Abstract
Amino acids are not only fundamental protein constituents but also serve as precursors for many essential plant metabolites. Although amino acid biosynthetic pathways in plants have been identified, pathway regulation, catabolism, and downstream metabolite partitioning remain relatively uninvestigated. Conversion of Thr to Gly and acetaldehyde by Thr aldolase (EC 4.1.2.5) was only recently shown to play a role in plant amino acid metabolism. Whereas one Arabidopsis thaliana Thr aldolase (THA1) is expressed primarily in seeds and seedlings, the other (THA2) is expressed in vascular tissue throughout the plant. Metabolite profiling of tha1 mutants identified a >50-fold increase in the seed Thr content, a 50% decrease in seedling Gly content, and few other significant metabolic changes. By contrast, homozygous tha2 mutations cause a lethal albino phenotype. Rescue of tha2 mutants and tha1 tha2 double mutants by overproduction of feedback-insensitive Thr deaminase (OMR1) shows that Gly formation by THA1 and THA2 is not essential in Arabidopsis. Seed-specific expression of feedback-insensitive Thr deaminase in both tha1 and tha2 Thr aldolase mutants greatly increases seed Ile content, suggesting that these two Thr catabolic enzymes compete for a common substrate pool.
INTRODUCTION
Animals rely on plants as dietary sources of amino acids that they cannot synthesize themselves. However, some essential amino acids are present at growth-limiting levels in the world's major field crops, including maize (Zea mays) (Lys, Trp, and Met), wheat (Triticum aestivum) (Lys), rice (Oryza sativa) (Lys, Ile, and Thr), soybean (Glycine max) (Met and Thr), and potato (Solanum tuberosum) (Ile, Met, and Cys). Four of these amino acids (Thr, Ile, Lys, and Met) are synthesized from Asp via a branched pathway with well-studied regulation by feedback inhibition and substrate competition (Coruzzi and Last, 2000). Targeted manipulation of this biosynthetic pathway has been used to increase the levels of Asp-derived amino acids (Galili et al., 2005) and may represent a way to improve the nutritional quality of crop plants. Although amino acid catabolism in plants remains relatively uninvestigated, altered regulation of amino acid breakdown is also an attractive target for plant metabolic engineering. For instance, inhibition of Lys ketoglutarate reductase, combined with upregulation of biosynthesis, greatly increases seed Lys levels (Zhu et al., 2001; Zhu and Galili, 2003).
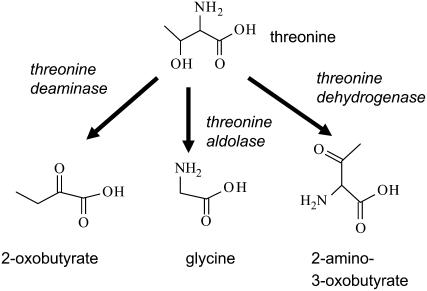
Three pathways for Thr catabolism are illustrated in Figure 1. Thr deaminase has been studied in plants as the committing enzyme leading to Ile synthesis (Wessel et al., 2000; Halgand et al., 2002; Garcia and Mourad, 2004). Thr dehydrogenase catalyzes Thr breakdown in animals and microbes (Epperly and Dekker, 1991; Edgar, 2002), but this enzymatic activity has not yet been confirmed in plants. Thr aldolase (EC 4.1.2.5), which catalyzes the reversible reaction Thr ↔ Gly + acetaldehyde, has been investigated in rats, yeast, and bacteria, where the reaction proceeds toward Gly + acetaldehyde (Monschau et al., 1997; Liu et al., 1998a, 1998b; House et al., 2001).
Figure 1.
Three Pathways for Thr Degradation.
Relatively little research has been done on Thr aldolase in plants, but it is likely that this plant enzyme also functions in Thr catabolism. In [α-13C]Gly labeling experiments of whole Arabidopsis thaliana, only labeled Gly and Ser accumulated, indicating that Gly is not converted directly to Thr by Thr aldolase in vivo (Prabhu et al., 1996, 1998). Metabolite profiling of Medicago truncatula cell cultures showed that the production of Gly from Thr, likely by Thr aldolase, is induced by treatment with methyl jasmonate (Broeckling et al., 2005). Seed Thr content was increased by the Arabidopsis tha1-1 Thr aldolase missense mutation (Jander et al., 2004). By contrast, overproduction of yeast Thr aldolase in Arabidopsis decreased Ile levels and increased Met levels, suggesting not only substrate competition between Thr aldolase and Thr deaminase but also upregulation of the Asp-derived amino acid pathway (Fernie et al., 2004).
l-allo-Thr, an isomer of l-Thr that has been detected in Arabidopsis metabolite profiling experiments (Fiehn et al., 2000), could also be a substrate for Thr aldolase. Both l-Thr and l-allo-Thr are substrates for low-specificity Thr aldolases found in yeast and bacteria (Liu et al., 1997, 1998a). Proteins purified from maize and mung bean (Vigna radiata) produced Gly from l-allo-Thr and l-Ser but not l-Thr (Masuda et al., 1980, 1982). However, this may indicate Ser hydroxymethyltransferase (SHMT), rather than Thr aldolase, activity. Similarly, purified SHMTs from rat, rabbit, human, and Eschericia coli cleave not only l-Ser but also l-allo-Thr in preference to l-Thr (Ogawa et al., 2000; Contestabile et al., 2001).
Bioinformatic analysis of the Arabidopsis genome shows two likely Thr aldolase genes with 67% amino acid sequence identity: THA1 and THA2. In previous work, we demonstrated that THA1 has Thr aldolase activity in vitro (Jander et al., 2004). Here, we confirm that THA2 is also a Thr aldolase. Furthermore, we use insertional mutations and promoter fusions to study the function of both Arabidopsis Thr aldolases and investigate their possible use in plant metabolic engineering.
RESULTS
Arabidopsis THA2 Is a Thr Aldolase
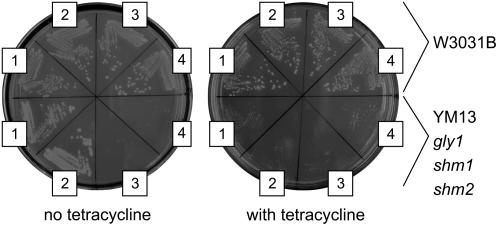
To confirm that THA2 encodes a Thr aldolase, a full-length cDNA was cloned into the tetracycline-repressible yeast vector pCM185. This plasmid was used to transform haploid yeast strains W3031B (control) and YM13 (gly1 shm1 shm2). YM13 is a Gly auxotroph due to mutations in Thr aldolase and both yeast SHMT genes (McNeil et al., 1994; Monschau et al., 1997). As in the case of THA1 (Jander et al., 2004), transformation with THA2 rescued the YM13 Gly auxotrophy. However, this does not occur when transgene expression was repressed with tetracycline (Figure 2).
Figure 2.
Cloned Arabidopsis THA1 and THA2 Thr Aldolases Relieve the Auxotrophy of Yeast Strain YM13 on Plates without Gly.
The top half of each plate is the W3031B wild-type GLY1;SHM1;SHM2 haploid strain; the bottom half is the isogenic Gly auxotrophic strain YM13 gly1;shm1;shm2. At the right, promoter expression is repressed with 10 μg/mL tetracycline. 1, THA1 clone; 2, THA2 clone; 3, pCM185 vector control; 4, no plasmid control.
