rDNA is then amplified to a final copy number that is approximately 200-fold higher than that of chromosomal sequences in the mature macronucleus. The resulting pD5H8-derived rDNA minichromosome effectively replaces endogenous rDNA due to an advantageous origin of replication (37) and confers paromomycin resistance to Tetrahymena transformants through a mutation in the 17S rRNA gene (7). IES-containing micronuclear DNA regions cloned into a unique NotI site in the 3′ untranscribed spacer region of pD5H8 rDNA undergo processing very similar to that of the elements in the chromosome (9, 18, 22, 39).
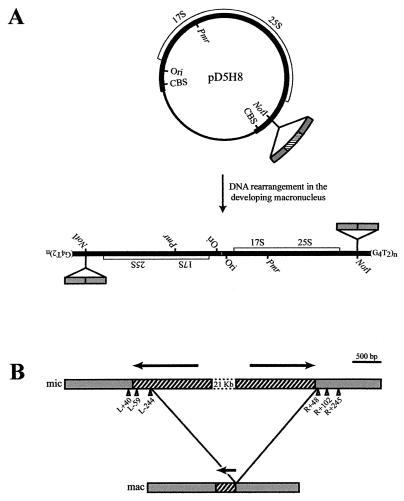
FIG. 1.
(A) rDNA-based in vivo rearrangement assay for T. thermophila. The pD5H8 processing vector consists of pUC18 plasmid DNA (thin black line) and Tetrahymena micronuclear rDNA (thick black line). The open box represents the 17S and 25S rRNA genes. The approximate positions of chromosome breakage sequences, the rDNA origin of replication (Ori), and a point mutation within the 17S rDNA (Pmr) conferring resistance to paromomycin are indicated. An IES (hatched box) and macronucleus-retained flanking sequences (gray boxes) are shown inserted into the unique NotI site in pD5H8. Upon transformation, pD5H8 and the cloned IES region undergo normal in vivo processing. (B) Tlr1 locus in the T. thermophila micronucleus (mic) and the predominant macronuclear (mac) product of programmed elimination. Hatched boxes depict the terminal regions of the Tlr1 element. Arrows represent the 825-bp terminal inverted repeat of Tlr1. Gray boxes depict Tlr1 flanking sequences. Positions of previously described elimination boundaries, including sites used in Tlr1 minor rearrangement variants, are indicated by arrowheads.
DNA isolation and analysis.
Tetrahymena transformants were propagated in PPYS-paromomycin at 30°C. Untransformed control cells were propagated in PPYS at 30°C. Whole-cell genomic DNA was isolated from 1 ml of overnight cultures by using protocol E from the Wizard genomic DNA purification kit (Promega, Madison, Wis.). DNA preparations from transformed and wild-type Tetrahymena as well as plasmid control DNA preparations were digested with NotI, fractionated by electrophoresis on 0.8% agarose gels, and blotted onto GeneScreen Plus membranes (NEN Life Science Products, Boston, Mass.). Following UV cross-linking and prehybridization, membranes were incubated with the appropriate hybridization probe and washed under high-stringency conditions as previously described (62). Washed membranes were exposed to X-ray film for 12 to 24 h.
The 869-bp Tlr1.L probe used for hybridization to pDTflank-series constructs was generated by PCR amplification with primers Tlr1L:25-47 and Tlr1L:894-872 and pHWT.cam as a template. The 518-bp Cyd1.R probe used for hybridization to pDCflank-series constructs was amplified from pCyd1X-B with primers Cyd1.585-607 and Cyd1.1103-1084. Following amplification, each probe was gel purified and radiolabeled with [α-32P]dATP by random priming (Roche Molecular, Indianapolis, Ind.).
PCR analysis of excision boundaries.
Rearrangement junctions from selected processed constructs were PCR amplified from transformant whole-cell DNA preparations. Each PCR product was generated with a primer from within Tlr1 flanking DNA for Tflank-series constructs or Cyd1X DNA for Cflank-series constructs in conjunction with a polylinker-specific primer. Primers C3 and R2 were used for Tflank.IRL and Tflank amplification reactions, and primers C3 and R3 were used for Tflank.IRR, Tflank.IN2, and Tflank.IN3 (Table 1). Primers C4 and Cyd.168-192 were used for the Cflank.IRL amplification reaction. The resulting PCR products were cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.) or into pGEM-T (Promega). Inserts were sequenced with vector primers or construct primer R3. The Cflank.IN2 rearrangement junction was PCR amplified with primers C4 and Cyd1:1103-1084, and the PCR product was sequenced directly.
RESULTS
The developmentally programmed deletion of the chromosomal Tlr1 element is diagrammed in Fig. 1B. In order to facilitate the description of Tlr1 elimination boundaries, we have devised a coordinate numbering system based on relative proximity to the terminal nucleotides of the Tlr1 inverted repeat. The predominant right boundary for Tlr1 deletion is located 48 bp outside the right end of the terminal inverted repeat (47, 60). In the current study, this elimination boundary is referred to as R+48, where the first nucleotide of the Tlr1 right flanking sequence is R+1 and the terminal right nucleotide of the Tlr1 inverted repeat is R−1. Additional minor rearrangement variants have also been observed at R+102 and R+245 (47, 60). The most common left elimination boundary resides within the left side of the Tlr1 inverted repeat (47, 60). This coordinate is designated L−244, where L−1 refers to the terminal left nucleotide of the Tlr1 element and L+1 refers to the first nucleotide of the Tlr1 left flanking sequence. Consequently, the leftmost 243 bp of the Tlr1 element is typically retained in the macronuclear genome. Less prevalent alternate rearrangements have elimination boundaries within the left side of the inverted repeat at L−59 and in the left flanking sequence at L+40 (47, 60).
The Tlr1 terminal inverted repeat is not required for programmed elimination.
The experiments described in this study were carried out with an in vivo DNA rearrangement assay (67) to monitor the ability of modified Tlr1 constructs to undergo programmed elimination, as described in Materials and Methods. IES-containing micronuclear DNA regions are cloned into an rDNA-based processing vector (pD5H8). When the constructs are introduced into developing macronuclei, the IES undergoes processing which closely mimics that of the endogenous chromosome (Fig. 1). Using the in vivo rearrangement assay, Patil and Karrer (48) previously demonstrated that the WT.cam construct, which consisted of the 825-bp Tlr1 terminal inverted repeat together with 893 bp of normal Tlr1 flanking DNA on the left and 831 bp on the right, underwent accurate and efficient programmed DNA elimination in Tetrahymena. Although that study established that the ∼22-kb Tlr internal region was not required in cis for developmentally programmed Tlr1 elimination, it did not determine whether the long terminal inverted repeat was required for rearrangement.
