Abstract
The proteins belonging to the Fur family are global regulators of gene expression involved in the response to several environmental stresses and to the maintenance of divalent cation homeostasis. The Mycobacterium tuberculosis genome encodes two Fur-like proteins, FurA and a protein formerly annotated FurB. Since in this paper we show that it represents a zinc uptake regulator, we refer to it as Zur. The gene encoding Zur is found in an operon together with the gene encoding a second transcriptional regulator (Rv2358). In a previous work we demonstrated that Rv2358 is responsible for the zinc-dependent repression of the Rv2358-zur operon, favoring the hypothesis that these genes represent key regulators of zinc homeostasis. In this study we generated a zur mutant in M. tuberculosis, examined its phenotype, and characterized the Zur regulon by DNA microarray analysis. Thirty-two genes, presumably organized in 16 operons, were found to be upregulated in the zur mutant. Twenty-four of them belonged to eight putative transcriptional units preceded by a conserved 26-bp palindrome. Electrophoretic mobility shift experiments demonstrated that Zur binds to this palindrome in a zinc-dependent manner, suggesting its direct regulation of these genes. The proteins encoded by Zur-regulated genes include a group of ribosomal proteins, three putative metal transporters, the proteins belonging to early secretory antigen target 6 (ESAT-6) cluster 3, and three additional proteins belonging to the ESAT-6/culture filtrate protein 10 (CFP-10) family known to contain immunodominant epitopes in the T-cell response to M. tuberculosis infection.
Mycobacterium tuberculosis is a human pathogen that infects and replicates within macrophages. This microorganism lives in phagosomes that fail to fuse with lysosomes and has adapted its lifestyle to survive and replicate in the changing environment within the endosomal system (20).
The long-recognized phenomenon of nutritional immunity, in which sequestration of iron and possibly other metals occurs as a nonspecific host response to infection (2), hints in general terms at the possibility of a keen competition between host and parasite for essential metal ions. A critical point is the bacterial ability to compete with the host for nutrients, and the acquisition of metal ions has important implications for intracellular survival.
Pathogenic bacteria respond to such limitations by inducing metabolic functions that overcome nutritional deficiencies and/or inducing virulence functions required for immediate survival and spread to subsequent anatomical sites of infection. The outcome of this competition between the host cell and the microorganism is certainly one of the most important factors determining the ability of pathogens to multiply and cause disease (41).
Metalloregulatory proteins sense the intracellular levels of specific metal ions and mediate a transcriptional response aimed at restoring homeostasis when these levels are altered. In prokaryotes, these transcriptional regulators are clustered in five distinct families: Fur (15), DtxR (34), MerR (6), SmtB/ArsR (7), and NikR (12). Typically, the reversible binding of metal ions to a metal-sensing site alters the conformation of the regulator, affecting its capability to bind to its operators. The affinity of the metal-sensing sites for a specific metal ion serves to set the intracellular concentration of the metal ion within the cell. The selectivity of the site is essential for ensuring that other metal ions do not interfere with this homeostasis mechanism (33).
The annotation of the M. tuberculosis genome sequence revealed the presence of two Fur-like proteins, FurA and a second protein, formerly annotated FurB (Rv2359). Since in this paper we show that this protein represents a zinc uptake regulator, we propose renaming it Zur (http://genolist.pasteur.fr/TubercuList/). FurA is a negative regulator of katG, and transcription of its structural gene is induced upon oxidative stress (28, 38, 48). The structural gene encoding Zur is cotranscribed with its upstream gene Rv2358, encoding a regulator of the SmtB/ArsR family (27).
Using Mycobacterium smegmatis as a model, we recently demonstrated that the transcription of this operon is regulated by Rv2358, which represses its transcription in the absence of zinc (8), suggesting a role of this protein and Zur in the regulation of zinc homeostasis. This finding is consistent with a simple model of derepression, in which Zn2+ binding by the sensor protein Rv2358 weakens the DNA binding affinity significantly, such that RNA polymerase can load and initiate transcription of the operon. The proposed Rv2358 DNA binding region contains an imperfect 12-2-12 inverted repeat, 5′-TTGACATGCATC-AT-CATGCATGTGAC-3′ (8), in agreement with other sites recognized by SmtB/ArsR-like regulators (7).
Zinc is an essential element for living organisms. It plays a vital role as a cofactor for numerous enzymes and DNA binding proteins and serves as a structural scaffold for several proteins. However, despite its physiological importance, it is toxic at high concentrations since it competes with other metals for binding to active centers of enzymes (3).
All bacteria tightly regulate zinc transport. In Escherichia coli, studies of two Zn-sensing metalloregulatory proteins (Zur and ZntR) have shown that these proteins switch off expression of the Zn2+ uptake machinery or switch on production of efflux pumps when free Zn2+ exceeds the extraordinarily low threshold of 0.5 fM (35). Zur regulates the high-affinity uptake system znuACB (37), while ZntR is involved in Zn2+ detoxification (7). In Bacillus subtilis, however, Zur regulates both zinc uptake (16) and zinc mobilization (1, 31).