In vitro assays with THA2 Thr aldolase that was partially purified from the transformed YM13 yeast strain were used to confirm the enzymatic activity. THA2-containing yeast extract was incubated with concentrations of l-Thr ranging from 1 to 50 mM, and the Thr aldolase activity was assayed as the production of acetaldehyde over time (Paz et al., 1965; Liu et al., 1998a). From these data we calculated that the apparent Km of THA2 for l-Thr is 3.8 ± 0.8 mM (mean ± sd of three independent measurements; see Supplemental Figure 1 online). This value is similar to those calculated for Arabidopsis THA1 (7.1 mM) and yeast Thr aldolase (4.3 mM) (Monschau et al., 1997; Jander et al., 2004).
Mutations in THA1 Have Recessive, Seed-Specific Effects on Thr
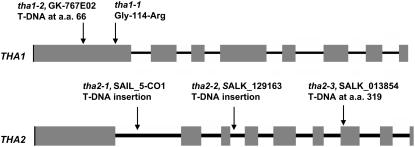
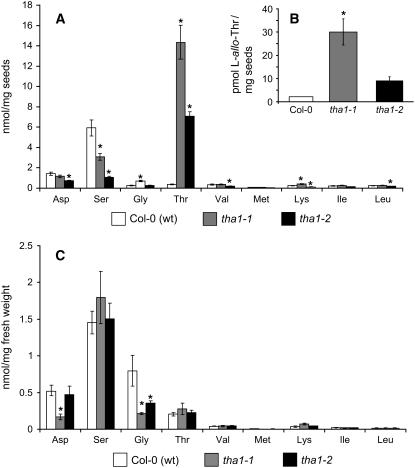
Although we were not able to detect tha1-1 enzymatic activity in vitro (Jander et al., 2004), partial activity in this missense mutant is suggested by the fact that yeast strain YM13 expressing tha1-1 grows at low (50 μM) Gly concentration, whereas untransformed YM13 does not (see Supplemental Figure 2 online). Therefore, we obtained line GK-767E02 (tha1-2) (Rosso et al., 2003), which carries a T-DNA insertion that truncates THA1 at amino acid 66 (Figure 3; see Supplemental Table 1 and Supplemental Figure 3A online). There was no THA1 mRNA detectable by RT-PCR of total RNA isolated from leaves and flowers of homozygous tha1-2 mutant lines, indicating a likely knockout mutation. THA2 transcription in these tissues was unaffected by the tha1-2 mutation (see Supplemental Figure 3A online). Like homozygous tha1-1 (see Supplemental Figure 3B online), the tha1-2 mutation causes a large increase in seed Thr content and relatively small decreases in most other seed amino acids (Figure 4A). Both complementation crosses between tha1-1 and tha1-2 and transformation with a genomic THA1 construct (see Supplemental Figure 4 online) showed that increased seed Thr levels are caused by mutations in this gene. Using a single-seed Thr assay, we determined that four out of 15 seeds from THA1-1/tha1-1 heterozygotes had elevated Thr content (see Supplemental Figure 5 online), close to the 25% that would be expected if the tha1-1 mutation had recessive, seed-specific effects.
Figure 3.
Location of Mutations in THA1 and THA2.
Introns are indicated by thin bars and exons by fat bars. DNA sequences of the T-DNA insertion junction sites are presented in Supplemental Table 1 online.
Figure 4.
Changes in the Free Seed Amino Acids Resulting from the tha1-1 and tha1-2 Mutations.
(A) Thr and metabolically related amino acids in seeds.
(B) l-allo-Thr in seeds.
(C) Free amino acid content in seedlings. Mean ± sd of five samples. * P < 0.01, Student's t test, comparing mutant and wild-type samples.
Since Thr aldolases from other organisms are able to act on l-allo-Thr, we determined whether the concentration of this nonprotein amino acid is also increased in tha1 mutants. In tha1-1 and tha1-2 seeds, the level of l-allo-Thr was increased from nearly undetectable levels to 30 and 10 pmol/mg, respectively (Figure 4B). However, from these results, it is not possible to determine whether l-allo-Thr is produced by isomerization of the overabundant l-Thr in these mutants or by other cellular processes. In the latter case, THA1 may play a role in the removal of l-allo-Thr.
Elevated Thr was not detected in vegetative tissue of 10-d-old tha1 seedlings with two true leaves. However, the Gly content was significantly lower in tha1-1 and tha1-2 mutant seedlings than in the Columbia (Col-0) wild type (Figure 4C). This suggests that there is a Gly deficit in the absence of synthesis by THA1. The enhanced Thr sensitivity of tha1-1 and tha1-2 is further evidence that THA1 is active in seedlings. The Thr concentration that inhibits root growth 50% (IC50) for wild-type seedlings (0.53 ± 0.02 mM) is significantly higher than the IC50 for tha1-1 (0.23 ± 0.01 mM) and tha1-2 (0.24 ± 0.01 mM) seedlings (see Supplemental Figure 6A online). The significantly decreased Ser levels in tha1 and tha2 seeds (Figure 4A) may indicate that this amino acid is used for seed Gly biosynthesis in the absence of THA1.
THA2 Function Is Essential for Arabidopsis
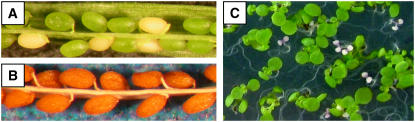
To study the role of THA2 in plant metabolism, we obtained three independent T-DNA insertions in this gene. PCR amplification and DNA sequencing confirmed insertions in the first intron (tha2-1), the third intron (tha2-2), and after amino acid 319 (tha2-3) (Figure 3; see Supplemental Table 1 online) in lines SAIL_5_C01, SALK_129163, and SALK_013854, respectively (Sessions et al., 2002; Alonso et al., 2003). It was not possible to isolate homozygous tha2-1 and tha2-2 mutants (see Supplemental Figure 3C online), suggesting that the gene is essential for the life of the plant. Examination of siliques on THA2/tha2-2 plants showed that 98 out of 399 developing seeds had an albino phenotype (Figure 5A), consistent with tha2-2 being a seed-specific recessive mutation. Crosses between tha2-1 and tha2-2 failed to complement, confirming that the phenotype results from mutations in the THA2 gene. Mature seeds from THA2/tha2-1 and THA2/tha2-2 plants all look phenotypically normal (Figure 5B), but 25% of the developing seedlings are albino, fail to mature beyond the cotyledon stage (Figure 5C), and do not accumulate any chlorophyll pigments (see Supplemental Figure 7 online). PCR-based genotyping showed that both albino seeds and albino seedlings are homozygous for T-DNA insertions in THA2, whereas their green siblings are either heterozygous or homozygous wild type (see Supplemental Figure 3D online). It was not possible to rescue the tha2 albino phenotype by adding the metabolically related amino acids Met, Lys, Lys + Met, Thr, Ile, or Gly to the agar plates on which the seeds were germinated. Similarly, seeds germinating in liquid culture were not rescued by the addition of acetaldehyde. Spraying flowering-stage THA2/tha2 heterozygotes with Thr, Ile, Met, Gly, or acetaldehyde failed to prevent the formation of albino seeds.
Figure 5.
Seed and Seedling Phenotypes of tha2 Mutants.
(A) Homozygous tha2-2 seeds on heterozygous THA2/tha2-2 plants are albino, whereas THA2/tha2-2 heterozygous and THA2 wild-type siblings are green.
(B) Mature mutant and wild-type seeds look identical.
(C) Homozygous tha2-2 seedlings are albino and do not progress beyond the cotyledon stage. THA2/tha2-2 heterozygous and THA2 wild-type siblings develop normally.