For many transposable elements, terminal inverted repeats are critical structures for element excision during transposition (4, 13, 35, 55, 57, 58). In addition, long inverted repeats are frequently unstable in prokaryotic and eukaryotic genomes, often leading to DNA deletion and/or recombination (5, 38, 40, 59). To determine whether the terminal inverted repeat is required for elimination of Tlr1, we generated a WT.cam derivative construct from which the entire right side of the Tlr1 inverted repeat was removed. This construct, designated Tflank.IRL, consisted of the leftmost 1,066 bp of the Tlr1 element, including the complete left side of the inverted repeat, within the context of the Tlr1 macronucleus-retained flanking DNA (Fig. 2A and B). Whole-cell DNA preparations from nine independent Tflank.IRL Tetrahymena transformant lines were digested with NotI to liberate the processed Tflank.IRL construct from pD5H8-derived rDNA, and the sizes of the resulting restriction fragments were determined by Southern blot hybridization (Fig. 2B). As expected, a 3.1-kb fragment was detected in the control lane containing NotI-digested pDTflank.IRL input plasmid. In contrast, all NotI fragments detected in Tflank.IRL transformant DNA preparations were less than ∼2.6 kb long, indicating that the construct underwent efficient programmed DNA elimination in vivo. The chromosomal Tlr1 locus was undetectable at the exposures used in this experiment, as judged by the lack of a signal in the negative control lane containing DNA from untransformed Tetrahymena cells.
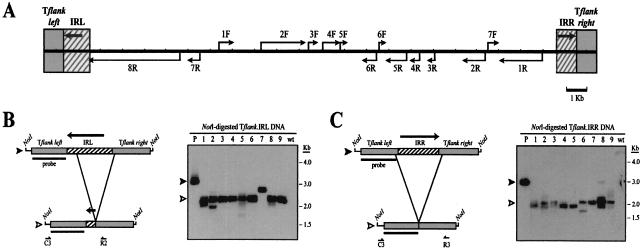
FIG. 2.
Either side of the Tlr1 terminal inverted repeat is sufficient to promote programmed DNA elimination within the context of normal Tlr1 flanking sequences. (A) Composite diagram of the micronuclear Tlr1 element indicating regions used to generate the Tflank.IRL and Tflank.IRR rearrangement constructs. The hatched boxes represent sequences from the terminal regions of the element, and the gray boxes represent Tlr1 flanking sequences. Thick arrows represent the Tlr1 terminal inverted repeat, and thin arrows indicate Tlr open reading frames. (B and C) (Left panels) Tflank.IRL (B) and Tflank.IRR (C) rearrangement constructs (filled arrowheads) and their in vivo processed macronuclear products (open arrowheads). Boxes represent the Tlr1 regions highlighted in panel A, and thin lines represent the vector polylinker included in the NotI fragments. Southern blotting probe Tlr1-L is depicted as a bar. Positions of primers used for PCR amplification of rearrangement junctions are indicated below the processed constructs. (Right panels) Southern blot analysis of Tflank.IRL (B) and Tflank.IRR (C) transformant DNAs probed with Tlr1-L. Lane P contains NotI-digested plasmid DNA, lanes 1 to 9 contain NotI-digested whole-cell DNA from independent Tetrahymena transformant lines, and lane wt contains NotI-digested whole-cell DNA from untransformed Tetrahymena. Hybridization to the chromosomal copy of Tlr1 was not detected at this level of exposure because the rDNA was amplified ∼200-fold.
Eight of the nine Tflank.IRL transformants analyzed contained a predominant 2.3-kb NotI fragment (Fig. 2B), corresponding to the size expected if the normal Tlr1 elimination boundaries were utilized on the construct. Three of the transformants in Fig. 2B had NotI fragments that were different in size from the major fragment. This result was not unexpected and may reflect the variability of the boundaries in the chromosomal deletion (Fig. 1B).
To map the precise deletion boundaries of the most prevalent rearrangement, processed products were PCR amplified from two independent Tflank.IRL transformant lines, and the nucleotide sequence spanning each elimination junction was determined. In order to avoid amplification of the macronuclear Tlr1 locus, the oligonucleotide set used to prime these reactions was specific for the rDNA rearrangement construct (Fig. 2B). In both cell lines, the major right boundary for Tflank.IRL rearrangement was located within 5 bp of the predominant R+48 junction utilized by chromosomal Tlr1. The minor variations in junctional positioning likely were a consequence of the sequence microheterogeneity normally observed at the elimination boundaries of Tlr1 and other Tetrahymena IESs (2, 47, 60). The left elimination boundaries of the products mapped precisely to the L−244 major boundary of chromosomal Tlr1 elimination or 30 bp to the right, at L−274 (Table 2).
TABLE 2.
Sequences of the boundaries of Tlr deletionsa
| Construct | Boundary
|
|
|---|---|---|
| Left | Right | |
| Major chromosomal | AGTTTCTAAAL-244gtttctcacg | tcaagttgccR+48AAAAAAGATT |
| Minor chromosomal | ATATTCTACAL-59taaaattaaa | caagttgccaR+49AAAAAGATTA |
| Minor chromosomal | TTAATTTATAL+40taaattaaaa | aagttgccaaR+50AAAAGATTAA |
| Minor chromosomal | TATATTTTTAL-204tatatttctt | atgtaccctaR+245CTTTCTTTTG |
| Minor chromosomal | TTTTCTCAAGL-236tttctaaagt | aaagcttacgR+102AGGAATCATA |
| Tflank.IRL ln1 | TTTTTAAAATL−274ttctcatttt | ttgccaaaaaR+53AGATTAATTA |
| Tflank.IRL ln2 | AGTTTCTAAAL−244gtttctcacg | ttgccaaaaaR+53AGATTAATTA |
| Tflank.IRR ln3 | ATTTGCTACTL+3cgtgagagaa | gtaccctactR+247TTCTTTTGAA |
| Tflank.IRR ln6 | TTTAAAATAAL+146taaataaatt | aagttgccaaR+50AAAAGATTAA |
| Tflank.IN2 ln4 | CAAAATTATAL+55ttttttaatt | caagttgccaR+49AAAAAGATTA |
| Tflank.IN2 ln6 | AAATTAAAAGL+64caaaattata | gccaaaaaagR+55ATTAATTATC |
| Tflank.IN3 ln2 | AAAATTATATL+53tttttaattt | ttgccaaaaaR+53AGATTAATTA |
| Tflank.IN3 ln4 | TCTTTGATTAL+77tagaaattaa | aagttgccaaR+50AAAAGATTAA |
| Cflank.IRL ln3 | TCTCATTTTAL−304tgaaaagttt | gtttcttttaR+266ATTCAATAGC |
| Cflank.IRL ln3 | AATTTTTAAAL−272atttctcatt | aatagctaaaR+280GTATGAACGT |
| Cflank.IRL ln6 | TTCTAAAGTTL-245tctcacgttt | gtttcttttaR+266ATTCAATAGC |
| Cflank.IN2 ln3 | ACTACTACTAL+206ttaatagtca | agtaaaactaR+134AATCCAGAAC |
Macronucleus-retained sequences are shown in uppercase type, and deleted sequences are shown in lowercase type. Coordinates indicate the position of the last nucleotide deleted. Ambiguous elimination boundaries lying within direct repeats are underlined. In these instances, we designated the nucleotide to the immediate right of the repeat as the boundary.
