Abstract
The effective size of the HIV population in vivo, although critically important for the prediction of appearance of drug-resistant variants, is currently unknown. To address this issue, we have developed a simple virus population model, within which the relative importance of stochastic factors and purifying selection for genetic evolution differs over, at least, three broad intervals of the effective population size, with approximate boundaries given by the inverse selection coefficient and the inverse mutation rate per base per cycle. Random drift and selection dominate the smallest (stochastic) and largest (deterministic) population intervals, respectively. In the intermediate (selection–drift) interval, random drift controls weakly diverse populations, whereas strongly diverse populations are controlled by selection. To estimate the effective size of the HIV population in vivo, we tested 200 pro sequences isolated from 11 HIV-infected patients for the presence of a linkage disequilibrium effect which must exist only in small populations. This analysis demonstrated a steady-state virus population of 105 infected cells or more, which is either in or at the border of the deterministic regime with respect to evolution of separate bases.
One of the most striking properties of HIV is the extent of genetic variation within the virus population in a single infected individual. A much debated issue is the degree to which the variation is controlled by deterministic (Darwinian) as opposed to stochastic effects (1). The most universal deterministic force is purifying selection caused by the fitness difference between genetic variants. The random nature of mutations and random genetic drift due to sampling of progenitor alleles (Fig. 1a) are omnipresent stochastic factors. The relative importance of deterministic and stochastic factors for virus evolution depends essentially on the size of the virus population (the number of productively infected cells). Random factors can be neglected, and deterministic theory applied, only if the population is sufficiently large. If both the population size and the fitness difference are small, selection becomes negligible compared with random drift, and “neutral” theory rules (2).
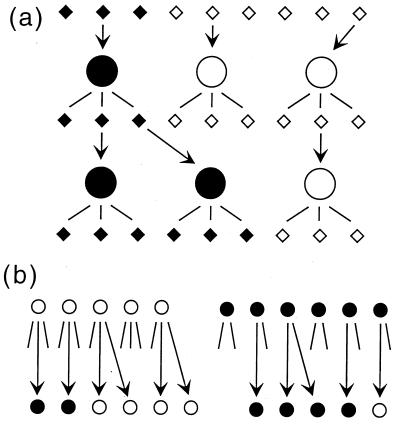
Figure 1.
(a) Random drift of genetic composition because of sampling of infecting virions. Circles denote productively infected cells; small diamonds represent free virus particles. Two genetic variants of the virus are shown as black or white. (b) A virus population model including the factors of evolution: random drift, selection, and mutation. Two consecutive generations of infected cells are shown. Lines radiating from circles denote virions produced by infected cells, some of which (shown by arrows) infect new cells. A cell infected with mutant virus (black circle) leaves fewer infectious progeny than the wild type (white circle).
Whether the steady-state HIV population in an infected individual is large enough to follow deterministic laws is currently unknown. Although existing estimates of the population size, 107 to 108 HIV RNA-positive cells (3), are much greater than the inverse mutation rate [0.4⋅10−5 to 4⋅10−5 mutations per base per cycle (4)] and are, therefore, consistent with deterministic evolution, one could imagine a few scenarios in which the effective size of the virus population is smaller than the total size. For example, not all RNA-producing cells may produce virus that can reach a target cell.
The difficulty of testing for a low population size is that the test must be model-insensitive. One could construct an enormous number of population models based on potentially important factors of evolution, including selection for diversity, coselection (epistasis) at different loci, etc. Therefore, we decided to find a striking qualitative effect that could exist only at low population sizes and that would not be affected by any kind of selection. Such an effect was predicted by Fisher (5) and Muller (6), who realized that fixation of advantageous mutations in a small population can occur at only one site at a time. These authors proposed that sexual reproduction and recombination are mechanisms that evolved to counteract this effect and, therefore, to accelerate the overall progress of evolution (7). Later, Maynard Smith estimated that that sex accelerates the speed of evolution in a broad window of population sizes around and above the inverse mutation rate (equation 11 in ref. 8). At one time point, the effect (5, 6) can be observed as almost complete (9) linkage disequilibrium between pairs of loci. By definition, linkage disequilibrium means that the frequencies at which the four possible genetic variants at two loci (haplotypes) found in the population are not equal to products of the corresponding one-locus frequencies—i.e., that loci do not segregate independently (9, 10).
Below, we introduce a simple genetic model for virus populations, summarize results of basic stochastic evolution theory which will be reviewed in detail elsewhere, describe the linkage disequilibrium test, and apply it to two sequence databases, of pro (11) and env (12).