In this work we describe an M. tuberculosis zur knockout mutant and characterize its phenotype. Moreover, using DNA microarrays we identify 32 genes upregulated in the mutant, 24 of which (belonging to eight putative transcriptional units) are directly regulated by Zur, which was shown to be a zinc-sensing transcriptional repressor. In contrast to Rv2358, which binds its operator in the absence of zinc, Zur binds its operator in the presence of this metal.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All experiments were performed with M. tuberculosis H37Rv. Bacteria were grown in either liquid Middlebrook 7H9 medium or solid Middlebrook 7H10 medium (Difco) supplemented with ADN (2% glucose, 5% bovine serum albumin, 0.85% NaCl) and 0.05% Tween 80. Liquid cultures were grown in roller bottles at 37°C with gentle rotation (∼10 rpm). Plates were incubated at 37°C in sealed plastic bags.
M. smegmatis strains were grown in liquid Middlebrook 7H9 or solid Middlebrook 7H10 medium (Difco) 7H10 supplemented with Middlebrook oleic acid-albumin-dextrose-catalase at 37°C. For studies of promoter regulation mediated by zinc, M. smegmatis strains were grown on Sauton medium treated with Chelex 100 resin (Sigma) as previously described (27).
E. coli strains JM109 and HB101 were grown in Luria broth (Difco) at 37°C. When required, antibiotics were added at the following concentrations: kanamycin, 50 μg ml−1; ampicillin, 100 μg ml−1; streptomycin, 20 μg ml−1; hygromycin, 100 μg ml−1 (E. coli); kanamycin, 20 μg ml−1; streptomycin, 20 μg ml−1; hygromycin, 100 μg ml−1 (M. tuberculosis and M. smegmatis).
Bioinformatics.
The complete M. tuberculosis H37Rv genome sequence is available at http://genolist.pasteur.fr/TubercuList/.
DNA alignment was performed with the ClustalW program (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page = /NPSA/npsa_clustalwan.html).
The WebLogo program (http://weblogo.berkeley.edu/) was used to build a Zur consensus sequence logo (43).
DNA manipulations.
All recombinant DNA techniques were performed by standard procedures, with E. coli HB101 used as the initial host. DNA restriction and modifying enzymes were obtained from New England Biolabs and used according to the manufacturer's suggestions.
Construction of an M. tuberculosis zur knockout strain.
To disrupt zur, we cloned a 783-bp PCR fragment containing the entire gene in the suicide vector pSM270, a vector containing both sacB (conferring sucrose sensitivity) and a cassette conferring streptomycin resistance (24). The sequences of the primers used for amplification are the following: upper primer (furB1), 5′-TCCCCGCACCACCGCC-3′; lower primer (furB2), 5′-GCGCCGAACGTGCCCT-3′. The zur gene was disrupted by introducing a 1.7-kb cassette conferring hygromycin resistance into the unique BspI restriction site internal to the zur gene. The construct was electroporated into M. tuberculosis H37Rv with selection for hygromycin followed by selection for sucrose resistance, which would result from the loss of the plasmid backbone containing sacB. Hygromycin- and sucrose-resistant colonies were analyzed for streptomycin sensitivity to confirm the loss of the plasmid and analyzed by Southern blotting to confirm zur disruption.
Electroporation of M. tuberculosis.
Bacteria were grown in 30 ml of Middlebrook 7H9 to reach mid-exponential phase. The culture was centrifuged at 5,000 × g for 5 min at room temperature and washed twice with 1 volume of sterile 10% glycerol. The pellet was then resuspended in 1 ml of 10% glycerol, centrifuged, and resuspended again in 800 μl of 10% glycerol. Next, 50 μl of concentrated cells was mixed with 2 μg of DNA. Samples were transferred to 0.2-cm gap cuvettes (Eppendorf) and electroporated with the Electroporator 2510 (Eppendorf) (capacitance, 10 μF; voltage, 12.5 kV cm−1; resistance, 600 Ω). After the pulse, the cells were diluted in 1 ml of 7H9, incubated for 24 h at 37°C, and finally plated on selective solid medium.
Determination of growth inhibition by disk diffusion assay.
M. tuberculosis strains were grown to early exponential phase, and 100 μl of culture containing 3 × 106 CFU was spread on 20-ml 7H10 plates. Paper disks containing 10 μl of the inhibitory reagent were placed on the top of the agar. Stock concentrations were the following: EDTA, 0.5 M; plumbagin, 5 M; diamide, 2 M; hydrogen peroxide, 13%; cumene hydroperoxide, 40 mM; sodium dodecyl sulfate (SDS), 10%. The diameter of the inhibition zone was measured after 15 days of incubation at 37°C. Plumbagin was dissolved in 96% ethanol, and an experiment with ethanol only was performed as a negative control.