Taken together, the data collected from Tflank.IRL processing indicate that the left terminus of Tlr1, together with normal flanking sequences, is sufficient to elicit efficient programmed DNA deletion in vivo. Furthermore, the predominant deletion junctions utilized in the processed constructs appear to be accurate to within approximately 30 bp of the elimination boundaries of chromosomal Tlr1. Thus, the long inverted repeat structure does not appear to be required in cis for developmentally programmed deletion of the Tlr1 element.
To determine whether the elimination activity of Tflank.IRL was dependent on the orientation of the Tlr1 terminal sequence, we generated a second WT.cam derivative from which the left side of the inverted repeat was removed. The resulting construct, designated Tflank.IRR, contained the rightmost 854 bp of the Tlr1 element, including the complete right side of the terminal inverted repeat, embedded within the Tlr1 flanking sequences (Fig. 2A and C). Southern blot analysis of in vivo processed Tflank.IRR is shown in Fig. 2C. Like Tflank.IRL, Tflank.IRR consistently underwent efficient programmed DNA elimination, indicating that the Tlr1 right terminus is also sufficient for programmed rearrangement at this site. Sequence analysis of processed constructs from two independent transformant lines revealed that, in each case, the Tlr1 sequence included in Tflank.IRR was completely eliminated (Table 2). This finding is consistent with previous observations that the entire right terminal region of chromosomal Tlr1 is consistently eliminated from the developing macronuclear genome.
As discussed above, the left elimination boundary most frequently observed for chromosomal Tlr1 is located within the left side of the inverted repeat at L−244 (Fig. 1B). Since this region was not included (in its normal orientation) in the Tflank.IRR input construct, the data suggest that the sequences in the vicinity of this preferred left junction are not required for efficient elimination activity. The left elimination boundaries sequenced from Tflank.IRR transformants were located at L+3 and L+146 (Table 2), indicating that sequences within left flanking DNA can serve as junction sites for programmed Tlr1 deletion. Indeed, a left boundary variant for chromosomal Tlr elimination was previously observed at L+40 (Fig. 1B and Table 2). However, DNA from the left side of the inverted repeat may contribute to the overall precision of Tlr1 elimination, since processed Tflank.IRR constructs detected on the Southern blot in Fig. 2C were more heterogeneous in size than processed Tflank.IRL products (Fig. 2B) and chromosomal Tlr1 rearrangements. It should be noted that the right elimination boundaries sequenced from Tflank.IRR transformants were located at R+50 (Table 2), 2 nucleotides outside the predominant right junction for chromosomal Tlr1, and at R+247 (Table 2), 2 nucleotides outside a previously observed Tlr1 right boundary variant (Fig. 1B) (47, 60). Thus, Tflank.IRR rearrangement appears to utilize normal right elimination boundaries.
Tlr1 flanking sequences are insufficient for programmed rearrangement.
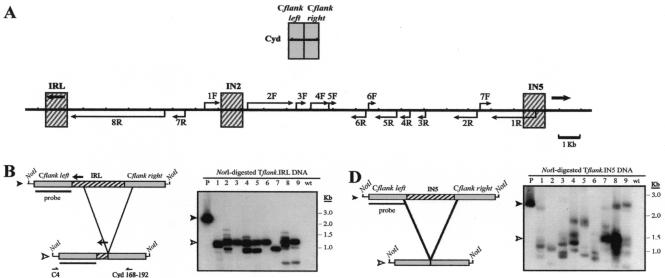
Since most of the Tlr family members identified to date appear to be embedded within long stretches of micronucleus-limited DNA (62), it is possible that programmed deletion of Tlr elements is a consequence of their targeted insertion into regions of the germ line genome that are predestined for elimination. As discussed above, at least 48 bp of flanking DNA is normally deleted from the developing macronucleus along with the Tlr1 element (Fig. 1B). To determine whether elimination of this sequence is dependent on the presence of Tlr1 or whether the DNA adjacent to Tlr1 comprises a small, independent IES, we generated a WT.cam derivative construct from which all Tlr1 DNA was removed, leaving 48 bp of non-Tlr1, micronucleus-limited DNA between the normal flanking sequences (Fig. 3A and B). Southern hybridization analysis of DNA from Tflank transformants revealed that this “empty site” construct did not undergo detectable elimination in vivo (Fig. 3B). Constructs were PCR amplified from five Tetrahymena transformant DNA preparations, and the absence of elimination activity for Tflank was confirmed by direct sequencing of the resulting products (data not shown). These results indicate that Tlr1 flanking sequences alone are insufficient to elicit DNA elimination. Thus, Tlr1 flanking DNA does not appear to contain an autonomous IES.
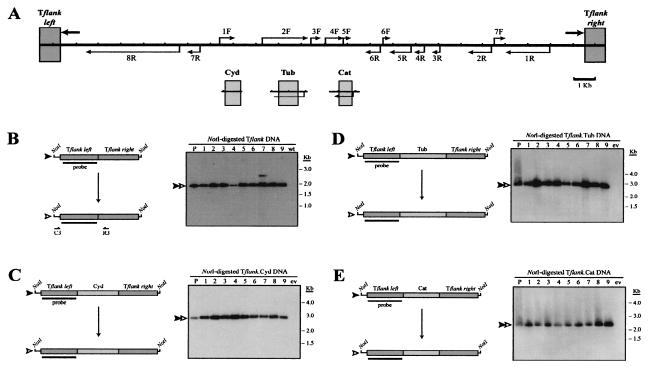
FIG. 3.
Developmentally programmed DNA elimination requires IESs. (A) Composite diagram of the micronuclear Tlr1 element, the T. thermophila Cyd and α-tubulin (Tub) loci, and the Cat gene from E. coli transposon Tn9 indicating the DNA fragments used to generate control constructs. Thick arrows represent the Tlr1 terminal inverted repeat, and thin arrows indicate open reading frames. (B through E) (Left panels) Tflank, Tflank.Cyd, Tflank.Tub, and Tflank.Cat constructs (filled arrowheads) and their in vivo processed macronuclear products (open arrowheads). Boxes represent the DNA regions highlighted in panel A, and thin lines represent the vector polylinker included in the NotI fragments. Southern blotting probe Tlr1-L is depicted as a bar. Positions of primers used for PCR amplification of processed Tflank are indicated below the constructs. (Right panels) Southern blot analysis of Tflank (B), Tflank.Cyd (C), Tflank.Tub (D), and Tflank.Cat (E) transformant DNAs probed with Tlr1-L. Lane P contains NotI-digested plasmid DNA, lanes 1 to 9 contain NotI-digested whole-cell DNA from independent Tetrahymena transformant lines, lane wt contains NotI-digested whole-cell DNA from untransformed Tetrahymena, and lane ev contains NotI-digested whole-cell DNA from Tetrahymena transformed with pD5H8 containing no insert.