One-Locus Model of Virus Populations
We start from the simplest one-locus, two-allele model of a virus population, including the factors of random drift, purifying selection, and mutation. We neglect the effects of coselection (epistasis) and consider the evolution of separate bases. We assume that each base can be one of two genetic variants, and that the relative difference in fitness between the two variants, the selection coefficient, s, is small. We will use the terms “wild type” and “mutant” to denote the more- and the less-fit variants, respectively, in a given selective environment. The assumed absence of coselection among several bases means that small relative differences in fitness between haplotypes are additive over these bases. In this case, a sufficiently large population, once in linkage equilibrium, will maintain it—i.e., different bases will evolve independently (9, 13). [If the fitness differences are not small, the exact condition of the absence of coselection depends on whether generations are continuous or discrete (13).]
According to the model, a virus population is represented by a number of productively infected cells, N (Fig. 1b), some of which are infected by the wild-type virus and some by the mutant virus. Each cell produces a fixed number of infectious virus particles and then dies. A small relative difference in the productivity between the two virus variants, s, accounts for selection (Fig. 1b). From all the virions produced by a generation of cells, a number of virions equal to the number of infected cells is randomly chosen to infect a new generation of cells. Therefore, in this model, the total number of infected cells does not change between generations. When infecting a new cell, a genome can mutate with a small probability, μ, to the opposite genetic variant. For HIV in vivo, the mutation rate per base per cycle is in the interval μ = (0.4–4)⋅10−5, depending on the substitution (4).
Most details of the present model, including nonoverlapping generations of cells, fixed burst sizes, and the point of the replication cycle at which mutation occurs, are of no consequence when large time scales are considered. By contrast, such assumptions as two variants per base and the absence of coselection, of selection for diversity, and of recombination are essential. For most bases in pro and env, only two variants can be found in the respective databases (11, 12), justifying the first assumption. Effects of recombination and coselection will be discussed in detail below. Because the model does not include selection for diversity, it is not directly applicable to such genes as env or, probably, gag, but is expected to be a good approximation for, e.g., pro, in which, as can be inferred from the prevalence of synonymous substitutions, purifying selection is the dominant type of selection (14). Most importantly, the linkage equilibrium test is expected to be robust with respect to the population model, as we confirm below by repeating it for three different models, with and without purifying selection, and with coselection.
How To Calculate Stochastic Evolution
In this model, the frequency of mutants in the population will, in general, change slightly between consecutive generations. The change is a combined effect of (i) selection due to the difference in productivity, (ii) random drift due to random choice of infecting virions, and (iii) mutation. The aim of an evolution theory is, given the present state, predict the future—i.e., given an initial mutant frequency, calculate its value at any other time. Although this time dependence is random and cannot be predicted in the true sense, it is possible (and useful) to calculate the probability of finding the mutant frequency, at a given time, within a specified interval of values. The simplest way to approach the problem is to simulate the time dependence of the mutant frequency (many times), following the rules of the model and using a pseudorandom number generator (“Monte-Carlo” method). The diffusion approach based on the Fokker–Planck (forward Kolmogorov) equation is a more general technique, which was used in gas kinetics before it was applied to genetics (2, 5). Discrete methods of probability theory are the most general and cumbersome; they must be used, if one wishes to study long segments of genome (see refs. in ref. 15). In the present work, we used both the Monte-Carlo method and the diffusion approach and checked that they agreed with each other and with the original results on stochastic evolution (2, 5, 16).
Three Regimes of Evolution
Most of the ongoing debate on the effective HIV population size implies, as a self-evident matter, that there are only two intervals of population size in which either random drift or selection dominates (1, 17). In fact, the leading factors and observable behavior of evolution differ significantly in three broad intervals of population size (“regimes”), with boundaries given by the inverse selection coefficient and the inverse mutation rate. (The biological meaning of the larger boundary is that, at this point, one mutation occurs, on average, in the entire population per generation.) For example, for a substitution with the selection coefficient s = 0.01, and for a mutation rate in vivo μ ≈10−5 (4), selection is negligible if there are less than 1/s = 102 infected cells (the neutral limit), and random drift is a small correction if there are more than 1/μ = 105 cells (the deterministic limit). The crossover between the two limits occurs very gradually over a broad interval of population sizes. In this “selection–drift regime”, weakly diverse populations are controlled mostly by random drift, whereas highly diverse populations are “almost deterministic” and are controlled by selection. The characteristic copy number of the minority allele separating the “weakly” and the “highly” diverse is also the inverse selection coefficient (in our example, 1/s = 102 cells). The existence of the border in the gene copy number separating stochastic and deterministic behavior was noticed by Maynard Smith (8).