RNA extraction.
Appropriate strains were inoculated in 30 ml of Middlebrook 7H9, grown to mid-exponential phase, centrifuged at 4,500 × g for 5 min at room temperature, and frozen on dry ice. RNA extraction was performed as previously described (24). The frozen cell pellets were suspended in 1 ml of TRIzol reagent (Gibco-BRL) and transferred to 2-ml screw cap tubes containing 0.5 ml of 0.1-mm-diameter zirconia/silica beads (BioSpec Products). Cells were disrupted with two 30-s pulses in a Mini-Bead-Beater (BioSpec Products). After 5 min of incubation at room temperature, samples were centrifuged at maximum speed for 45 s and the supernatants were transferred to 2-ml Heavy Phase Lock Gel I tubes (Eppendorf) containing 300 μl of chloroform-isoamyl alcohol (24:1), inverted rapidly for 15 s, and incubated for 2 min. The samples were centrifuged for 5 min, and the aqueous phase was added to 270 μl of isopropanol. After the addition of 270 μl of a high-salt solution (0.8 M Na citrate, 1.2 M NaCl), samples were incubated overnight at 4°C and finally centrifuged at maximum speed for 10 min at 4°C. The RNA pellets were washed with 1 ml of 75% ethanol, centrifuged for 5 min, and air dried. RNA pellets were resuspended in 0.1 ml of DNase I 1× buffer containing 4 units of DNase I (Ambion). After 30 min of incubation at 37°C, the RNA was finally purified with an RNeasy column (QIAGEN).
RT-PCR.
Reverse transcription was performed with random primers using murine leucoblastoma virus retrotranscriptase (MULV-RT) (Applied Biosystems). Briefly, 500 ng of RNA was denatured at 98°C for 2 min in the presence of the appropriate volume of water and then chilled on ice. The RNA sample was used to prepare 25 μl of annealing mixture (5.5 mM MgCl2, 0.55 mM [each] deoxynucleoside triphosphates [dNTPs], 0.25 mmol random hexamer; 32 U of MULV, 10 U of RNase inhibitor, and 1× reaction buffer [Applied Biosystems]). Samples were then incubated at 25°C for 10 min, at 45°C for 50 min, and finally at 95°C for 5 min to allow the annealing of the random hexamers. Quantitative PCR was performed with SYBR green master mix (Applied Biosystems). After 10 min at 95°C to activate the enzyme, 40 amplification cycles were performed with an Applied Biosystems 7700 Prism spectrofluorometric thermal cycler (Perkin-Elmer) under the following conditions: 1 min of denaturation at 95°C, 30 s of annealing at 64°C, and 30 s of extension at 72°C. Fluorescence was measured during the annealing step and plotted automatically for each sample. Results were normalized to the amount of sigA mRNA, as previously described (23). RNA samples that had not been reverse transcribed were included in all experiments to exclude significant DNA contamination. For each sample, melting curves were performed to confirm the purity of the products. Sequences of the primers for quantitative RT-PCR are available upon request.
Preparation of labeled cDNA.
Fluorescently labeled cDNA copies of total RNA were prepared by direct incorporation of fluorescent nucleotide analogues during a first-strand reverse transcription reaction. Each 25.5-μl labeling reaction mixture included 1.8 μg of RNA; 172.5 ng/μl of a random hexamer mix (Invitrogen); 0.5 mM (each) dATP, dGTP, and dCTP; 0.02 mM dTTP; 10 mM dithiothreitol (DTT); and 200 U of RT (Superscript II; Invitrogen) in a 1× reaction buffer provided by the enzyme manufacturer plus 1.5 nmol of either Cy3-dUTP or Cy5-dUTP (Amersham Pharmacia Biotech). The RNA and random hexamers were preheated to 98°C for 2 min and snap cooled on ice before the addition of the remaining reaction components. The RT reaction was allowed to proceed for 10 min at 25°C followed by 90 min at 42°C. The Cy3- and Cy5-labeled products were purified in pairs with the CyScribeGFX purification kit (Amersham Biosciences) and concentrated in Microcon YM-30 centrifugal filter devices (Millipore).
Microarray hybridization and data analysis.