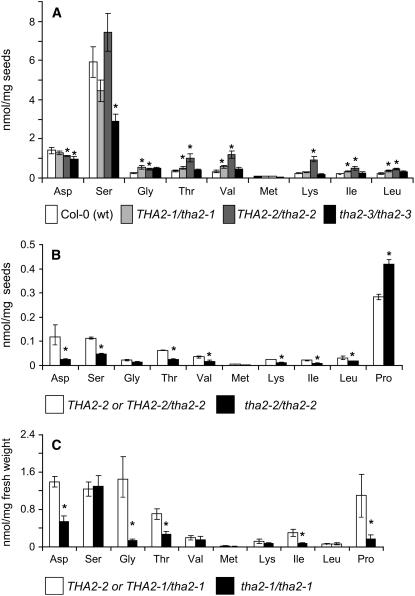
Analysis of seed amino acid content in pooled seeds from THA2/tha2-1 and THA2/tha2-2 heterozygotes showed moderate increases in the levels of Thr and branched-chain amino acids (Figure 6A), perhaps indicating haplo-insufficiency for Thr aldolase in these lines. The levels of all amino acids except Pro were decreased in homozygous tha2-2 albino seeds and 10-d-old seedlings (Figures 6B and 6C). However, this general decrease in free amino acids could be a secondary effect resulting from the absence of photosynthesis. Unlike the tha2-1 and tha2-2 mutations, the tha2-3 T-DNA insertion, which truncates the final 36 amino acids of THA2, is not lethal. Homozygous tha2-3 mutants (see Supplemental Figure 3E online) do not show any visible phenotypic differences compared with wild-type plants. Furthermore, analysis of the seed amino acid content of tha2-3 homozygotes showed no significant Thr increase compared with wild-type Col-0 (Figure 6A). In the crystal structure of Thermotoga maritima Thr aldolase, the C terminus is free in solution (Kielkopf and Burley, 2002). Therefore, it is possible that 36–amino acid truncation of THA2 does not have a significant effect on the overall structure and function of the protein.
Figure 6.
Amino Acid Content of tha2 Seeds and Seedlings.
(A) THA2/tha2-1 and THA2/tha2-2 heterozygotes have slightly elevated levels of Thr and branched-chain amino acids in the seeds. Thr and Ile content in homozygous tha2-3/tha2-3 plants is not significantly different from the wild type.
(B) and (C) Most free amino acids are decreased in albino seeds (B) and seedlings (C) relative to wild-type or heterozygous seedlings.
Mean ± sd of five samples. *P < 0.01, Student's t test.
A THA2/tha2-2 heterozygote was used as the female parent in crosses to both tha1 alleles. In the F2 generation, tha1 THA2/tha2-2 progeny were identified by herbicide resistance (sulfadiazine and kanamycin) and PCR-based genotyping. No F2 progeny homozygous for both tha1 and tha2 mutations were identified. The seed Thr content of the tha1-1 THA2/tha2-2 and tha1-2 THA2/tha2-2 plants was increased 55- and 25-fold relative to Col-0 (see Supplemental Figure 8 online). This increase is similar to that observed in the tha1-1 and tha1-2 parent lines in the same experiment. There were no consistent, significant changes in the abundance of other seed amino acids.
Metabolic Profiling of Thr Aldolase Mutants
To investigate the broader metabolic changes that are induced by Thr aldolase mutations, we measured amino acids, sugars, and organic acids by gas chromatography–mass spectrometry (GC-MS) of flowers, leaves, stems, siliques, and seeds from Col-0 wild-type, tha1-1, tha1-2, THA2-1/tha2-1, and THA2-2/tha2-2 plants. For the amino acids that were detected, the GC-MS data (see Supplemental Table 3 online) confirmed the results obtained previously by HPLC. The tha1-1 and tha1-2 mutations caused 120- and 50-fold increases in seed Thr, respectively. Free Gly content of most vegetative tissue in tha1 mutants showed a moderate decrease, generally <50%, despite no significant changes in the Thr levels. Both THA2-1/tha2-1 and THA2-2/tha2-2 heterozygotes had significant alteration in the amino acid profiles of most tissue types. Haplo-insufficiency is suggested by the fact that lower amounts of Gly, a product of Thr aldolase reaction, were detected in all tissues of THA2/tha2 heterozygotes. In vegetative tissue, branched-chain amino acids (Ile, Leu, and Val) showed increases in THA2/tha2 compared with the wild type in all tissue types except in stems, where there was a decrease.
Consistent changes were also seen in the levels of organic acids and sugars in the tha1 and THA2/tha2 mutant lines (see Supplemental Table 3 online). Citrate and isocitrate levels in seeds and flowers of all four mutants were significantly lower than in the Col-0 wild type. Similarly, levels of most sugars were significantly decreased in flowers and siliques of THA2/tha2 heterozygotes. The two tha1 mutants exhibited strikingly different sugar profiles. Overall sugar levels were significantly decreased in tha1-1 but not tha1-2 flowers. Glucose, mannose, fructose, and galactose were significantly (threefold to sixfold) increased in tha1-2 leaves but not in tha1-1 leaves. These differences may result from partial function of the tha1-1 missense mutation.
THA1 and THA2 Have Different Expression Patterns
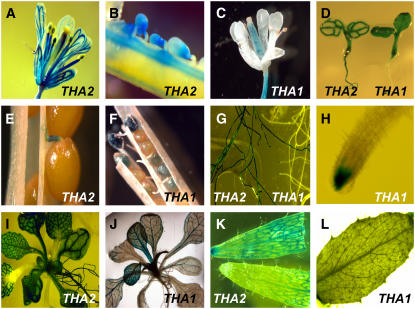
THA1 and THA2 promoter fusions to β-glucuronidase (GUS) were used to study the expression patterns of the two genes (Figure 7). The THA1 promoter shows expression primarily in seeds and young seedlings, which is consistent with the phenotypic effects that we observed in these growth stages. By contrast, the THA2 promoter drives GUS expression in all vascular tissues of the plant, including the flowers and the funiculus. Unlike THA1, which is expressed in seeds almost all the way to maturity (Figure 7F), THA2 is expressed only at an early developmental stage (Figure 7B). As the seeds mature, expression shifts to the attachment point of the funiculus and eventually disappears. In roots, THA1 expression can only be detected at the root tips, but THA2 expression occurs in the entire root (Figures 7G and 7H). Analysis of publicly available microarray data with Genevestigator (Zimmermann et al., 2004) showed an expression pattern that is consistent with the GUS staining pattern (Figure 7), with highest expression of THA1 in the developing seeds and THA2 in the flowers and shoot apex (see Supplemental Figure 9 online).
Figure 7.
GUS Promoter Fusions Show THA1 and THA2 Expression Patterns.
Expression of THA1-GUS and THA2-GUS promoter fusions in flowers and developing seeds ([A] to [C]), seedlings (D), almost-mature seeds ([E] and [F]), roots ([G] and [H]), 2-week-old rosettes ([I] and [J]), and cauline leaves ([K] and [L]).