Given the initial conditions and rules of the population model, we can simulate a typical random time dependence of the mutant frequency. In the model described above, independent of initial conditions, the population sooner or later arrives at a dynamic steady state. The way this transition occurs and properties of the steady state can be used, in principle, to determine the interval of population size. We illustrate this point for three important types of initial conditions: (i) 100% wild type, (ii) 100% mutant, (iii) 50%–50%. The respective experiments are the accumulation of mutants, the reversion of a mutant population (fixation of an advantageous allele), and growth competition between two virus variants.
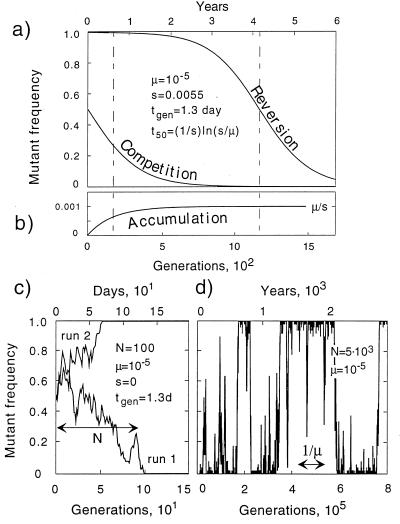
Fig. 2 a and b shows results of the three simulated experiments in the deterministic limit for a realistic set of parameters. The mutant frequency in the steady-state population approaches μ/s (in our example, 10−3). The time scale of the transition to steady state is proportional to the inverse selection coefficient for all three experiments, but it has an additional large factor, ln(s/μ), in the case of reversion. In the opposite limit of small populations (neutral regime), “growth competition” between two alleles yields a ragged curve that depends on a random simulation run (Fig. 2c). One of two competitors is always driven to extinction, which happens at an average generation number equal to the population size. The extinct allele reappears later because of mutation. Fig. 2d shows a “reversion” or “accumulation” experiment (which, in this regime, is the same) on a much longer time scale than used in Fig. 2c. The extinct allele is generated at random moments but then becomes extinct again because of random drift. Eventually, the allele succeeds in taking over the entire population, and the other allele becomes extinct. The resulting time dependence of the mutant frequency resembles a corrupted telegraph signal, switching back and forth between two genetically uniform states (Fig. 2d).
Figure 2.
Time dependence of mutant frequency. (a and b) In the deterministic limit, N ≫ 1/μ, for two initial compositions: 100% mutant and 50%–50% (a); 100% wild type (b). Notation: μ, the mutation rate per base per cycle; s, the selection coefficient (relative fitness difference); and tgen, the time per generation. The value of the selection coefficient, s, is chosen to have a 50% reversion at time t50 = 4 years after infection. Values of μ and tgen are the average values (4, 23, 25, 26). (c and d) In the neutral regime, N ≪ 1/s, for two representative Monte-Carlo runs in the growth competition experiment (c); and the accumulation/reversion experiment on a long time scale (d). The latter time dependence was obtained at μ = 10−3 and N = 50 and then rescaled along the horizontal axis to correspond to the values of μ and N shown.
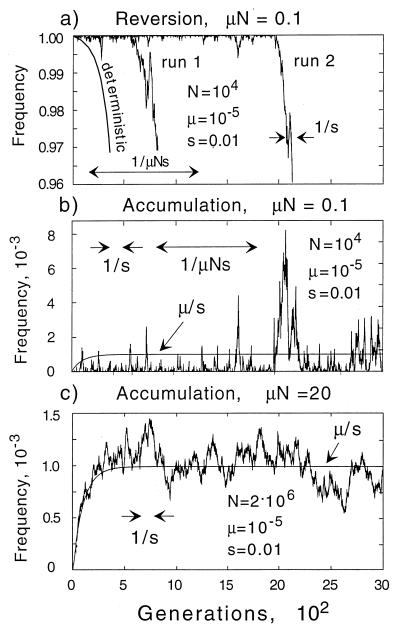
The intermediate “selection–drift” regime (1/s < N < 1/μ) has features similar to both adjoining regimes. Because selection dominates in a highly diverse population, the growth competition simulation does not differ significantly from the deterministic case (Fig. 2a), except for some small fluctuations. Reversion, however, is delayed by a random time interval as compared with the deterministic limit (Fig. 3a). There are two reasons for the delay: (i) it takes many generations to produce a single copy of wild-type genome; (ii) a typical wild-type clone is lost because of random drift soon after its birth, just as in the neutral regime. There exists an approximate critical size, equal to the inverse selection coefficient (in our example, 1/s = 102 cells), above which random drift yields to selection. If the clone passes through the critical size bottleneck, it will grow rapidly and in an “almost deterministic” fashion (Fig. 3a). Similar processes and similar time/size scales appear in the accumulation experiment (Fig. 3b), except that the size of 1/s = 102 cells is now the typical maximum size to which a mutant clone can grow before it is checked by selection. Most clones become extinct even before reaching this size; only a very few clones exceed it. As in the neutral regime, the population is uniform most of the time.