M. tuberculosis oligoarrays were obtained from the Center for Applied Genomics, International Center for Public Health (Newark, NJ). These microarrays consisted of 4,295 70-mer oligonucleotides representing 3,924 open reading frames from M. tuberculosis strain H37Rv and 371 unique open reading frames from strain CDC1551 that are not present in the H37Rv strain's annotated gene complement. Prior to hybridization, microarray slides were incubated at 42°C for 1 h in prehybridization buffer (2.8% bovine serum albumin, 0.1% SDS), washed twice in MilliQ water for 2 min and then in isopropanol for two additional minutes, and then air dried. Probes were applied to the array in 10 μl of hybridization solution (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% SDS, 25% formamide, and 0.5 mg/ml tRNA). Samples were first denatured by heating at 98°C for 2 min. Hybridization was carried out under a glass coverslip in a humidified slide chamber submerged in a 50°C water bath for approximately 18 h. The coverslip was removed by incubation for 1 min in wash buffer I (2× SSC, 0.1% SDS), and slides were then washed sequentially in buffer II (1× SSC, 0.05% SDS) and twice in buffer III (0.06× SSC) for 2 min at room temperature. Finally, slides were dried by centrifugation (100 × g, 2 min) and immediately scanned with a 428Array Scanner (Affimetrix). Hybridizations were performed with RNA extracted from four different biological samples. Each sample was hybridized twice through reverse labeling of the respective cDNAs. Fluorescence intensities of Cy3 and Cy5 dyes at each spot were quantified with ImaGene software, version 5.0 (BioDiscovery, Inc.), and data obtained from qualified spots on each chip were normalized with the print-tip Lowes implementation procedure included in GEPAS, version 1.1 (http://gepas.bioinfo.cnio.es/) (47). The expression ratio for the wild-type and mutant genes was determined from the normalized fluorescence intensity and was calculated as the average change of the different experiments. For data mining, significance analysis of microarrays (SAM) was applied (46). We accepted only genes which were up-regulated at least 1.7-fold, with a q value ≤1%. The q value is the equivalent of the P value after multiple-testing correction.
DNA-binding assays and fooprinting.
M. tuberculosis Zur, overexpressed in E. coli XL1-Blue and purified as described previously (27), was used in electrophoretic mobility shift assays (EMSA) and DNase I footprinting experiments. The recombinant Zur contains two zinc ions per protein monomer, as previously described (27). When zinc-free Zur was required, the purified protein was dialyzed in 50 mM EDTA, as previously described (27). In the EMSA, DNA fragments containing putative promoter regions were labeled with [γ32P]dATP by using Ready-To-Go T4 polynucleotide kinase (Amersham) and used as probes. Then, 10 μl of binding reaction mixture (20 mM Tris-HCl [pH 8.0], 50 mM KCl, 1 mM DTT, 50 μg/ml bovine serum albumin, 50 μg/ml salmon sperm DNA, 5% glycerol), containing 10 fmol of labeled probe, was incubated with purified Zur protein (800 ng, corresponding to 54 pmol of monomeric protein) for 20 min at room temperature. Reaction mixtures were loaded onto a nondenaturing 6% polyacrylamide gel containing 1× TA (40). Gels were run at 140 V at room temperature, dried, and exposed to Hyperfilm (Amersham). When EDTA was added to the binding reaction (final concentration, 400 μM), a running gel containing 1× Tris-acetate-EDTA was used.
For DNase footprinting experiments, pGEM-T Easy derivatives, containing the putative Zur-regulated promoters, were digested at one end of the cloned regions, radiolabeled with exonuclease-free Klenow (USB) and [α-32P]dCTP, and then digested with a second enzyme at the other end to excise the cloned fragment.
Binding reactions were performed as described for EMSA, with 50,000 cpm of the purified probe in a final volume of 50 μl. After 20 min of incubation at room temperature, 50 μl of a room temperature solution of 5 mM CaCl2 and 10 mM MgCl2 was added, followed by the addition of 0.015 to 0.030 U of RQ1 RNase-free DNase (Promega). The reaction mixtures were incubated for 5 min at 37°C. Reactions were stopped by the addition of 50 μl of stop solution (0.1 M EDTA, 0.8% SDS, 1.6 M CH3COONH4, 0.3 mg/ml salmon sperm DNA), and DNA was precipitated with 350 μl of ethanol, dried, and resuspended in loading buffer (1:2 0.1 M NaOH/formamide [vol/vol], 0.1% xylene cyanol, 0.1% bromophenol blue). Samples were loaded onto a 7 M urea-9% polyacrylamide sequencing gel. Maxam-Gilbert A+G sequencing reactions were performed as previously described (26).
5′ RACE.
For 5′ rapid amplification of cDNA ends (RACE), 1 μg of M. tuberculosis RNA and 1 μg of primers (reported in Table 1) were incubated at 70°C for 10 min and then at 42 to 50°C for 1 h in the presence of 1× avian myeloblastosis virus (AMV)-RT buffer, 1 mM dNTPs, 10 U of RNase inhibitor (Promega), and Durascript-enhanced AMV-RT (Invitrogen). Finally, the reaction was precipitated and incubated at 37°C for 30 min in the presence of 2 mM dATP and 18 U of terminal deoxynucleotidyl transferase (Amersham Biosciences) to add a poly(A) tail to the 3′ end, necessary for the annealing of RA1 primer in the next amplification reaction (Table 1). Samples corresponding to 100 ng of cDNA were used in the next PCRs. To improve assay sensitivity, the reaction products were used as templates for seminested PCRs, with RA2 and an internal oligonucleotide used as primers (Table 1).
TABLE 1.