Thr Aldolase Mutations Increase Substrate Availability for Thr Deaminase
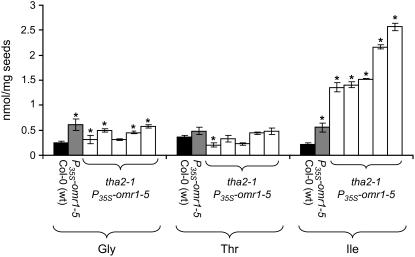
The inability to rescue tha2-1 and tha2-2 mutant growth with exogenous addition of amino acids suggested that lethality could be due to excess Thr rather than the absence of a downstream metabolite. To test this hypothesis, we transformed THA2/tha2-1 heterozygotes with T-DNA constructs that overproduce a feedback-insensitive form of Thr deaminase (the omr1-5 allele of OMR1) using the cauliflower mosaic virus 35S promoter (Mourad and King, 1995; Garcia and Mourad, 2004). Thr deaminase catalyzes the formation of α-ketobutyrate from Thr (Figure 1) as the first step in the pathway leading to Ile. Therefore, overproduction would be expected to reduce Thr content. PCR-based genotyping of the T2 generation was used to identify tha2-1/tha2-1 P35S-omr1-5 plants (see Supplemental Figure 3F online). Visual analysis of tha2-1/tha2-1 P35S-omr1-5 homozygotes showed some chlorosis, leaf curling, and misshapen flowers (see Supplemental Figure 10 online) but otherwise viable plants with normal seed set. The P35S-omr1-5 transgene in the THA2 background caused only a twofold increase in seed Ile content. By contrast, there was a 10-fold increase in seed Ile in the tha2-1 background (Figure 8), suggesting that Thr availability is limiting the formation of Ile via feedback-insensitive Thr deaminase in wild-type seeds. It is possible that P35S-omr1-5 expression rescues tha2-1/tha2-1 seeds and seedlings through the elimination of otherwise toxic amounts of Thr. However, experiments with P35S-omr1-5 and P35S-omr1-5 seedlings growing on agar showed only moderately increased resistance to exogenously added Thr (see Supplemental Figure 6B online). Since homozygous tha2-1 and tha2-2 mutations are lethal, we could not study transcript levels in the original T-DNA insertion lines. However, in the tha2-1/tha2-1 P35S-omr1-5 double mutants, we were unable to detect any THA2 transcript by RT-PCR (see Supplemental Figure 3F online), indicating that tha2-1 is a knockout mutation.
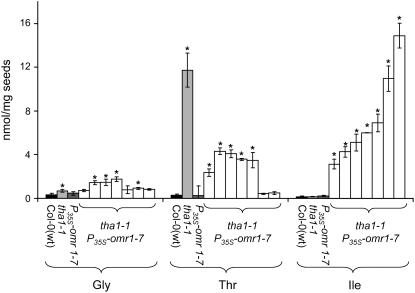
Figure 8.
The Lethal Effects of tha2-1 Thr Aldolase Mutations Are Rescued by Overexpression of Thr Deaminase (P35S-omr1-5), Which Also Causes Significantly Elevated Seed Ile Content.
Progeny from five independent transformation events are shown. Mean ± sd of five samples. *P < 0.01, Student's t test compared with Col-0 (wild type).
Further genetic analysis was used to demonstrate that rescue of tha2-1/tha2-1 albino seeds by P35S-omr1-5 depends entirely on the seed genotype. When pollen from tha2-1/tha2-1 P35S-omr1/P35S-omr1 plants was transferred onto THA2/tha2-1 pistils, no albino seeds were observed. This is consistent with P35S-omr1 contributed by the pollen rescuing the 50% of progeny seeds from this cross that are homozygous for tha2-1 mutation. Moreover, 50% of the individual seeds from the cross showed greatly increased Ile content (see Supplemental Figure 11A online), which is characteristic of tha2-1/tha2-1 P35S-omr1-5 seeds (Figure 8). In the reciprocal experiment to study maternal effects of P35S-omr1-5, tha2/tha2 P35S-omr1-5/+ plants were allowed to self-pollinate. Three progeny seed genotypes were expected: 25% tha2/tha2, 50% tha2/tha2 P35S-omr1-5/+, and 25% tha2/tha2 P35S-omr1-5/ P35S-omr1-5. If maternal-only P35S-omr1-5 expression does not provide rescue of the mutation, then the 25% of the progeny seeds that are tha2/tha2 should show an albino phenotype. Out of 193 progeny seeds that were examined, 46 were albino, not significantly different from the expected 25% (P < 0.05, χ2 test). Therefore, maternal expression of P35S-omr1-5 is not sufficient to rescue tha2/tha2 mutant seeds.
A different feedback-insensitive allele of Thr deaminase (P35S-omr1-7; Garcia and Mourad, 2004) was used to transform homozygous tha1-1 mutants. Greatly elevated Ile content was observed in tha1-1 P35S-omr1-7 double mutants (Figure 9), even though the seed Ile changes induced by the omr1-7 transgene itself are insignificant. The two double mutant lines with the highest seed Ile levels have the lowest Thr, suggesting competition for a common Thr pool between Thr aldolase and Thr deaminase. No visible defects beyond a mild chlorosis of the leaves were observed in tha1-1 P35S-omr1-7 double mutants. Similar to the case of tha2-1 P35S-omr1-5, seed-specific expression of P35S-omr1-7 in the tha1-1/tha1-1 genetic background resulted in greatly increased seed Ile content (see Supplemental Figure 11B online).
Figure 9.
Elevated Thr Caused by the tha1-1 Thr Aldolase Mutation Is Converted to Ile by Overexpression of Feedback-Insensitive Thr Deaminase (P35S-omr1-7).
Progeny from seven independent transformation events are shown. Mean ± sd of five samples. *P < 0.01, Student's t test compared with Col-0 (wild type).
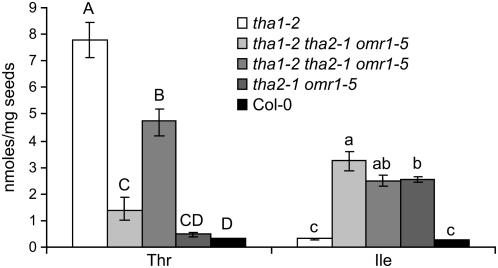
The tha1-2/tha1-2 and tha2-1/tha2-1 P35S-omr1-5 mutant lines were crossed, and homozygous tha1-2/tha1-2 tha2-1/tha2-1 P35S-omr1-5 triple mutant progeny were identified in the F2 generation (see Supplemental Figure 3G online). The homozygous triple mutants were viable, showing that Gly production from Thr by THA1 and THA2 is completely dispensable for Arabidopsis. The morphological defects of the triple mutants were similar to those of the tha2-1/tha2-1 P35S-omr1-5 double mutant shown in Supplemental Figure 10 online. At least in one case, seed Thr content of the triple mutant was significantly higher than that of the tha2-1/tha2-1 P35S-omr1-5 double mutant (Figure 10), indicating that the tha1-2 and tha2-1 knockout mutations have additive effects.
Figure 10.
Seed Thr and Ile Content of tha1-2/tha1-2 tha2-1/tha2-1 P35S-omr1-5 Triple Mutants (Two Independent Isolates) and the tha1-2/tha1-2 and tha2-1/tha2-1 P35S-omr1-5 Parent Lines.
Different letters indicate significant differences with Tukey-Kramer honestly significant difference test (P < 0.05). Mean ± sd of n = 3.
DISCUSSION
Promoter-GUS fusions indicate that the two Arabidopsis Thr aldolases have overlapping, though not identical expression patterns. The expression of THA1 in developing seeds is consistent with the observation that Thr content is greatly elevated in tha1 mutant seeds. THA2 is also expressed in seeds and seedlings, but the most readily apparent phenotype is a complete lack of chlorophyll. Two possible explanations for the different phenotypes are (1) Thr is compartmentalized, and the two enzymes have different functions; or (2) tha1 mutations cause a minor decrease in total Thr aldolase activity, whereas tha2 mutations cause a larger decrease that is lethal. In the latter case, one would have to assume that synthesis of THA1 cannot be upregulated to compensate for the lack of THA2 in these tissues.