Replacement of Tlr1 sequences with macronuclear or exogenous DNA abolishes programmed rearrangement activity.
Since Tflank failed to undergo detectable elimination in vivo, it seems likely that additional DNA is necessary to elicit programmed rearrangement at this site. As described above, the results obtained from Tflank.IRL and Tflank.IRR processing assays clearly demonstrated that either side of the Tlr1 inverted repeat is sufficient to meet this requirement. However, these experiments did not address the question of whether the elimination activity observed for the constructs was a property of the Tlr1 sequences or a general response to nonspecific DNA occupying this site. In order to distinguish between these two possibilities, we assembled chimeric constructs in which non-Tlr DNA fragments were inserted into the “empty” Tlr1 site in Tflank. The non-Tlr sequences included in these constructs consisted of a 936-bp segment of the macronuclear, nongenic Cyd1 locus or a 1,099-bp fragment from within the coding region of the T. thermophila α-tubulin gene. These constructs, designated Tflank.Cyd and Tflank.Tub, respectively, failed to undergo detectable DNA elimination in vivo (Fig. 3C and D).
It was previously shown that the abnormal presence of an IES in the parental macronucleus inhibits the deletion of the IES from the developing macronucleus (11). Thus, it was possible that the failure to eliminate the Cyd and Tub inserts was due to the presence of those sequences in the parental macronucleus. Therefore, a third control construct, Tflank.Cat, containing an 802-bp segment of the chloramphenicol acetyltransferase gene from bacterial transposon Tn9, was created and tested in the rearrangement assay. No deletion from this construct was detectable. These experiments demonstrate that rearrangement activity is not promoted by macronuclear DNA sequences or by foreign DNA placed between Tlr flanking sequences.
Multiple, nonoverlapping segments of the Tlr internal region are capable of stimulating DNA elimination within the context of normal Tlr1 flanking sequences.
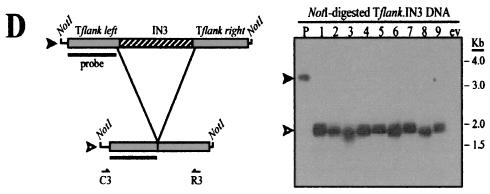
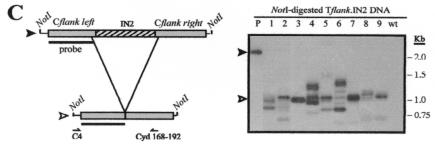
Taken together, the data suggest that DNAs from the left and right terminal regions of Tlr1 provide required signals for programmed deletion of the element that are not present in non-IES DNA. In order to determine whether additional sections of Tlr1 are also capable of inducing elimination, we generated Tflank-based rearrangement constructs that contained 642- to 1,317-bp DNA segments derived from different areas within the highly conserved Tlr internal region. Five arbitrarily selected, nonoverlapping portions from the Tlr inner core, designated IN1 to IN5, were inserted into the empty Tlr1 site in Tflank to give rise to Tflank.IN1 to Tflank.IN5 (Fig. 4). It should be noted that fragments IN1 to IN4 were amplified from Tlr internal clones that were probably not derived from the Tlr1 element. However, given the high degree of nucleotide conservation within the inner core of Tlr family members (21, 62), it is likely that the corresponding regions of Tlr1 contain nearly identical sequences. Southern hybridization analysis revealed that each of the five Tflank.IN constructs underwent efficient programmed elimination in transformed cell lines (Fig. 4B to F).
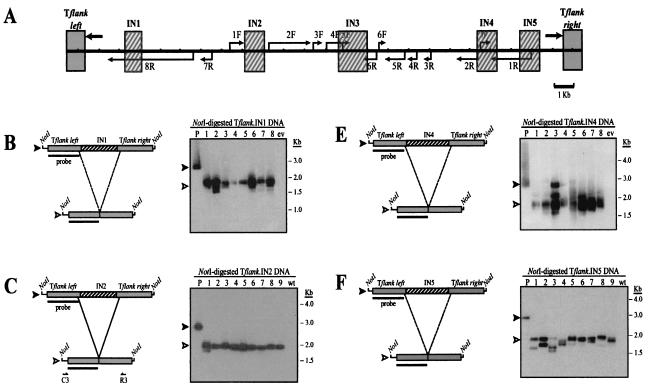
FIG. 4.
Internal fragments of Tlr elements promote programmed DNA elimination within the context of normal Tlr1 flanking DNA. (A) Composite diagram of the micronuclear Tlr1 element showing Tlr flanking sequences (gray boxes) and internal sequences (hatched boxes) used to generate Tflank.IN1 to Tflank.IN5 rearrangement constructs. Thick arrows represent the Tlr1 terminal inverted repeat, and thin arrows indicate Tlr open reading frames. (B through F) (Left panels) Tflank.IN1 to Tflank.IN5 constructs (filled arrowheads) and their in vivo processed macronuclear products (open arrowheads). Boxes represent the DNA regions highlighted in panel A, and thin lines represent the vector polylinker included in the NotI fragments. Southern blotting probe Tlr1-L is depicted as a bar. Positions of primers used for PCR amplification of processed Tflank.IN2 and Tflank.IN3 are indicated below the constructs. Deduced rearrangement activity of Tflank.IN1, Tflank.IN4, and Tflank.IN5 is represented by broken lines. (Right panels) Southern blot analysis of Tflank.IN1 to Tflank.IN5 transformant DNAs probed with Tlr1-L. Lane P contains NotI-digested plasmid DNA, lanes 1 to 8 (B and E) and 1 to 9 (C, D, and F) contain NotI-digested whole-cell DNA from independent Tetrahymena transformant lines, lane wt contains NotI-digested whole-cell DNA from untransformed Tetrahymena, and lane ev contains NotI-digested whole-cell DNA from Tetrahymena transformed with pD5H8 containing no insert.
An ∼2-kb rearranged product was consistently observed in cells transformed with Tflank.IN1 to Tflank.IN5 (Fig. 4B to F), corresponding approximately to the size predicted if the respective Tlr insertion sequence were completely eliminated from each construct. Processed constructs were PCR amplified from two Tflank.IN2 transformants and two Tflank.IN3 transformants. In all four Tflank.IN cell lines examined, the Tlr DNA sequences included in the input constructs were deleted in their entirety (Table 2).