Oligonucleotides used in this work
| Primer | Sequence | Purpose |
|---|---|---|
| Rv1 | 5′-TTGGTACCTGCGGCCGGTGACTTGG-3′ | EMSA of Rv2059 and rpmB2; cloning of Rv2059 promoter region |
| Rv2 | 5′-TTGGTACCGTCCGGTGACAAGGAT-3′ | EMSA of Rv2059 and rpmB2; cloning of Rv2059 promoter region |
| Rv0106-1 | 5′-CGGGATCCTACCGAAACCCACAGTG-3′ | EMSA and footprinting of Rv0106 and rpmB1; cloning of Rv0106 promoter region |
| Rv0106-2 | 5′-CGGGTACCTGACCTGCCACCAAT-3′ | EMSA of Rv0106; cloning of Rv0106 promoter region |
| Rv0282-1 | 5′-CGGGATCCCGCAACACCCTGGTC-3′ | EMSA of Rv0282; cloning of Rv0282 promoter region |
| Rv0282-2 | 5′-CGGGTACCCGCTGTCTCCTTCACC-3′ | EMSA of Rv0282; cloning of Rv0282 promoter region |
| PP1 | 5′-CGGGATCCTACGCATGACCGCTC-3′ | Cloning of Rv0280 promoter region |
| PP2 | 5′-CGGGATCCTGCGGTCGGCGCGTC-3′ | EMSA and footprinting of Rv0280 |
| PP4 | 5′-GGGGTACCGAATGCACCTCGGG-3′ | EMSA and footprinting of Rv0280; cloning of Rv0280 promoter region |
| Rv0282BAM | 5′-CGGGATCCCGGAATCCGAAGCCG-3′ | Footprinting of Rv0282 |
| Rv0282XBA | 5′-GCTCTAGAGCGACCAATCGACTC-3′ | Footprinting of Rv0282 |
| Rv2059BAM | 5′-CGGGATCCACGGCTTCGGCGATG-3′ | Footprinting of Rv2059 and rpmB2 |
| Rv0106-5 | 5′-GCTCTAGACTCCACGACCACCGTTC-3′ | Footprinting of Rv0106 and rpmB1 |
| Rv2059XBA | 5′-GCTCTAGAGCCCAGCAGGTCAGC-3′ | Footprinting of Rv2059 and rpmB2 |
| 3017c-F | 5′-TAGGATCCATTTGGTCGGTGTG-3′ | EMSA of Rv3017c |
| 3017c-R | 5′-GCTCTAGACCCGGCATGAGCCATC-3′ | EMSA of Rv3017c |
| 3019c-F | 5′-CAGGATCCCCAAGGTCAATAC-3′ | EMSA of Rv3019c |
| 3019c-R | 5′-AATCTAGACGCATAACCGGCCAT-3′ | EMSA of Rv3019c |
| F3612c | 5′-CGGGATCCAAAATGTGCACAATG-3′ | EMSA of Rv3612c |
| R3612c | 5′-GTTGTCGCGCATAGGTGAGCACAGC-3′ | EMSA of Rv3612c |
| F1195 | 5′-TTGGATCCAGATTGCACTTTGGCTC-3′ | EMSA of Rv1195 |
| R1195 | 5′-CTGGGTATGCATCACGAAAGAC-3′ | EMSA of Rv1195 |
| F2688c | 5′-TTGGATCCTTGCTCCGTATACAG-3′ | EMSA of Rv2688c |
| R2688c | 5′-GTTCCCACACGCGCCGATGCC-3′ | EMSA of Rv2688c |
| 0282-RT | 5′-GCGTCGCACTGGTCATGG-3′ | Retrotranscription of Rv0282 |
| 0106-RT | 5′-ACGATCCGGCCGACATTG-3′ | Retrotranscription of Rv0106 |
| L28-RT | 5′-CGTCGCGGTCGATGAC-3′ | Retrotranscription of rpmB1 and rpmB2 |
| Rv59-7 | 5′-GTGGTGGTCGGGATGGATG-3′ | Retrotranscription of Rv2059 |
| RA1 | 5′-GACCACGCGTATCGATGTCGAC(T)16V-3′ | 5′ RACE PCRs |
| RA2 | 5′-GACCACGCGTATCGATGTCGAC-3′ | 5′ RACE PCRs |
| 0282-4 | 5′-GTCAGCGCGGCGAAAC-3′ | 5′ RACE PCR of Rv0282 |
| 0282-3 | 5′-CCGGCGGACGTTTACG-3′ | 5′ RACE PCR of Rv0282 |
| 0106-4 | 5′-GGCGGTGCAGTCTGCG-3′ | 5′ RACE PCR of Rv0106 |
| 0106-3 | 5′-CAGCAGGTCGTCGCGG-3′ | 5′ RACE PCR of Rv0106 |
| L28-5 | 5′-AATGCGACGGCCCTC-3′ | 5′ RACE PCR of Rv2058c |
| L-28-4 | 5′-TATACCCTTCGTGGACAC-3′ | 5′ RACE PCR of Rv2058c |
| Rv59-4 | 5′-TGTTCTCGGCGGTGACAC-3′ | 5′ RACE PCR of Rv2059 |
| Rv59-5 | 5′-CCGCACGGATCCTGGT-3′ | 5′ RACE PCR of Rv2059 |
| 0105-5 | 5′-CTTGATGCGGCGGTCCTC-3′ | 5′ RACE PCR of Rv0105c |
| 0105-4 | 5′-GATTCCTTGGGCGCTGAC-3′ | 5′ RACE PCR of Rv0105c |
β-Galactosidase assays.