Figure 3.
Simulated dependence of the mutant frequency on time at population sizes N ≫ 1/s. (a) The beginning part of the reversion experiment in the selection–drift regime, 1/s ≪ N ≪ 1/μ. Two representative Monte-Carlo runs are shown. (b) The accumulation experiment in the selection–drift regime. (c) The accumulation experiment in the “almost deterministic” regime, N ≫ 1/μ. In all panels, smooth curves correspond to the deterministic limit (infinite N).
Linkage Disequilibrium Test
As the above discussion shows, the kinetics of appearance and disappearance of mutations depends strongly on the population size N. To estimate the effective HIV population size in vivo, we conducted a test based on the genetic variation at close pairs of highly diverse sites. As follows from the simulation examples above (Figs. 2 and 3), a site cannot preserve a high diversity indefinitely. Early in infection, the HIV population is almost uniform genetically or comprises a limited number of sequences, because of a transmission bottleneck and early competition between clones (12, 18–20). Therefore, highly diverse sites are sites that are caught in the act of “reversion” from mutant to wild type (14). Let us select two such bases and classify all sequences in the population into four groups (haplotypes): ab, Ab, aB, and AB, where the lower- and uppercase letters denote mutant and wild type, respectively, at a corresponding site. During reversion, the population starts from an almost uniform haplotype ab and arrives at an almost uniform haplotype AB. The two other haplotypes are transient. The idea of the test is that, deep in a stochastic regime, and given a limited sample size, one of the four haplotype groups will be empty at any time, because the time at which reversion ensues is random (Fig. 3a).
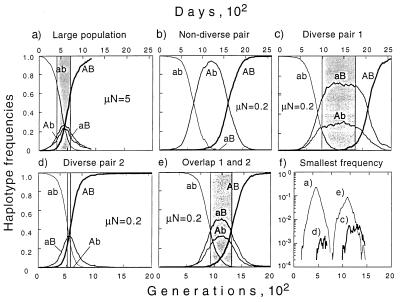
Two sites can be diverse at the same time only if they revert in approximately the same time frame. In the deterministic limit (Fig. 2a), the latter condition means that the selection coefficient must be similar for the two bases. A Monte-Carlo simulation of the time dependence of haplotype frequencies in an “almost deterministic case” is shown in Fig. 4a. Note that there exists a time interval (shaded) when all four haplotypes are well represented. Suppose now, that the population is deep in the selection–drift regime. Two sites revert typically at different random times, even if their selection coefficients are equal (Fig. 4b). Nearly simultaneous reversion can happen accidentally, according to one of two scenarios. In scenario I, independent mutations at two sites create two clones, Ab and aB. By chance, both clones pass through the critical size bottleneck (1/s, above) at approximately the same time. After the two clones outgrow the initial variant ab, they have to share the population, until another mutation within one of them generates a clone AB that succeeds in passing through the bottleneck (Fig. 4c). In scenario II, a mutation at one of two sites generates a clone, either Ab or aB. By chance, soon after the clone passes through the bottleneck, a second mutation within this clone generates clone AB (Fig. 4d). All pairs that we select must belong to one of these scenarios. In either scenario, the number of well represented subclones does not exceed three at any time point (Fig. 4 c and d). The fourth subclone can be abundant only if the second mutation in scenario I occurs unusually early—i.e., only if scenarios I and II overlap (Fig. 4e).
Figure 4.