The data indicate that multiple, nonoverlapping interior regions of Tlr elements, as well as the terminal regions of Tlr1, are capable of stimulating efficient DNA deletion from within normal Tlr1 flanking sequences. Since the Tflank.Cyd, Tflank.Tub, and Tflank.Cat constructs discussed above failed to undergo detectable rearrangement, our results strongly suggest that Tlr elements contain redundant signals that target programmed elimination activity. Blastn analysis of the Tlr segments used to generate the Tflank.IN1 to Tflank.IN5 constructs and the Tflank.IRL and Tflank.IRR constructs failed to reveal regions of significant sequence similarity, indicating that the functionally redundant elimination targeting signals within Tlr DNA are highly divergent at the primary nucleotide level.
Tlr sequences induce elimination when inserted into macronuclear DNA that is not normally associated with programmed rearrangement.
The experiments presented thus far have focused on the ability of Tlr segments to undergo programmed elimination within the context of normal Tlr1 flanking sequences. Since Tlr DNA appears to provide the primary targeting signal for the elimination on Tflank-series constructs, we investigated whether the deletion of Tlr sequences required specific flanking sequences. Cyd1 is a nongenic, single-copy locus in the Tetrahymena genome that is not associated with programmed DNA rearrangement, as determined by Southern blot analysis (J. Wuitschick, P. Lindstrom, and K. Karrer, unpublished data). Inspection of the Cyd1 nucleotide sequence indicates that it is approximately 25% G+C and has little protein-coding potential. Tlr fragments IRL, IN2, and IN5 described above were inserted into an ∼1.2-kb HindIII fragment of the Cyd1 locus to give rise to the chimeric constructs Cflank.IRL, Cflank.IN2, and Cflank.IN5, respectively (Fig. 5). Southern hybridization analysis of whole-cell DNA preparations from Tetrahymena transformed with these constructs revealed that each of the Tlr fragments underwent programmed DNA elimination from within the Cyd1 flanking sequences (Fig. 5B to D). Control constructs comprised of the Tub and Cat fragments flanked by Cyd1 DNA or the Cyd1 HindIII fragment with no insert did not undergo detectable rearrangement in vivo (data not shown), indicating that elimination activity was targeted to Cflank.IRL, Cflank.IN2, and Cflank.IN5 by Tlr sequences.
FIG. 5.
Tlr DNA is eliminated from within flanking sequences that are not normally associated with DNA rearrangement. (A) Diagram of the Tetrahymena Cyd1 locus and a composite of the Tlr1 element. Boxed areas indicate regions included in the Cflank.IRL, Cflank.IN2, and Cflank.IN5 constructs. Thick arrows represent the Tlr1 terminal inverted repeat, and thin arrows indicate open reading frames. (B through D) (Left panels) Cflank.IRL, Cflank.IN2, and Cflank.IN5 constructs (filled arrowheads) and their in vivo processed macronuclear products (open arrowheads). Boxes represent the regions highlighted in panel A, and thin lines represent the vector polylinker included in the NotI fragments. Southern blotting probe Cyd1.R is depicted as a bar. Positions of primers used for PCR amplification of processed Cflank.IRL and Cflank.IN2 are indicated below the constructs. Deduced rearrangement activity of Cflank.IN5 is represented by thick lines. (Right panels) Southern blot analysis of Cflank.IRL, Cflank.IN2, and Cflank.IN5 transformant DNAs. Lane P contains NotI-digested plasmid DNA, lanes 1 to 9 contain NotI-digested whole-cell DNA from independent Tetrahymena transformant lines, and lane wt contains NotI-digested whole-cell DNA from untransformed Tetrahymena.
A prominent ∼1.3-kb rearranged product was detected in eight of the nine Cflank.IRL transformant lines analyzed. Sequence analysis of processed constructs from three of these lines revealed that, in each case, the left elimination boundary was located inside IRL, within 60 bp of the major L−244 boundary used for chromosomal Tlr1 rearrangement (Table 2). This result suggests that the left side of the Tlr1 inverted repeat contains sequences capable of specifying the location of a Tlr1 elimination boundary. Right elimination boundaries in these constructs were located 266 to 280 bp to the right of the IRL insert, within Cyd1 DNA (Table 2).
The Cflank.IN2 and Cflank.IN5 processed products detected on Southern blots in Fig. 5C and D were considerably more heterogeneous in size than the Cflank.IRL products or the Tflank.IN2 and Tflank.IN5 products. However, most of the Cflank.IN2 transformants produced a NotI fragment of ∼1.0 to 1.2 kb. Since PCR-amplified junction fragments were unstable in bacterial vectors, the PCR fragment produced by amplification of DNA from the transformant in Fig. 5C, lane 3, was sequenced directly. The results showed that the IN2 fragment was deleted along with 134 bp of Cyd flanking DNA from the left and 206 bp from the right.
Notably, Cflank.IN2 and Cflank.IN5 were the only constructs studied here that did not contain at least one normal junction site used in programmed deletion of chromosomal Tlr1. Thus, specific flanking sequences are not absolutely required for the elimination of Tlr1. However, the greater variability observed in these rearrangements might be attributed to the lack of regular elimination boundaries. In contrast, the relative consistency in the sizes of Cflank.IRL rearrangement products (Fig. 5B) suggests that, while not required for the initiation of DNA elimination, sequences from the vicinity of one regular elimination boundary can add considerable precision to a programmed DNA rearrangement event in Tetrahymena.
DISCUSSION
Tlr elements, along with thousands of other diverse DNA segments, are reproducibly eliminated from the differentiating somatic macronucleus of Tetrahymena. In this report, we examined the internal sequence requirements for developmentally regulated deletion of the Tlr1 element. Seven nonoverlapping Tlr segments were shown to be capable of undergoing efficient programmed elimination within the context of normal Tlr flanking sequences. In addition, three of these (all of the fragments tested) underwent elimination when inserted into DNA that is not normally associated with genomic rearrangement in Tetrahymena. Since the complete removal of element sequences abolished DNA elimination activity in constructs, as did replacement with macronucleus-retained DNA or Escherichia coli transposon Tn9 DNA, our findings provide strong evidence that Tlr DNA contains redundant cis-acting signals that target the elements for programmed elimination from the differentiating macronuclear genome.
The Tlr fragments analyzed in vivo were derived from widely separated regions of element family members, including the left and right termini of Tlr1 as well as five arbitrarily selected segments of the 22-kb inner core. Detailed comparisons of these sequences failed to reveal common structural features or obvious regions of shared nucleotide similarity, suggesting that the redundant internal signals involved in Tlr1 rearrangement may be highly divergent at the primary sequence level. In view of the fact that each of the different Tlr DNA fragments analyzed in this study elicited efficient elimination activity, we find it probable that elimination targeting signals are distributed throughout most, or perhaps all, regions of this family of putative mobile genetic elements.
Tlr elements may not be the only micronucleus-limited sequences that contain internally redundant elimination targeting signals. Preliminary data obtained from construct-based deletion analyses of the M element suggest that this well-studied 0.6- to 0.9-kb IES is composed of multiple internal signals that act independently of one another to promote programmed DNA elimination (reviewed in reference 14). It should be noted that the M element bears no detectable sequence or structural similarities to Tlr elements. Thus, functionally redundant elimination targeting signals within micronucleus-limited regions may be a common characteristic of diverse Tetrahymena IESs.