Promoter regions of the Rv0106, Rv2059, Rv0280 and Rv0282 genes were obtained by PCR using M. tuberculosis H37Rv DNA as the template and specific primers (Table 1). PCR fragments were digested with the appropriate restriction enzymes and cloned in the shuttle vector pJEM15 or in the integrative vector pSM128 (14, 45), upstream of a promotorless lacZ reporter gene. Independent cultures of M. smegmatis mc2155 and mcJF3, a zur mutant strain, were transformed with recombinant plasmids and grown in Sauton medium at 37°C to an optical density at 600 nm of approximately 0.8, as previously described (27). The cells were recovered and disrupted by sonication. β-Galactosidase activity was measured on cellular extracts as previously described (45).
Microarray data accession number.
Microarray data have been deposited in the Gene Expression Omnibus public database, http://www.ncbi.nlm.nih.gov/geo, under accession number GSE 5815.
RESULTS
Construction of a zur mutant in M. tuberculosis.
In order to assess the physiological role of Zur, its structural gene was disrupted in M. tuberculosis H37Rv by the insertion of a cassette conferring hygromycin resistance, as described in Materials and Methods. The resulting strain (ST129) shows a growth curve and colony morphology indistinguishable from the wild-type parental strain (data not shown).
In the M. tuberculosis genome, zur lies downstream of Rv2358, and these two genes are cotranscribed (8). In order to rule out the possibility that zur inactivation could modify Rv2358 expression, we quantified Rv2358 mRNA in the mutant and in the wild-type parental strain by using sigA mRNA as internal invariant control (23). No difference in Rv2358 expression was detected in the two strains (data not shown).
Characterization of the zur mutant phenotype.
Proteins of the Fur family function in processes ranging from the acid shock response (18) to oxygen radical detoxification (13), metal uptake (8), and toxin and virulence factor production (32). To characterize the phenotype of the zur mutant, ST129 and the wild-type parental strain H37Rv were exposed to different stressing agents and conditions. In particular, we exposed the two strains to different oxidative compounds such as hydrogen peroxide, plumbagin, diamide, and cumene hydroperoxide; the chelating agent EDTA; the detergent SDS; and the heavy metals cadmium and cobalt by disk diffusion assay on agar plates. No significant differences between the two strains were observed (data not shown). We also tested the requirement of different metals (Fe, Mn, Zn, and Mg) for growth, but also in this case we could not observe any difference between the two strains (data not shown). Finally, the zur mutant and wild-type strains were used to infect C57BL/6 mice, and the bacterial loads in lungs, spleen, and liver were monitored for 250 days. No difference between the two strains was detected, showing that, at least in this model, Zur is not required for virulence (G. M. Rodriguez et al., unpublished results).
Identification of Zur-regulated genes.
In order to identify Zur-regulated genes, mutant strain ST129 and its parental strain H37Rv were grown to mid-exponential phase in Middlebrook 7H9 supplemented with ADN (see Materials and Methods). RNA was extracted, and expression profiles were compared with DNA microarrays. A total of 32 genes included in 16 putative transcriptional units were shown to be up-regulated in ST129 compared to H37Rv (Table 2). No genes that were down-regulated in ST129 were identified. Six representative genes (Rv0106, Rv0280 [ppe3], Rv1857 [modA], Rv2058c [rpmB2], Rv2059, and Rv3017c [esxQ]) found in the microarray analysis to be transcribed at higher levels in the zur mutant than in the wild type were chosen for validation, and their expression was measured by quantitative RT-PCR using sigA as an invariant internal control. In support of the gene expression profiling data, the mRNA levels of all selected genes were clearly higher in the zur mutant strain relative to H37Rv (Table 2). Quantitative RT-PCR was also used to measure Rv0282 expression. This gene is adjacent to other induced genes, but no information about its expression was retrieved from the DNA microarray experiments for technical reasons. As shown in Table 2, this gene was induced 5.9-fold in the zur mutant strain.
TABLE 2.