Both THA1 and THA2 are expressed in the vasculature of Arabidopsis leaves (Figures 7I and 7J). This expression pattern suggests a possible role in phloem loading or amino acid transport for Thr aldolase. The expression of THA2 at the attachment point of the funiculus in developing seeds (Figure 7B) may indicate a gatekeeper function in the processing of metabolites as they are transported into the seeds. The more uniform expression of THA1 throughout seed development (Figures 7C and 7F) indicates a broader, though nonessential, metabolic function during seed filling. Seed-specific expression of feedback-insensitive Thr deaminase contributed by the pollen in crosses counteracts the effects of both the tha2-1 and the tha1-2 mutations in a similar manner as whole-plant expression (see Supplemental Figure 11 online). Therefore, metabolic communication between maternal and seed tissue is not necessary to explain the observed rescue of tha1 and tha2 phenotypes by Thr deaminase.
The rescue of tha2-1 mutations by P35S-omr1-5 suggests that the absence of Gly is not the proximal cause of lethality. However, at this point, we cannot rule out the possibility that tha2 P35S-omr1-5 double mutants survive because the plants synthesize Gly by an as yet unknown pathway downstream of Thr deaminase. Production of Gly by THA1 is not necessary for the observed phenotypic rescue because tha1-2/tha1-2 tha2-1/tha2-1 P35S-omr1-5 triple mutants are viable and have a visible phenotype that is similar to tha2-1/tha2-1 P35S-omr1-5 double mutants. It is conceivable that acetaldehyde produced by Thr aldolase plays an essential role in plant metabolism. However, this seems unlikely due to the generally phytotoxic effects of acetaldehyde (Kursteiner et al., 2003; Tsuji et al., 2003; Fujishige et al., 2004; Hirayama et al., 2004).
It is tempting to speculate that the lethal albino phenotype of homozygous tha2/tha2 seeds results from excess Thr in the affected tissues, perhaps via feedback inhibition of the Asp-derived amino acid pathway (Coruzzi and Last, 2000). However, our inability to rescue tha2 mutants with exogenous Met, Lys, and other amino acids suggests otherwise. The 14 mM Thr observed in the perfectly normal-looking and viable tha1-1 seeds (Figure 4) shows that mature seeds can tolerate a considerable amount of excess Thr without deleterious effects. By contrast, the IC50 for Arabidopsis root growth inhibition by exogenous Thr is 0.53 mM, though active transport may make the concentration in roots higher than in the surrounding medium. Similar to effects of tha2 mutations described here, it has been suggested that knockout of the Lys biosynthetic enzyme DHDPS2 (dihydrodipicolinate synthase 2) causes the accumulation of toxic levels of Thr in Arabidopsis (Sarrobert et al., 2000). However, there is also as yet no proven mechanism by which this happens. It is also possible that a toxic metabolite derived from excess Thr contributes to plant lethality. One candidate compound might be l-allo-Thr, but exogenous l-allo-Thr is not significantly more toxic than l-Thr for root growth of Arabidopsis seedlings (V. Joshi, unpublished data).
In addition to Thr aldolase, at least two other plant enzymes, SHMT and glyoxylate aminotransferase, can produce Gly. There is quite considerable metabolic flux through Gly as a substrate for SHMT in the photorespiratory cycle. Therefore, one possible function of Thr aldolase is that it contributes to maintaining Gly homeostasis in plants at times of transition in the use of the photorespiratory cycle. Several lines of evidence suggest that Thr aldolase activity contributes to such Gly homeostasis in Arabidopsis: (1) Gly levels are decreased in tha1 seedlings (Figures 4C); (2) Gly levels are decreased in vegetative tissue of THA2/tha2 heterozygotes (see Supplemental Figure 7 online); (3) Ser, which can be interconverted with Gly by SHMT, is increased in tha1 seed (Figure 4A); and (4) THA1 mRNA levels are increased eightfold in seedlings of an Arabidopsis isocitrate lyase mutant (Cornah et al., 2004). Conversely, the reduced levels of isocitrate and citrate in seeds and flowers of both tha1 and THA2/tha2 mutants (see Supplemental Table 3 online) may indicate increased production of Gly via isocitrate lyase and glyoxylate.
Although feedback-insensitive Thr deaminase alone significantly increases Ile in vegetative tissue (Garcia and Mourad, 2004), there is little or no effect on seed Ile content (Figures 8 and 9). However, if P35S-omr1 is combined with tha1 or tha2 mutations, seed Ile is greatly elevated (Figure 9). These results show that Thr aldolase and Thr deaminase compete for a common pool of Thr in developing seeds and that Thr availability is limiting for Ile formation. Similarly, Thr availability is rate-limiting for Ile formation in Arabidopsis leaves, as was demonstrated previously with metabolic precursor feeding studies (Lee et al., 2005). By reducing Thr aldolase and increasing Thr deaminase activity, we have redirected Thr flux from Gly toward α-ketobutyrate and Ile. Even in the absence of Thr deaminase overproduction, THA2 haplo-insufficiency increases the levels of branched-chain amino acids (see Supplemental Figure 7 online). Concomitant increases in Val and Leu are likely due to coregulation of the linked pathways leading to the formation of these amino acids. Similarly, production of Ile, Leu, and Val was coordinately regulated in interspecific tomato (Solanum lycopersicum) introgression lines (Schauer et al., 2006) and Arabidopsis mutants that were selected to have increased seed Ile content (Jander et al., 2004).
Thr, Lys, Met, and other limiting essential amino acids are commonly added as supplements to livestock feed. By providing an optimal ratio of amino acids, rather than increasing total protein content, it is possible to improve animal growth without having an excess of nonlimiting amino acids, which would be degraded to ammonia by livestock and excreted as an environmental pollutant (Pfefferle et al., 2003). Our results suggest that inhibiting Thr aldolase function in the seeds of crop plants could be used to increase Thr content. Indeed, comparison of EST data from several crops, including maize, rice, and soy (see Supplemental Figure 12 online), shows likely Thr aldolases whose expression could be manipulated to improve grain quality. The relatively specific effects of Thr aldolase mutations on seed Thr content make this enzyme an attractive target for plant genetic engineering. Increased production of Ile, another essential amino acid, can be limited by the availability of Thr as a substrate. By combining Thr aldolase mutations with overproduction of Thr deaminase, we demonstrated that seed Ile levels are greatly increased with few deleterious metabolic effects on the growth of the plant.
METHODS
Plant Material
Wild-type Arabidopsis thaliana ecotype Col-0 is from the ABRC (http://www.arabidopsis.org/abrc/). Thr aldolase T-DNA insertion mutants, all in the Col-0 background, were obtained from publicly available collections. Lines SALK_129163 (tha2-2) and SALK_013854 (tha2-3) are from the SIGnAL collection (http://signal.salk.edu/) (Alonso et al., 2003). Line SAIL_5_C01 (tha2-1) is from the Syngenta Arabidopsis Insertion Library (Sessions et al., 2002). Segregating SALK_129163 (CS6934), segregating SAIL_5_C01 (CS6960), and homozygous SALK_013854 (CS6935) insertion lines are available from the ABRC. GK-767E02 (tha1-2) is from the GABI-KAT collection (Rosso et al., 2003) and can be obtained from the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/). The Col-0 tha1-1 mutant line (Jander et al., 2004) will be made available by the authors but requires a material transfer agreement from the Monsanto Company. Transgenic Arabidopsis lines and bacterial T-DNA vectors with P35S-omr1 constructs were kindly supplied by G. Mourad (Purdue University, Fort Wayne, IN) and require a material transfer agreement.