How are the elimination targeting signals within Tlr elements and other Tetrahymena IESs recognized? All known micronucleus-limited sequences are strikingly A+T rich (77 to 86% A+T); however, their A+T content does not differ appreciably from that of nongenic, macronucleus-retained DNA (63). Thus, an IES recognition system based solely on nucleotide composition seems unlikely.
A compelling model to account for the specific recognition of the seemingly disparate internal signal sequences within IESs involves the participation of RNA intermediates. Mochizuki et al. (44) recently demonstrated that ∼26 to 31 nucleotide RNAs accumulate in Tetrahymena cells exclusively during the developmental stages preceding DNA elimination. These RNA molecules preferentially hybridize to micronuclear DNA on genomic Southern blots, suggesting that they are homologous to IES regions. It has been suggested that the small RNA molecules might provide sequence specificity for DNA elimination by guiding heterochromatin factors, such as Pdd1p and Pdd3p, to complementary regions of the developing macronuclear genome. Thus, the elimination targeting signals within Tlr DNA might be pairing substrates for small RNAs. This hypothesis is consistent with the findings that mammalian heterochromatin protein 1, which contains a chromodomain and binds K9-MeH3, like Pdd1p (36), requires an RNA component for its localization to heterochromatin regions in vivo (42).
In eukaryotes, small RNA molecules can be generated by RNAi, a gene-silencing phenomenon that functions through the fragmentation of double-stranded RNA molecules (reviewed in references 25 and 69). There is strong evidence to suggest that the small RNAs implicated in DNA elimination are products of a Tetrahymena RNAi-like pathway involving TWI1, a piwi-related gene of the argonaute family. TWI1-knockout cells fail to accumulate small RNAs during macronuclear development and exhibit severe defects in programmed DNA elimination (44). Interestingly, argonaute and other components of the RNAi machinery were recently demonstrated to be critical for heterochromatin formation in the fission yeast Schizosaccharomyces pombe (24, 56). Nongenic, bidirectional transcription of Tetrahymena IES regions is known to occur during early macronuclear development (10). Although it is currently unclear how this transcription is induced and whether it is restricted to micronucleus-limited regions of the genome, the transcriptional activity appears to be an important initiating event for programmed DNA elimination, since a brief pulse with the RNA polymerase inhibitor actinomycin D perturbs IES excision. It has been proposed (44) that double-stranded RNAs formed from bidirectional IES transcripts might be primary targets of the Tetrahymena RNAi machinery and thus the initial source of the small RNA molecules implicated in the elimination process.
In Tetrahymena, there appears to be a strong correlation between sequence copy number and DNA elimination. Tlr elements and, consequently, each of the Tlr segments included in our in vivo rearrangement constructs, are moderately repetitive in the Tetrahymena genome. Similarly, most other IES regions described to date have been demonstrated by Southern blot analyses to belong to families of multicopy sequences (1, 12, 28, 29, 32, 34, 53, 60, 61, 64). In most cases, all copies of these repeated sequences are eliminated from the differentiating macronucleus. Furthermore, the mature macronuclear genome is largely depleted of repetitive sequences (28, 32, 68). In view of these observations, we propose that the repeated character of micronucleus-limited sequences may play an important role in promoting their efficient programmed elimination. It is currently unknown whether the nongenic, bidirectional transcripts identified by Chalker and Yao (10) are specific for the IES or whether the entire genome is transcribed, as suggested in the model of Mochizuki et al. (44). If the transcription is specific, the repetition of a sequence may induce its transcription by an unknown mechanism. If the entire micronuclear genome is transcribed, repetition of a sequence may raise the concentration of that sequence above a level required for its function as a scan RNA. It is important to note that multicopy sequences are thought to be the primary targets of RNAi pathways in several organisms (reviewed in references 25 and 69); therefore, a relationship between sequence repetition and RNAi may not be unique to Tetrahymena.
Some characterized IESs in Tetrahymena appear to be single copy, as determined by Southern blot analysis (26). Thus, sequence repetition may not be an absolute requirement for programmed DNA elimination. Alternatively, these apparent single-copy IESs may contain redundant sequence elements that are too short to be detected at conventional hybridization stringencies. For example, we recently isolated a previously uncharacterized micronucleus-limited DNA region that appeared to be a single-copy sequence by Southern hybridization but was shown by sequence analysis to share an ∼75-bp block of significant nucleotide similarity with a known Tetrahymena IES (J. D. Wuitschick and K. M. Karrer, unpublished data). We predict that a larger database of germ line-limited DNA would reveal additional examples of short, multicopy sequence blocks located within “unique” IESs.
Programmed deletion of repeated sequences would, in theory, provide an effective surveillance mechanism to rid the transcriptionally active macronuclear genome of potentially harmful transposons and viruses. Yao (65) has pointed out that at least some germ line-limited sequences may play functional roles in genetic processes, such as mitosis, that are restricted to the micronucleus. Whereas the micronucleus undergoes conventional mitosis, the macronucleus is highly unusual among eukaryotic nuclei in that it divides through an amitotic process that does not involve chromosome segregation or condensation (19). Thus, centromeric DNA and matrix or scaffold attachment regions are predicted to be dispensable in the macronucleus (14). Although neither class of sequence has yet been characterized in Tetrahymena, it seems likely, based on analogies from other eukaryotic systems, that these regions of the micronuclear genome may belong to families of repeated DNA sequences (8, 16, 27, 50, 51, 54). Since it is apparently unnecessary for the macronucleus to retain these sequences, the deletion of repeated sequences that are essential in diploid organisms would be allowed in the Tetrahymena macronucleus. This activity might facilitate the evolution of a system for recognizing and eliminating repeated sequences.
In addition to the apparent elimination targeting sequences located within micronucleus-limited DNA, macronucleus-destined sequences flanking IESs are also thought to play a role in programmed DNA rearrangement. These flanking sequences have been demonstrated to contain cis-acting signals involved in delineating the boundaries of several T. thermophila IESs, including Tlr1 (22, 23, 39). Although programmed elimination of the chromosomal M, R, and mse2.9 elements is 100% efficient, deletion of these low-copy-number elements from processing vectors is less efficient. These IESs appear to be dependent on cis-acting signals in the flanking DNA, since engineered deletions or mutations in these flanking sequences have been demonstrated to diminish or completely abolish construct-based elimination activity. Nonetheless, it has been shown that flanking sequences from different IESs can substitute for one another in construct rearrangements (14, 18; Wuitschick and Karrer, unpublished). For multicopy Tlr DNA, which undergoes efficient elimination from the chromosome (62) and robust elimination from processing vectors, there appears to be no requirement for specific cis-acting sequences in the flanking DNA. Each of the three different Tlr fragments analyzed underwent efficient, albeit variable, elimination from between flanking sequences that are not associated with chromosomal rearrangements.