Genes under Zur transcriptional controla
| Locusb | Gene | q value (%)c | Fold inductiond | Gene product/function |
|---|---|---|---|---|
| Rv0105c | rpmB1 | 0 | 2.7 | Probable 50S ribosomal protein L28 RpmB1 |
| Rv0106 | 0 | 8.1 (39.6) | Similar to low-affinity zinc transporter YciC | |
| Rv0232 | 0 | 1.9 | Probable transcriptional regulator (TetR family) | |
| Rv0280 | ppe3 | 0 | 9.6 (2.3) | PPE family protein |
| Rv0281 | 0 | 3.6 | Unknown; possibly O-methyltransferase involved in polyketide biosynthesis | |
| Rv0282e | ND (5.9) | Unknown; membrane protein similar to ESX-1 secretion system member | ||
| Rv0283 | 0 | 4.3 | Unknown; membrane protein similar to ESX-1 secretion system member | |
| Rv0284 | 0 | 3.0 | Unknown; membrane protein, contains putative FtsK/SpoIIIE family protein domain similar to ESX-1 secretion system member | |
| Rv0285 | pe5 | 0 | 3.4 | PE family protein |
| Rv0286 | ppe4 | 0 | 3.3 | PPE family protein |
| Rv0287 | esxG | 0 | 3.0 | ESAT-6-like protein |
| Rv0288 | esxH | 0 | 3.2 | ESAT-6-like protein |
| Rv0289 | 0 | 3.1 | Unknown; similar to ESX-1 secretion system member | |
| Rv0290 | 0 | 3.7 | Unknown; similar to ESX-1 secretion system member | |
| Rv0291 | mycP3 | 0 | 3.1 | Peptidase of subtilase family/membrane-anchored mycosin, similar to ESX-1 secretion system member |
| Rv0292 | 0 | 3.3 | Unknown; transmembrane protein similar to ESX-1 secretion system member | |
| Rv1195 | pe11 | 0 | 2.4 | PE family protein |
| Rv1857 | modA | 0 | 1.9 (3.3) | Lipoprotein involved in transport of molybdenum into the cell |
| Rv1870c | 0.6 | 2.1 | Unknown | |
| Rv2055c | rpsR2 | 0 | 12.8 | Probable ribosomal protein S18 RpsR2 |
| Rv2056c | rpsN2 | 0 | 22.5 | Probable ribosomal protein S14 RpsN2 |
| Rv2057c | rpmG1 | 0 | 23.3 | Probable ribosomal protein L33 |
| Rv2058c | rpmB2 | 0 | 30.1 (125) | Probable 50S ribosomal protein L28 RpmB2 |
| Rv2059 | 0 | 5.8 (22.5) | Similar to periplasmic metal binding proteins of ABC transport systems | |
| Rv2060 | 0 | 6.3 | Similar to ZnuB, ABC-type Mn2+/Zn2+ transport systems, permease components | |
| Rv2990c | 0 | 4.1 | Unknown | |
| Rv3017c | esxQ | 1.0 | 2.1 (2.8) | ESAT-6-like protein |
| Rv3019c | esxR | 0 | 3.9 | ESAT-6-like protein |
| Rv3020c | esxS | 0 | 2.5 | ESAT-6-like protein |
| Rv3022c | ppe48 | 0 | 2.2 | PPE family protein |
| Rv3229c | 0 | 2.9 | Involved in lipid metabolism, possible fatty acid desaturase | |
| Rv3612c | 0 | 2.1 | Unknown; up-regulated after 4 h and 24 h of starvation |
Genes were included in the table if their q value was ≤1 and fold induction was ≥1.9.
Genes are annotated as described by the Pasteur Institute on TUBERCULIST (http://genolist.pasteur.fr/TubercuList/). Genes downstream of a putative Zur box are shown in boldface type.
False discovery rate (probability that the gene was falsely called, calculated by SAM).
mRNA levels in the zur mutant strain ST129 divided by those in the wild-type strain H37Rv. Values in parentheses represent the fold induction obtained by quantitative RT-PCR. ND, not determined.
Included in the table based on results obtained by RT-PCR, as no data were obtained from DNA microarray experiments.
The products of Zur-dependent genes identified by this approach included a group of ribosomal proteins, three putative metal transporters, all of the proteins belonging to early secretory antigen target 6 (ESAT-6) cluster 3, and three additional proteins belonging to the ESAT-6/culture filtrate protein 10 (CFP-10) family.
Characterization of Zur DNA binding activity.
To determine which genes among those identified by DNA microarray analysis are direct targets for Zur-mediated repression, we analyzed the sequences of the regions upstream of the genes shown in Table 2. This analysis revealed the presence of a conserved AT-rich palindromic sequence upstream of eight genes (rpmB1, Rv0106, Rv0280, Rv0282, Rv2059, rpmB2, esxQ, and Rv3019c) (Fig. 1 and Table 2), suggesting a common regulatory mechanism. Interestingly, all of these genes were located immediately upstream of other induced genes (Table 2). It is noteworthy that two couples of these genes, rpmB1-Rv0106 and rpmB2-Rv2059, are adjacent in the genome and oriented divergently; in their intergenic region are thus present two conserved palindromic sequences.