Computer simulation of the reversion at two sites in the selection–drift regime (1/s ≪ N ≪ 1/μ). The four curves in a–e are frequencies of four haplotypes. A and B denote the first and second site, A and a denote wild type and mutant. The selection coefficient, s, is equal for the two sites. Parameter values: all panels are obtained at s = 0.1, N = 5,000, and rescaled along the time axis to correspond to s = 0.01. The mutation rate μ = 10−3 in panel a and 4⋅10−5 in b–f. Runs like that in panel b are the most frequent, pattern c is less frequent, and patterns d and e are least frequent. Gray shading shows the time interval in which both sites of a pair have mutant frequency in the interval 25–75%. Panel f shows the time dependence of the smallest haplotype frequency for the four runs a and c–e.
To test the pro data (11) for the missing haplotype effect, we selected pairs of bases (4 pairs total) such that the mutant frequency at both bases was in the interval 25–75%. For each pair, we defined four haplotypes according to consensus or anticonsensus variant at each base. (It is of no consequence for the test whether the consensus corresponds to the wild type or mutant.) Numbers of sequences in each haplotype are given in Table 1: all four haplotypes are present in three pairs of the four studied. Note that, far into the selection–drift regime, one of four haplotypes must be missing for all four pairs. The existence of a single pair with three haplotypes is within sampling fluctuations. We repeated the same test on env sequences (12). Again, of six pairs, only two were missing one of the haplotypes (Table 1). Therefore, the effective population of HIV must be either in the deterministic regime or, at least, at its border. This qualitative conclusion is expected to be rather model-independent because it is based only on the universal expectation that a small population is genetically uniform at a base for most of the time. Indeed, whatever the selective conditions may be, minority alleles are expected to appear very infrequently and be very soon cleared by random drift.
Table 1.
Distribution of sequences among four haplotypes for a few highly diverse pairs of sites in the HIV genome
| Site nos. | Pair
|
Sample size | |||
|---|---|---|---|---|---|
| a–a | a–c | c–a | c–c | ||
| pro | |||||
| 21, 174 | 1 | 6 | 6 | 1 | 14 |
| 21, 201 | 4 | 3 | 3 | 4 | 14 |
| 114, 209 | 0 | 6 | 14 | 3 | 23 |
| 174, 201 | 2 | 5 | 5 | 2 | 14 |
| env | |||||
| −17, −2 | 3 | 4 | 3 | 5 | 15 |
| −2, 3 | 0 | 6 | 4 | 5 | 15 |
| −17, 3 | 1 | 6 | 3 | 5 | 15 |
| 3, 10 | 0 | 4 | 5 | 6 | 15 |
| −2, 10 | 2 | 4 | 3 | 6 | 15 |
| −17, 10 | 2 | 5 | 3 | 5 | 15 |
Three samples (of 14, 23, and 15 sequences) were isolated from three HIV-infected individuals. pro and env (V3 region) sequences were obtained from refs. 11 and 12, respectively. The GenBank accession nos. for ref. 12 are M84240–M84314. Letters “a” and “c” in the first line denote consensus and anticonsensus, with respect to the sample consensus. Site numbers in the first column for pro are standard nucleotide positions; for env, they are codon positions counted from the GPG crown of the V3 loop (the first G is numbered 0).
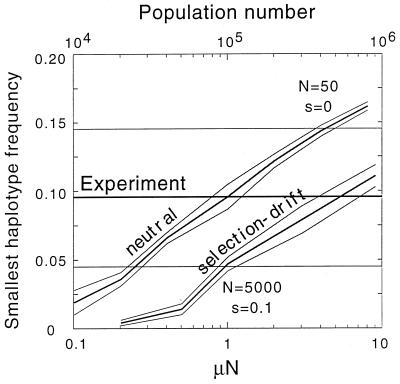
To obtain a more quantitative estimate of the population size, we calculated the least abundant haplotype frequency and compared it with the observed data. A complete linkage disequilibrium effect is expected only in the limit of very small μN. At finite μN, there will be a finite quantity of the fourth (least represented) haplotype in the population. In Fig. 4f, we compare the time dependence of the least represented haplotype frequencies between representative Monte-Carlo runs (Fig. 4 a–e). We averaged the least-represented haplotype frequency over a few hundred runs at different N (Fig. 5 and legend). The average experimental value was obtained by combining data on pro and env (Table 1). We used the mutation rate μ = 10−5, which is the log intermediate between the rate of transitions A → G or C → T and the rate of opposite transitions (4). We found that the population size is certainly (P = 0.05) larger than 9⋅104 infected cells, with the most likely value larger than 5⋅105 infected cells (Fig. 5).
Figure 5.