We suggest a model to account for these observations that is consistent with previous studies on the role of flanking sequences in DNA rearrangement. We propose that the specialized chromatin structure that contains the Pdd proteins and probably Twi1p complexes is nucleated by the IES. This heterochromatin spreads until it reaches sequences that halt or slow the assembly of the specialized chromatin. These are the cis-acting sequences that define the boundary of the IES. They probably do not require the binding of specific proteins for their function but may be particularly strong nucleosome positioning sequences, sequences that produce a bend in the DNA, and so forth. Various DNA fragments are expected to contain sequences that serve this function better than others. The variability in processing of the Cflank.IN2 and Cflank.IN5 constructs suggests that the Cyd sequences are relatively weak delineators of rearrangement boundaries. Thus, the spread of the specialized chromatin in these constructs is highly variable, with a correspondingly high level of heterogeneity in the deletion boundaries.
The mechanism by which Tlr elements have proliferated within the micronuclear genome is currently unclear. However, the ability of Tlr sequences to target their own programmed elimination in the absence of specific flanking DNA has important implications for the notion that programmed DNA rearrangement has the selective advantage of removing transposons and/or viruses from the somatic genome. It suggests that the transposition of Tlr family members to different germ line chromosomal loci would not prevent their subsequent elimination from the developing macronucleus.
Acknowledgments
We thank Janet Partridge for critically reviewing the manuscript.
This work was supported by grant MCB-9974885 from the National Science Foundation to K.M.K. J.D.W. was supported by fellowships from the Arthur J. Schmitt Foundation, the Richard W. Jobling Trust Fund, and the Marquette University Graduate School.
REFERENCES
- 1.Austerberry, C. F., C. D. Allis, and M. C. Yao. 1984. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl. Acad. Sci. USA 81:7383-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austerberry, C. F., R. O. Snyder, and M. C. Yao. 1989. Sequence microheterogeneity is generated at junctions of programmed DNA deletions in Tetrahymena thermophila. Nucleic Acids Res. 17:7263-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Benito, M. I., and V. Walbot. 1997. Characterization of the maize mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17:5165-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi, X., and L. F. Liu. 1996. DNA rearrangement mediated by inverted repeats. Proc. Natl. Acad. Sci. USA 93:819-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunk, C. F., and R. K. Conover. 1985. Elimination of micronucleus-specific DNA sequences early in anlagen development. Mol. Cell. Biol. 5:93-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruns, P. J., A. L. Katzen, L. Martin, and E. H. Blackburn. 1985. A drug-resistant mutation in the ribosomal DNA of Tetrahymena. Proc. Natl. Acad. Sci. USA 82:2844-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cambareri, E. B., R. Aisner, and J. Carbon. 1998. Structure of the chromosome VII centromere region in Neurospora crassa: degenerate transposons and simple repeats. Mol. Cell. Biol. 18:5465-5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker, D. L., A. La Terza, A. Wilson, C. D. Kroenke, and M. C. Yao. 1999. Flanking regulatory sequences of the Tetrahymena R deletion element determine the boundaries of DNA rearrangement. Mol. Cell. Biol. 19:5631-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chalker, D. L., and M. C. Yao. 2001. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 15:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalker, D. L., and M. C. Yao. 1996. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 16:3658-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherry, J. M., and E. H. Blackburn. 1985. The internally located telomeric sequences in the germ-line chromosomes of Tetrahymena are at the ends of transposon-like elements. Cell 43:747-758. [DOI] [PubMed] [Google Scholar]
- 13.Colloms, S. D., H. G. van Luenen, and R. H. Plasterk. 1994. DNA binding activities of the Caenorhabditis elegans Tc3 transposase. Nucleic Acids Res. 22:5548-5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coyne, R. S., D. L. Chalker, and M. C. Yao. 1996. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 30:557-578. [DOI] [PubMed] [Google Scholar]
- 15.Coyne, R. S., M. A. Nikiforov, J. F. Smothers, C. D. Allis, and M. C. Yao. 1999. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell 4:865-872. [DOI] [PubMed] [Google Scholar]
- 16.Craig, J. M., W. C. Earnshaw, and P. Vagnarelli. 1999. Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp. Cell Res. 246:249-262. [DOI] [PubMed] [Google Scholar]
- 17.Duharcourt, S., and M. C. Yao. 2002. Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell 1:293-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fillingham, J. S., D. Bruno, and R. E. Pearlman. 2001. Cis-acting requirements in flanking DNA for the programmed elimination of mse2.9: a common mechanism for deletion of internal eliminated sequences from the developing macronucleus of Tetrahymena thermophila. Nucleic Acids Res. 29:488-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frankel, J. 2000. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 62:27-125. [DOI] [PubMed] [Google Scholar]
- 20.Gaertig, J., and M. A. Gorovsky. 1992. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc. Natl. Acad. Sci. USA 89:9196-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gershan, J. A., and K. M. Karrer. 2000. A family of developmentally excised DNA elements in Tetrahymena is under selective pressure to maintain an open reading frame encoding an integrase-like protein. Nucleic Acids Res. 28:4105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Godiska, R., C. James, and M. C. Yao. 1993. A distant 10-bp sequence specifies the boundaries of a programmed DNA deletion in Tetrahymena. Genes Dev. 7:2357-2365. [DOI] [PubMed] [Google Scholar]
- 23.Godiska, R., and M. C. Yao. 1990. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell 61:1237-1246. [DOI] [PubMed] [Google Scholar]
- 24.Hall, I. M., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewa. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 25.Hannon, G. J. 2002. RNA interference. Nature 418:244-251. [DOI] [PubMed] [Google Scholar]
- 26.Heinonen, T. Y., and R. E. Pearlman. 1994. A germ line-specific sequence element in an intron in Tetrahymena thermophila. J. Biol. Chem. 269:17428-17433. [PubMed] [Google Scholar]
- 27.Heslop-Harrison, J. S., M. Murata, Y. Ogura, T. Schwarzacher, and F. Motoyoshi. 1999. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell 11:31-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howard, E. A., and E. H. Blackburn. 1985. Reproducible and variable genomic rearrangements occur in the developing somatic nucleus of the ciliate Tetrahymena thermophila. Mol. Cell. Biol. 5:2039-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huvos, P. E., M. Wu, and M. A. Gorovsky. 1998. A developmentally eliminated sequence in the flanking region of the histone H1 gene in Tetrahymena thermophila contains short repeats. J. Eukaryot. Microbiol. 45:189-197. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahn, C. L., and L. A. Klobutcher. 2002. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 56:489-520. [DOI] [PubMed] [Google Scholar]