Growth Conditions
Plants were grown on Cornell Mix with Osmocoat fertilizer (Landry et al., 1995) in 20 × 40 cm nursery flats in Conviron growth chambers. Photosynthetic photon flux density was 200 μmol m−2 s−1, the photoperiod was 16:8 day:night, the temperature was 23°C, and the relative humidity was 50%. For growth on agar, 1× Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) with or without 1% sucrose was used, and the plates were placed in the same growth chambers as the soil-grown plants. Arabidopsis seeds were surface-sterilized by incubation for 1 min in 70% ethanol, followed by shaking in 50% commercial bleach (CleanAll Products; 5.25% sodium hypochlorite) with 0.01% Triton X-100 for 10 min. After five washes with sterile water, seeds were suspended in 0.1% Phytagar (Invitrogen). Seeds were cold-stratified by incubating at 4°C in the dark for 3 d.
To measure seedling Thr sensitivity, seeds were plated on MS agar with 0, 0.5, 1, 2, or 4 mM l-Thr. Root length was measured after 10 d. The IC50, defined as the concentration at which the root length gets inhibited by 50%, was calculated by nonlinear regression using JMP 6 software (SAS Institute).
For experiments to rescue tha2 homozygous seedlings, Met, Lys, Lys + Met, Thr, Ile, Gly, and Arg were added to MS agar plates at 100 μM and 1 mM concentrations. Acetaldehyde was added at 0, 10, 50, and 100 μM concentration to seeds germinating in 1× MS liquid cultures. For attempts to rescue the albino phenotype of developing seeds, flowering plants were sprayed and drenched with 100 μM and 1 mM Met, Lys, Lys + Met, Thr, Ile, and Gly at 2-d intervals for 10 d before and after flowering.
Genetic Crosses and Generation of Transgenic Plants
Crosses between Arabidopsis lines were performed as described (Weigel and Glazebrook, 2002). For complementation of the tha1-1 mutation, the THA1 gene, including the 5′ and 3′ untranslated regions, was amplified from genomic DNA of Col-0 using the primers gTHA1f and gTHA1r (see Supplemental Table 2 online) and cloned into pCAMBIA3301 (http://www.cambia.org/) and subsequently transferred to Agrobacterium tumafaciens strain GV3101 (Koncz and Schell, 1986). Plants were transformed using the floral dip method (Clough and Bent, 1998). T1 and T2 transformants were selected by spraying the seedlings twice with 300 μM glufosinate ammonium (1:200 dilution of Finale; Farnam Companies), and T3 seeds were used for amino acid analysis. For promoter-reporter fusions, the promoter regions, 2529 bp for THA1 and 1818 bp for THA2, were amplified using the specific primers promTHA1F and promTHA1R, and promTHA2F and promTHA2R (see Supplemental Table 2 online), respectively. The resulting PCR products were moved into pDONR201 by BP clonase reactions (Invitrogen). The promoters were then transferred into the destination vector pBGWFS7 (Karimi et al., 2002) using the LR clonase reaction. Binary vectors were introduced into A. tumefaciens strain GV3101 (Koncz and Schell, 1986) and subsequently used in plant transformation (Clough and Bent, 1998). Sequences of all the constructs were confirmed by PCR amplification and DNA sequencing before transformation using the Big Dye Terminator v.3.0 cycle sequencing kit (Applied Biosystems; ABI PRISM). T1 populations were selected by spraying with glufosinate ammonium (Finale; Farnam Companies), and the T2 generation was used to screen for GUS expression.
Yeast Experiments
Yeast strains W3031B (MATα ura3-1 trp1-1 ade2-1 his3-11,-15 leu2-3,-112 can1-100) and YM13 (W3031B shm1∷HIS3 shm2∷LEU2 gly1∷URA3) (McNeil et al., 1994; Monschau et al., 1997) were kindly supplied by A. Bognar (University of Toronto, Canada). Yeast strains were grown on YPD medium (1% yeast extract, 2% peptone, and 2% dextrose) or on SD medium (0.67% Bacto-yeast nitrogen base and 2% dextrose) supplemented, where appropriate, with 750 μg/mL Gly, 30 μg/mL Leu, and 20 μg/mL each uracil, Trp, adenine, and His.
Primers CDTHA2F and CDTHA2R (see Supplemental Table 2 online) were used to amplify full-length THA2 cDNA from ABRC clone S51 for cloning into pCM185 (Gari et al., 1997) using the BamHI and PstI restriction sites of the plasmid. Yeast was transformed by the lithium acetate method (Ausubel et al., 1998). Complementation of the Trp auxotrophy was used to select for transformation of strains W3031B and YM13 with pCM185 and a derived plasmid construct carrying the THA2 Thr aldolase. The correct THA2 cDNA sequence was verified by PCR amplification from yeast and DNA sequencing. Thr aldolase activity in yeast extracts was measured as described previously (Jander et al., 2004).
Verification of T-DNA Insertions in THA1 and THA2 by PCR
T-DNA insertion sites were confirmed using T-DNA insertion–specific primers (Lba1 for SALK and Lba3 for Syngenta lines) and flanking THA1 or THA2 sequence-specific primers. Amplified products were sequenced to determine actual site of insertion. For genotyping T-DNA insertions, gene-specific primers (tha1-2 P1, tha1-2 P2, tha2-1 P1, tha2-1 P2, tha2-2 P1, tha2-2 P2, tha2-3 P1 and tha2-3 P2; see Supplemental Table 2 online) that flank the insertion site were used to detect the wild-type allele, and THA1 or THA2-specific primers in combination with a T-DNA primer (LBa1 or LBb1 for SALK, Lb3 for Syngenta, and GABI SEQ for GABI-KAT lines; see Supplemental Table 2 online) were used to identify the mutant alleles. T-DNA insertions were also selected by growth on herbicide-containing MS agar plates: SALK lines with 30 μg/mL kanamycin, GABI-KAT line with 11.25 mg/L sulfadiazine, and SAIL lines with 50 mg/L glufosinate. T-DNA insertions in SAIL lines growing on soil were selected by spraying with 300 μM glufosinate ammonium (Finale; Farnam Companies).
Amino Acid Assays
For analysis of amino acids, 10 mg of plant tissue was frozen in liquid nitrogen in 2-mL tubes and ground to fine powder with 3-mm steel beads using a Harbil model 5G-HD paint shaker. Ground tissue was taken up in 20 mM HCl (10 μL per mg of tissue), the extracts were centrifuged at 14,000 rpm for 20 min at 4°C, and the supernatant was derivatized with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate (Cohen and van Wandelen, 1997) using an AccQ-Fluor reagent kit (Waters). During derivatization, 5-μL extracts were mixed with 35 μL borate buffer, and the reaction was initiated by the addition of 10 μL 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate reagent followed by immediate mixing and incubation for 10 min at 55°C. Ten microliters of each sample were injected onto a 3.9 × 150-mm Nova-Pak C18 column (Waters) using a Waters 2690 pump system, and the data were recorded using Waters' Empower Software. L-Norleucine was used as an internal standard. Eluted amino acid derivatives were detected using a Waters model 2475 multi λ fluorescence detector with an excitation wavelength of 250 nm and an emission wavelength of 395 nm. Solvent A (containing sodium acetate and triethylamine at pH 5.05) was purchased premixed from Waters; Solvent B was acetonitrile:water (60:40). The gradient used was 0 to 0.01 min, 100% A; 0.01 to 0.5 min, linear gradient to 3% B; 0.5 to 12 min, linear gradient to 5% B; 12 to 15 min, linear gradient to 8% B; 15 to 45 min, 35% B; 45 to 49 min, linear gradient to 35% B; 50 to 60 min, 100% B. Flow rate was 1.0 mL min−1. Standard curves were prepared using amino acids purchased from Sigma-Aldrich.