Dependence of the average frequency of the least-represented haplotype on the population number. Crossover from the deterministic, N ≫ 1/μ, to a stochastic, N ≪ 1/μ, regime is shown for two values of the selection coefficient: s = 0.1, and s ≪ 10−5. Only simulation runs in which the genetic composition at each site is in the interval 25–75% in a time interval (cf. panels c–e in Fig. 4) are used for averaging, with different runs weighed accordingly to the length of this time interval (i.e., same selection criterion as in experiment). Thick lines are the average; thin lines show the 95%-confidence region. Dependence on μN was obtained by varying μ at a fixed population size: N = 5⋅103 for the selection–drift regime, and N = 50 for the neutral regime. Population numbers shown in the upper horizontal axis correspond to the fixed mutation rate μ = 10−5. The thick horizontal line and shaded band are the experimental average and the 95%-confidence region obtained from data in Table 1.
We repeated the above calculation for the neutral limit, which takes place if the selection coefficient is less than the mutation rate 10−5. In this case, the selection–drift regime does not exist, and the neutral regime at N < 105 crosses over directly to the deterministic, selectionless regime at N > 105. The results are shown in Fig. 5. They predict a population of more than 2⋅104 infected cells with P = 0.05, which has the same order as the estimate obtained above for the selection–drift model. This result confirms the model-independent nature of the linkage disequilibrium test. A more complete confirmation of model-independence would require consideration of a few more population models, especially those including selection for diversity.
In principle, recombination during HIV replication could produce the missing fourth haplotype even in a small population. The fact that recombination occurs in vivo is well documented (21, 22). Therefore, before drawing final conclusions, we have to include recombination into our calculations. Obviously, recombination does not affect the genetic composition of separate sites, but only redistributes already existing alleles among different sequences. Therefore, to obtain a diverse pair of sites, two clones, Ab and aB, still have to be generated within the ab population by two independent point mutations as described above (Fig. 4c). The role of recombination is to generate clone AB from clones Ab and aB. All four clones will be present at one time point only if this event happens early, before clone ab disappears (Fig. 4e). In other words, the effect of recombination is equivalent to the effect of a second point mutation, Ab → AB or aB → AB. Therefore, results shown in Fig. 5 apply to the case with recombination as well, except that the point mutation rate, μ, has to be replaced by the effective mutation rate, which includes both point mutations and recombination, as given by μeff = μ + (1/8)〈4fAbfaB〉rLfcoinf, where r is the recombination rate per base per cycle, L is the distance between the two sites, fAb and faB are frequencies of corresponding single mutants, fcoinf is the (small) fraction of double-infected cells among all productively infected cells; 〈 … 〉 denotes averaging over random trials; and the prefactor 1/8 is the combined probability of having a heterozygous pair of proviruses, packing a heterozygous pair of genomes into a virion, and producing the recombinant AB rather than ab.
Let us estimate parameters in the above equation. The average recombination rate for HIV in vivo can be estimated as r = 4⋅10−5 per base per cycle (21). The average pair separation from data in Table 1 is L = 71. Since direct data on the coinfection frequency are not available, we estimate parameter fcoinf indirectly, from the tempo of T cell turnover and from the number of productively infected cells in an average individual. As follows both from the kinetics of T cells in humans and from the T cell turnover rate in simian immunodeficiency virus (SIV)-infected animals, 2–5% of T cells in an infected individual are replaced daily, which correspond to, at least, 109 cells per day in an individual with 200 CD4+ cells per μl of blood (23, 24). In an average individual, ≈4⋅107 cells are productively infected at any one time (3). The average lifetime of a productively infected cell is 1–2 days (23, 25, 26). Assuming that most of the T cells produced pass through a phase permissive for virus replication, we obtain that, after a permissive cell is generated, it has less than 3% chance to be infected before it dies. We assume also that infected cells are randomly chosen among permissive cells, and that the time a cell remains permissive is 0.5 day or longer. From the Poisson distribution, we obtain that the fraction of double-infected cells among all infected cells cannot exceed fcoinf = (2 day/0.5 day)⋅(3%/2) ≈ 6%, even if superinfection resistance is absent. Using the cited values of r, L, and fcoinf and estimating the average 〈4fAbfaB〉 (which cannot exceed 1) as approximately 0.5, from the above formula for μeff we obtain that the existence of recombination increases the effective mutation rate by less than a factor of 2. The presence of superinfection resistance can only lower this value. Therefore, recombination does not significantly alter estimates of the effective population size. Note that the calculation of the doubly infected cell frequency involved a few assumptions which, although plausible, are not derived from direct data. A direct quantitation of doubly infected cells would be very useful not only for finding out the effective population size but, in a more general sense, for evaluating the importance of recombination in HIV infection.