- 32.Karrer, K. M. 1983. Germ line-specific DNA sequences are present on all five micronuclear chromosomes in Tetrahymena thermophila. Mol. Cell. Biol. 3:1909-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karrer, K. M. 2000. Tetrahymena genetics: two nuclei are better than one. Methods Cell Biol. 62:127-186. [DOI] [PubMed] [Google Scholar]
- 34.Katoh, M., M. Hirono, T. Takemasa, M. Kimura, and Y. Watanabe. 1993. A micronucleus-specific sequence exists in the 5′-upstream region of calmodulin gene in Tetrahymena thermophila. Nucleic Acids Res. 21:2409-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleckner, N., R. M. Chalmers, D. Kwon, J. Sakai, and S. Bolland. 1996. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr. Top. Microbiol. Immunol. 204:49-82. [DOI] [PubMed] [Google Scholar]
- 36.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 37.Larson, D. D., E. H. Blackburn, P. C. Yaeger, and E. Orias. 1986. Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell 47:229-240. [DOI] [PubMed] [Google Scholar]
- 38.Lewis, S., E. Akgun, and M. Jasin. 1999. Palindromic DNA and genome stability. Further studies. Ann. N. Y. Acad. Sci. 870:45-57. [DOI] [PubMed] [Google Scholar]
- 39.Li, J., and R. E. Pearlman. 1996. Programmed DNA rearrangement from an intron during nuclear development in Tetrahymena thermophila: molecular analysis and identification of potential cis-acting sequences. Nucleic Acids Res. 24:1943-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, C. T., W. H. Lin, Y. L. Lyu, and J. Whang-Peng. 2001. Inverted repeats as genetic elements for promoting DNA inverted duplication: implications in gene amplification. Nucleic Acids Res. 29:3529-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madireddi, M. T., R. S. Coyne, J. F. Smothers, K. M. Mickey, M. C. Yao, and C. D. Allis. 1996. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87:75-84. [DOI] [PubMed] [Google Scholar]
- 42.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 43.Martindale, D. W., C. D. Allis, and P. J. Bruns. 1982. Conjugation in Tetrahymena thermophila. A temporal analysis of cytological stages. Exp. Cell Res. 140:227-236. [DOI] [PubMed] [Google Scholar]
- 44.Mochizuki, K., N. A. Fine, T. Fujisawa, and M. A. Gorovsky. 2002. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110:689-699. [DOI] [PubMed] [Google Scholar]
- 45.Nikiforov, M. A., M. A. Gorovsky, and C. D. Allis. 2000. A novel chromodomain protein, Pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol. 20:4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Partridge, J. F., K. S. Scott, A. J. Bannister, T. Kouzarides, and R. C. Allshire. 2002. cis-Acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 12:1652-1660. [DOI] [PubMed] [Google Scholar]
- 47.Patil, N. S., P. M. Hempen, R. A. Udani, and K. M. Karrer. 1997. Alternate junctions and microheterogeneity of Tlr1, a developmentally regulated DNA rearrangement in Tetrahymena thermophila. J. Eukaryot. Microbiol. 44:518-522. [DOI] [PubMed] [Google Scholar]
- 48.Patil, N. S., and K. M. Karrer. 2000. A developmentally regulated deletion element with long terminal repeats has cis-acting sequences in the flanking DNA. Nucleic Acids Res. 28:1465-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seifert, H. S., E. Y. Chen, M. So, and F. Heffron. 1986. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 83:735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh, L., S. G. Panicker, R. Nagaraj, and K. C. Majumdar. 1994. Banded krait minor-satellite (Bkm)-associated Y chromosome-specific repetitive DNA in mouse. Nucleic Acids Res. 22:2289-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, X., J. Wahlstrom, and G. Karpen. 1997. Molecular structure of a functional Drosophila centromere. Cell 91:1007-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 53.Tsao, N. N., S. G. Tsao, and R. E. Pearlman. 1992. A micronucleus-limited sequence family in Tetrahymena thermophila: organization and sequence conservation. Dev. Genet. 13:75-79. [DOI] [PubMed] [Google Scholar]
- 54.van Drunen, C. M., R. G. Sewalt, R. W. Oosterling, P. J. Weisbeek, S. C. Smeekens, and R. van Driel. 1999. A bipartite sequence element associated with matrix/scaffold attachment regions. Nucleic Acids Res. 27:2924-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Luenen, H. G., S. D. Colloms, and R. H. Plasterk. 1994. The mechanism of transposition of Tc3 in C. elegans. Cell 79:293-301. [DOI] [PubMed] [Google Scholar]
- 56.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 57.Vos, J. C., and R. H. Plasterk. 1994. Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J. 13:6125-6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vos, J. C., H. G. van Luenen, and R. H. Plasterk. 1993. Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev. 7:1244-1253. [DOI] [PubMed] [Google Scholar]
- 59.Waldman, A. S., H. Tran, E. C. Goldsmith, and M. A. Resnick. 1999. Long inverted repeats are an at-risk motif for recombination in mammalian cells. Genetics 153:1873-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wells, J. M., J. L. Ellingson, D. M. Catt, P. J. Berger, and K. M. Karrer. 1994. A small family of elements with long inverted repeats is located near sites of developmentally regulated DNA rearrangement in Tetrahymena thermophila. Mol. Cell. Biol. 14:5939-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White, T. C., M. R. el-Gewely, and S. L. Allen. 1985. Eliminated sequences with different copy numbers clustered in the micronuclear genome of Tetrahymena thermophila. Mol. Gen. Genet. 201:65-75. [DOI] [PubMed] [Google Scholar]
- 62.Wuitschick, J. D., J. A. Gershan, A. J. Lochowicz, S. Li, and K. M. Karrer. 2002. A novel family of mobile genetic elements is limited to the germline genome in Tetrahymena thermophila. Nucleic Acids Res. 30:2524-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wuitschick, J. D., and K. M. Karrer. 1999. Analysis of genomic G + C content, codon usage, initiator codon context and translation termination sites in Tetrahymena thermophila. J. Eukaryot. Microbiol. 46:239-247. [DOI] [PubMed] [Google Scholar]
- 64.Yao, M. C. 1982. Elimination of specific DNA sequences from the somatic nucleus of the ciliate Tetrahymena. J. Cell Biol. 92:783-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao, M. C. 1996. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet. 12:26-30. [DOI] [PubMed] [Google Scholar]
- 66.Yao, M. C., and M. A. Gorovsky. 1974. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma 48:1-18. [DOI] [PubMed] [Google Scholar]
- 67.Yao, M. C., and C. H. Yao. 1989. Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol. Cell. Biol. 9:1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yokoyama, R. W., and M. C. Yao. 1982. Elimination of DNA sequences during macronuclear differentiation in Tetrahymena thermophila, as detected by in situ hybridization. Chromosoma 85:11-22. [DOI] [PubMed] [Google Scholar]
- 69.Zamore, P. D. 2001. RNA interference: listening to the sound of silence. Nat. Struct. Biol. 8:746-750. [DOI] [PubMed] [Google Scholar]