GUS Staining
Different plant tissues from transgenic plants carrying THA1 promoter:GUS and THA2 promoter:GUS were incubated in GUS assay solution (50 mM sodium phosphate, pH 7.2, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, 20% methanol, and 2 mM 5-bromo-4-chloro-β-glucuronide) at 37°C for 12 to 16 h as described previously (Jefferson et al., 1987). Slight vacuum was applied to facilitate substrate infiltration. Chlorophyll-containing tissue was cleared in 70% ethanol for photographic analysis. Samples were examined and photographed using an SZX12 microscope (Olympus) equipped with a JENOTCX ProgRes C14 digital camera (Olympus).
Metabolite Profiling
Metabolite analysis by GC-MS was performed as described previously (Roessner-Tunali et al., 2003), with minor modifications. Arabidopsis tissue (50 mg of seeds and 100 mg of leaves, flowers, siliques, and stem) from Col-0 (wild type), tha1-1, tha1-2, tha2-1, and tha2-2, harvested at same developmental stages, was homogenized using a ball mill precooled with liquid nitrogen. Chloroform extraction was done twice only in case of seeds. After centrifugation, 200 and 150 μL of the polar phase was reduced to dryness in vacuum for seeds and other tissue types, respectively. Reduced residues were redissolved in and derivatized for 120 min at 37°C (in 40 μL of 20 mg mL−1 methoxyamine hydrochloride in pyridine), followed by a 30-min treatment with 70 μL N-methyl-N-(trimethylsilyl)trifluoroacetamide at 37°C. Seven microliters of a retention time standard mixture (0.029% [v/v] n-dodecane, n-pentadecane, n-nonadecane, n-docosane, n-octacosane, n-dotracontane, and n-hexatriacontane dissolved in pyridine) was added prior to trimethylsilylation. The GC-MS system used comprised an AS 2000 autosampler, a GC 8000 gas chromatograph, and a Voyager quadrupole mass spectrometer (ThermoFinnigan). Both chromatograms and mass spectra were evaluated using the MASSLAB program (ThermoQuest), and normalization of metabolite levels with respect to wild-type levels was performed as described previously (Roessner et al., 2001).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are THA1 (At1g08630), THA2 (At3g04520), and OMR1 (At3g10050).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table 1. Sequences of the Junction Sites of THA1 and THA2 T-DNA Insertion Lines.
Supplemental Table 2. PCR Primers Used in the Experiments.
Supplemental Table 3. GC-MS Metabolite Profiling of Thr Aldolase Mutants.
Supplemental Figure 1. Lineweaver-Burke Plot Showing Apparent Km for Thr of THA2.
Supplemental Figure 2. Growth of Yeast Expressing Arabidopsis tha1-1 on Medium Containing 50 μM Gly.
Supplemental Figure 3. PCR to Confirm Zygosity and RT-PCR to Confirm Gene Expression of Thr Aldolase Mutants.
Supplemental Figure 4. Complementation of the tha1-1 Amino Acid Phenotype by Transformation.
Supplemental Figure 5. Thr Levels in Single Seeds of THA1/tha1 Heterozygotes.
Supplemental Figure 6. Thr IC50 Determination for Wild-Type, tha1, and P35S-omr1 Seedlings.
Supplemental Figure 7. Chlorophyll Content of tha2 Mutant Seedlings.
Supplemental Figure 8. Amino Acid Content of tha1 THA2/tha2 Double Mutants.
Supplemental Figure 9. Digital RNA Gel Blot of THA1 and THA2 Expression in Arabidopsis.
Supplemental Figure 10. Photographs of tha2-1/tha2-1 P35S-omr1-5 Rosette and Flowers.
Supplemental Figure 11. Seed Amino Acid Changes Resulting from Seed-Specific Expression of P35S-omr1-5 in the tha2/tha2 and tha1/tha1 Genetic Backgrounds.
Supplemental Figure 12. DNA Sequence Comparison of Thr Aldolases from Arabidopsis and Selected Crop Plants.
Supplementary Material
Acknowledgments
We thank G. Mourad for Thr deaminase overexpression constructs and A. Bognar for yeast strain YM13. This research was funded by National Science Foundation Grants MCB-0416567 and DBI-0453331.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Georg Jander (gj32@cornell.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1998). Current Protocols in Molecular Biology. (Hoboken, NJ: John Wiley and Sons).
- Broeckling, C.D., Huhman, D.V., Farag, M.A., Smith, J.T., May, G.D., Mendes, P., Dixon, R.A., and Sumner, L.W. (2005). Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J. Exp. Bot. 56 323–336. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, S.A., and van Wandelen, C. (1997). Amino acid analysis of unusual and complex samples based on 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate derivitization. In Techniques in Protein Chemistry VIII, J.W. Crabb, ed (New York: Academic Press), pp. 185–196.
- Contestabile, R., Paiardini, A., Pascarella, S., di Salvo, M.L., D'Aguanno, S., and Bossa, F. (2001). L-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur. J. Biochem. 268 6508–6525. [DOI] [PubMed] [Google Scholar]
- Cornah, J.E., Germain, V., Ward, J.L., Beale, M.H., and Smith, S.M. (2004). Lipid utilization, gluconeogenesis, and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279 42916–42923. [DOI] [PubMed] [Google Scholar]
- Coruzzi, G.M., and Last, R.L. (2000). Amino acids. In Biochemistry and Molecular Biology of Plants, R.B. Buchanan, W. Gruissem, and R. Jones, eds (Rockville, MD: American Society of Plant Physiology Press), pp. 358–410.