Four additional points should be made. (i) We have assumed that selection coefficients at two sites, sA and sB, are equal. As we checked numerically, at μN = 1, the difference between sA and sB by a factor of 2 makes a variable pair much less likely to appear (in 4.6% of 1,800 Monte-Carlo runs vs. 66% of 50 runs at sA = sB), because the two sites tend to revert in different time frames. The least haplotype frequency averaged over the runs with variable pairs does not change much (0.051 ± 0.013 vs. 0.044 ± 0.009). (ii) Coselection enhances linkage disequilibrium, leading to underestimation of the population size by the above test. We modified the test to assume either strong positive selection (the fitness difference between AB and Ab/aB is twice as large as between Ab/aB and ab), or strong negative coselection (Ab/aB and AB have the same fitness). We found that both positive and negative types of coselection lower the least haplotype frequency at μN ≈1 and, respectively, elevate the above estimate of the population size, by an order of magnitude. (iii) We have assumed that the initial virus population (at steady state) is 100% ab—i.e., mutant at both bases. In principle, there may be a small, undetectable admixture of the other three haplotypes that can later amplify due to selection. Can this effect increase the abundance of the fourth haplotype in a stochastic regime, μN ≪ 1? The answer is negative. For example, if haplotypes Ab and aB preexist in small quantities, a simulation in the selection–drift regime at μN = 1 shows that the average frequency of the fourth haplotype either stays approximately the same or even declines, depending on whether the initial quantities of Ab and aB are similar or differ significantly. The corresponding “fourth haplotype” frequencies at different initial admixtures of Ab and aB are 0.047 ± 0.005 at 0% and 0%; 0.061 ± 0.018 at 1% and 5%; and 0.029 ± 0.005 at 2% and 2%. (iv) We have assumed that a typical HIV population is a single “well-stirred pot” of infected cells and far-travelling virus particles. In principle, the population could consist of several or even many weakly connected subpopulations, each contributing to total virus load. The average haplotype frequencies measured in the experiment would be then affected by the time overlap between reversion processes in different subpopulations. This could explain the presence of all four haplotypes. At the same time, if the probability for a cell being infected by a virion from another subpopulation were smaller than the mutation rate ≈10−5, subpopulations would be genetically isolated, and the effective population size would be that of a separate subpopulation and, possibly, very small. Two separate facts argue against this scenario. First, the rapid flow of virus particles into blood shows that a considerable part of virions travel far from their producing cells, as opposed to being trapped locally (27). Second, visualization of separate HIV genetic variants in spleen by selective labeling shows that, although infected cells, indeed, concentrate in separate islands, most of islands are shared by different variants (28). Therefore, the speckled pattern is likely to result from a nonuniform supply of permissive cells (i.e., in germinal centers), rather than from a cell-to-cell mode of infection spread, as suggested by some authors (26, 29).
Our results differ from recent estimates of the effective population size by Leigh-Brown (1). By applying a “neutrality test” proposed by Tajima (17) to data on genetic variation in env, a portion of which was available to us and used here (12), this author concluded that the virus population is within the neutral regime and estimated an effective population as small as 1,000 or even 100 infected cells. Possible reasons for the discrepancy between this estimate and our result are as follows. (i) As shown by Waterson (15), the neutral model predicts that the average number of diverse sites in a sample of sequences grows, roughly, as a logarithm of the sample size. The “neutrality test” (17) checks whether samples of different size agree with this dependence within the predicted statistical interval. Common sense suggests that, to select a theoretical model based on a quantitative prediction, one must show that this prediction is statistically distinct from predictions of alternative models. (ii) The tested average dependence on the sample size is very weak, and the predicted statistical error is extremely large—of the same order of magnitude as the average (15).
Attempts to use the absolute number of segregating sites in a sample of sequences to measure the effective population size (1, 30) are also based on the a priori assumption that the neutral model applies. The presence of selection may decrease the number of segregating sites. If one uses the neutral model formalism in which the number of segregating sites is proportional to the population size, this effect will be misinterpreted as a small population size. Recent work (31, 32) allows us to hope that the coalescent method, which is efficient for study of evolution at multiple loci in the neutral model (30), will be in the future generalized to account for effects of selection.
Our conclusion about a relatively weak role of stochastic effects is restricted to evolution of separate bases in the steady-state HIV population in an individual. There are other aspects of HIV evolution in which randomness is expected to be important. This includes multiple substitutions, for which the deterministic “floor” of the effective population size is much larger than 105. Even for separate substitutions, random factors enter the picture at the level of transmission between individuals, due to random sampling of infecting inoculum from the infection source and genetic difference between individuals, such as pseudorandom variation of MHC I subtypes. Genetic bottlenecks created by highly active antiviral therapy are another potential source of stochastic effects. Further study of HIV populations in vivo will be needed to sort out these complex issues.
Acknowledgments
We thank J. Felsenstein for helpful comments and discussion. This work was supported by Grant R35 CA 44385 from the National Cancer Institute. J.M.C. was a Research Professor of the American Cancer Society.
Footnotes
A Commentary on this article begins on page 10559.
References
- 1.Leigh-Brown A J. Proc Natl Acad Sci USA. 1997;94:1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimura M. Population Genetics, Molecular Evolution, and the Neutral Theory. Selected papers. Chicago: Univ. of Chicago Press; 1994. [Google Scholar]
- 3.Haase A T, Henry K, Zupancic M, Sedgewick G, Faust R A, Melroe H, Cavert W, Gebhard K, Staskus K, Zhang Z-Q, et al. Science. 1996;274:985–989. doi: 10.1126/science.274.5289.985. [DOI] [PubMed] [Google Scholar]
- 4.Mansky L M, Temin H M. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher R A. The Genetical Theory of Natural Selection. Oxford: Clarendon; 1958. [Google Scholar]
- 6.Muller H J. Am Nat. 1932;66:118–128. [Google Scholar]
- 7.Felsenstein J. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maynard Smith J M. J Theor Biol. 1971;30:319–335. doi: 10.1016/0022-5193(71)90058-0. [DOI] [PubMed] [Google Scholar]
- 9.Lewontin R C. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill W G, Robertson A. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]
- 11.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmes E C, Zhang L Q, Simmonds P, Ludlam C A, Brown A J L. Proc Natl Acad Sci USA. 1992;89:4835–4839. doi: 10.1073/pnas.89.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felsenstein J. Genetics. 1965;52:349–363. doi: 10.1093/genetics/52.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouzine, I. M. & Coffin, J. M. (1999) J. Virol. 73, in press. [DOI] [PMC free article] [PubMed]
- 15.Waterson G A. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 16.Haldane J B S. Trans Camb Philos Soc. 1924;23:19–41. [Google Scholar]
- 17.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Brown A J L, Simmonds P. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delwart E L, Sheppard H W, Walker B D, Goudsmit J, Mullins J I. J Virol. 1994;68:6672–6683. doi: 10.1128/jvi.68.10.6672-6683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S L, Schaker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu W-S, Temin H M. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 22.Robertson D L, Sharp P M, McCutchan F E, Hahn B H. Nature (London) 1995;374:124–126. doi: 10.1038/374124b0. [DOI] [PubMed] [Google Scholar]
- 23.Ho D D, Neumann A U, Perelson A S, Chen W, Leonard J M, Markowitz M. Nature (London) 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- 24.Mohri H, Bonhoeffer S, Monard S, Perelson A S, Ho D D. Science. 1998;279:1223–1227. doi: 10.1126/science.279.5354.1223. [DOI] [PubMed] [Google Scholar]
- 25.Wei X, Ghosh S, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, et al. Nature (London) 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 26.Haase A T. Annu Rev Immun. 1998;17:625–656. doi: 10.1146/annurev.immunol.17.1.625. [DOI] [PubMed] [Google Scholar]
- 27.Rouzine I M, Coffin J M. In: Origin and Evolution of Viruses. Domingo E, Webster R, Holland J, editors. London: Academic; 1999. pp. 225–262. [Google Scholar]
- 28.Reinhart T A, Rogan M J, Amedee A M, Murphey-Corb M, Rausch D M, Eiden L E, Haase A T. J Virol. 1998;72:113–120. doi: 10.1128/jvi.72.1.113-120.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman Z, Feinberg M B, Paul W E. Proc Natl Acad Sci USA. 1998;95:6314–6319. doi: 10.1073/pnas.95.11.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigo A G, Felsenstein J. In: Molecular Evolution of HIV. Crandall K, editor. Baltimore: Johns Hopkins Univ. Press; 1999. , in press. [Google Scholar]
- 31.Neuhauser C, Krone S M. Genetics. 1997;145:519–534. doi: 10.1093/genetics/145.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krone S M, Neuhauser C. Theor Popul Biol. 1997;51:210–237. doi: 10.1006/tpbi.1997.1299. [DOI] [PubMed] [Google Scholar]