- Edgar, A.J. (2002). Molecular cloning and tissue distribution of mammalian L-threonine 3-dehydrogenases. BMC Biochem. 3 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epperly, B.R., and Dekker, E.E. (1991). L-threonine dehydrogenase from Escherichia coli. Identification of an active site cysteine residue and metal ion studies. J. Biol. Chem. 266 6086–6092. [PubMed] [Google Scholar]
- Fernie, A.R., Trethewey, R.N., Krotzky, A.J., and Willmitzer, L. (2004). Metabolite profiling: From diagnostics to systems biology. Nat. Rev. Mol. Cell Biol. 5 763–769. [DOI] [PubMed] [Google Scholar]
- Fiehn, O., Kopka, J., Trethewey, R.N., and Willmitzer, L. (2000). Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 72 3573–3580. [DOI] [PubMed] [Google Scholar]
- Fujishige, N., Nishimura, N., Iuchi, S., Kunii, T., Shinozaki, K., and Hirayama, T. (2004). A novel Arabidopsis gene required for ethanol tolerance is conserved among plants and archaea. Plant Cell Physiol. 45 659–666. [DOI] [PubMed] [Google Scholar]
- Galili, G., Amir, R., Hoefgen, R., and Hesse, H. (2005). Improving the levels of essential amino acids and sulfur metabolites in plants. Biol. Chem. 386 817–831. [DOI] [PubMed] [Google Scholar]
- Garcia, E.L., and Mourad, G.S. (2004). A site-directed mutagenesis interrogation of the carboxy-terminal end of Arabidopsis thaliana threonine dehydratase/deaminase reveals a synergistic interaction between two effector-binding sites and contributes to the development of a novel selectable marker. Plant Mol. Biol. 55 121–134. [DOI] [PubMed] [Google Scholar]
- Gari, E., Piedrafita, L., Aldea, M., and Herrero, E. (1997). A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13 837–848. [DOI] [PubMed] [Google Scholar]
- Halgand, F., Wessel, P.M., Laprevote, O., and Dumas, R. (2002). Biochemical and mass spectrometric evidence for quaternary structure modifications of plant threonine deaminase induced by isoleucine. Biochemistry 41 13767–13773. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., Fujishige, N., Kunii, T., Nishimura, N., Iuchi, S., and Shinozaki, K. (2004). A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell Physiol. 45 703–711. [DOI] [PubMed] [Google Scholar]
- House, J.D., Hall, B.N., and Brosnan, J.T. (2001). Threonine metabolism in isolated rat hepatocytes. Am. J. Physiol. Endocrinol. Metab. 281 E1300–E1307. [DOI] [PubMed] [Google Scholar]
- Jander, G., Norris, S.R., Joshi, V., Fraga, M., Rugg, A., Yu, S., Li, L., and Last, R.L. (2004). Application of a high-throughput HPLC-MS/MS assay to Arabidopsis mutant screening; evidence that threonine aldolase plays a role in seed nutritional quality. Plant J. 39 465–475. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Kielkopf, C.L., and Burley, S.K. (2002). X-ray structures of threonine aldolase complexes: Structural basis of substrate recognition. Biochemistry 41 11711–11720. [DOI] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Kursteiner, O., Dupuis, I., and Kuhlemeier, C. (2003). The Pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiol. 132 968–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, L.G., Chapple, C.C., and Last, R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., Martin, M.N., Hudson, A.O., Lee, J., Muhitch, M.J., and Leustek, T. (2005). Methionine and threonine synthesis are limited by homoserine availability and not the activity of homoserine kinase in Arabidopsis thaliana. Plant J. 41 685–696. [DOI] [PubMed] [Google Scholar]
- Liu, J.Q., Dairi, T., Itoh, N., Kataoka, M., Shimizu, S., and Yamada, H. (1998. a). Gene cloning, biochemical characterization and physiological role of a thermostable low-specificity L-threonine aldolase from Escherichia coli. Eur. J. Biochem. 255 220–226. [DOI] [PubMed] [Google Scholar]
- Liu, J.Q., Ito, S., Dairi, T., Itoh, N., Kataoka, M., Shimuzu, S., and Yamada, H. (1998. b). Gene cloning, nucleotide sequencing, purification, and characterization of low-specificity L-threonine aldolase from Pseudomonas sp. NCIMB strain 10558. Appl. Environ. Microbiol. 64 549–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.Q., Nakata, S., Dairi, T., Misono, T., Shimuzu, S., and Yamada, H. (1997). Gly1 gene of Saccharomyces cerevisiae encodes a low-specificity L-threonine aldolase that catalyzes cleavage of L-allo-threonine and L-threonine to glycine: Expression of the gene in Escherichia coli and purification and characterization of the enzyme. Eur. J. Biochem. 245 289–293. [DOI] [PubMed] [Google Scholar]
- Masuda, T., Ozaki, H., and Tai, K. (1982). Reexamaination of threonine aldolase activity in supernatant solutions of different organisms. Nippon Nogeikagaku Kaishi 56 549–552. [Google Scholar]
- Masuda, T., Yoshino, M., Nishizaki, I., Tai, A., and Ozaki, H. (1980). Purification and properties of allo-threonine aldolase EC-4.1.2.5 from maize Zea mays seedlings. Agric Biol Chem 44 2199–2202. [Google Scholar]
- McNeil, J.B., McIntosh, E.M., Taylor, B.V., Zhang, F.R., Tang, S., and Bognar, A.L. (1994). Cloning and molecular characterization of three genes, including two genes encoding serine hydroxymethyltransferases, whose inactivation is required to render yeast auxotrophic for glycine. J. Biol. Chem. 269 9155–9165. [PubMed] [Google Scholar]
- Monschau, N., Stahmann, K.P., Sahm, H., McNeil, J.B., and Bognar, A.L. (1997). Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: A key enzyme in glycine biosynthesis. FEMS Microbiol. Lett. 150 55–60. [DOI] [PubMed] [Google Scholar]
- Mourad, G., and King, J. (1995). L-O-Methylthreonine-resistant mutant of Arabidopsis defective in isoleucine feedback regulation. Plant Physiol. 107 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F.A. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15 473–497. [Google Scholar]
- Ogawa, H., Gomi, T., and Fujioka, M. (2000). Serine hydroxymethyltransferase and threonine aldolase: Are they identical? Int. J. Biochem. Cell Biol. 32 289–301. [DOI] [PubMed] [Google Scholar]
- Paz, M.A., Blumenfeld, O.O., Rojkind, M., Henson, E., Furfine, C., and Gallop, P.M. (1965). Determination of carbonyl compounds with N-methyl benzothiazolone hydrazone. Arch. Biochem. Biophys. 109 548–559. [DOI] [PubMed] [Google Scholar]
- Pfefferle, W., Mockel, B., Bathe, B., and Marx, A. (2003). Biotechnological manufacture of lysine. Adv. Biochem. Eng. Biotechnol. 79 59–112. [DOI] [PubMed] [Google Scholar]
- Prabhu, V., Chatson, K.B., Abrams, G.D., and King, J. (1996). 13C nuclear magnetic resonance detection of interactions of serine hydroxymethyltransferase with C1-tetrahydrofolate synthase and glycine decarboxylase complex activities in Arabidopsis. Plant Physiol. 112 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu, V., Chatson, K.B., Lui, H., Abrams, G.D., and King, J. (1998). Effects of sulfanilamide and methotrexate on 13C fluxes through the glycine decarboxylase/serine hydroxymethyltransferase enzyme system in Arabidopsis. Plant Physiol. 116 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner, U., Luedemann, A., Brust, D., Fiehn, O., Linke, T., Willmitzer, L., and Fernie, A. (2001). Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13 11–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner-Tunali, U., Hegemann, B., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., and Fernie, A.R. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133 84–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K., and Weisshaar, B. (2003). An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 53 247–259. [DOI] [PubMed] [Google Scholar]
- Sarrobert, C., Thibaud, M.C., Contard-David, P., Gineste, S., Bechtold, N., Robaglia, C., and Nussaume, L. (2000). Identification of an Arabidopsis thaliana mutant accumulating threonine resulting from mutation in a new dihydrodipicolinate synthase gene. Plant J. 24 357–367. [DOI] [PubMed] [Google Scholar]
- Schauer, N., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24 447–454. [DOI] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, H., Meguro, N., Suzuki, Y., Tsutsumi, N., Hirai, A., and Nakazono, M. (2003). Induction of mitochondrial aldehyde dehydrogenase by submergence facilitates oxidation of acetaldehyde during re-aeration in rice. FEBS Lett. 546 369–373. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2002). Arabidopsis: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Wessel, P.M., Graciet, E., Douce, R., and Dumas, R. (2000). Evidence for two distinct effector-binding sites in threonine deaminase by site-directed mutagenesis, kinetic, and binding experiments. Biochemistry 39 15136–15143. [DOI] [PubMed] [Google Scholar]
- Zhu, X., and Galili, G. (2003). Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., Tang, G., Granier, F., Bouchez, D., and Galili, G. (2001). A T-DNA insertion knockout of the bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase gene elevates lysine levels in Arabidopsis seeds. Plant Physiol. 126 1539–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.