FIG. 1.
ClustalW alignment of AT-rich palindromic regions upstream of eight up-regulated genes in mutant strain ST129. The numbers on the right indicate positions with respect to the putative translational start codon.
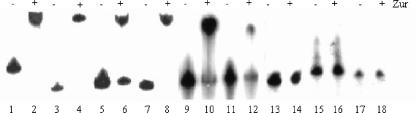
In order to determine whether this palindromic sequence is indeed recognized by Zur, we performed EMSA using γ-32P-labeled PCR-amplified DNA fragments. As shown in Fig. 2 (lanes 1 to 12), EMSA clearly demonstrated that Zur was able to retard the migration of the six DNA fragments containing the palindromic sequence (two palindromic sequences in the case of fragments representing the rpmB1-Rv0106 and rpmB2-Rv2059 intergenic regions). However, Zur was unable to bind DNA fragments corresponding to the upstream region of two genes (Rv1195 and Rv3612c) induced in the zur mutant strain but lacking the palindromic sequence, as well as the promoter region of an unrelated gene (Rv2688c) (Fig. 2, lanes 13 to 18). The specificity of Zur binding to fragments corresponding to the upstream regions of Rv0280 and Rv0282, and to the rpmB1-Rv0106 and rpmB2-Rv2059 intergenic region, was further investigated in competition experiments in which a 10- to 1,000-fold excess of unlabeled DNA fragments was used.
FIG. 2.
EMSA. Migration of different DNA fragments representing the upstream regions of Rv0280 (lanes 1 and 2), rpmB1-Rv0106 (lanes 3 and 4), Rv0282 (lanes 5 and 6), rpmB2-Rv2059 (lanes 7 and 8), Rv3017c (lanes 9 and 10), Rv3019c (lanes 11 and 12), Rv1195 (lanes 15 and 16), Rv3612c (lanes 17 and 18), and Rv2688c (lanes 13 and 14), used as a negative control, in the absence (−) or in the presence (+) of Zur.
As gel shift was completely inhibited by a 1,000-fold excess (10 pmol) of unlabeled specific fragment in presence of 54 pmol of Zur (corresponding to 27 pmol of dimeric protein), we were able to estimate that about 40% of purified protein was active in DNA binding (data not shown).
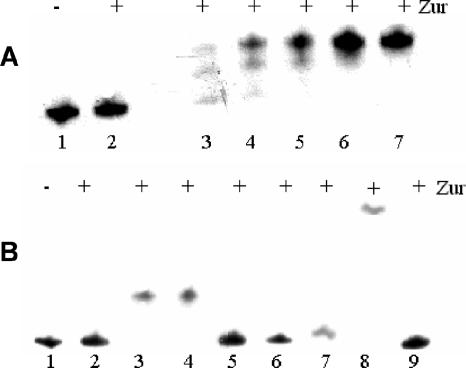
As previously demonstrated (27), purified Zur protein contains two zinc ions per monomer. To better define the role of zinc in the ability of Zur to bind its target sequence, EMSA were performed in the presence of EDTA: Zur, purified and dialyzed against 50 mM EDTA, was unable to retard the migration of a DNA fragment containing the Rv0282 upstream region in presence of 400 μM EDTA (Fig. 3A, lane 2). However, Zur binding activity was recovered by subsequent addition of zinc ions in a dose-effect manner (Fig. 3A, lanes 3 to 7), demonstrating that Zur binding activity is zinc dependent.
FIG. 3.
EMSA with the Rv0282 upstream region. (A) Dose-response with increasing zinc concentrations. The purified Zur protein (800 ng) was incubated with the DNA fragment containing the Rv0282 upstream region (10 fmol) in the presence of increasing concentrations of ZnCl2 in 400 μM EDTA. Binding reactions were loaded onto a nondenaturing polyacrylamide gel. Lane 1, negative control (no Zur); lane 2, no ZnCl2; lane 3, 100 μM ZnCl2; lane 4, 250 μM ZnCl2; lane 5, 500 μM ZnCl2; lane 6, 1 mM ZnCl2; lane 7, 10 mM ZnCl2. (B) Binding of Zur to the DNA fragment containing the Rv0282 upstream region in the presence of 1 mM concentrations of different metal divalent cations. Lane 1, negative control (no Zur); lane 2, no metal cations; lane 3, Zn2+; lane 4, Mn2+; lane 5, Fe2+; lane 6, Cu2+; lane 7, Co2+; lane 8, Cd2+; lane 9, Ni2+.
The DNA binding ability of certain metal-dependent repressors can be activated in vitro by several different transition metals. The influence of manganese, iron, copper, cadmium, and nickel on Zur DNA binding is shown in Fig. 3B. Zinc, cadmium, and manganese were able to promote the ability of Zur to retard the migration of the DNA fragment containing the Rv0282 upstream region. The reason for the strong reduction in mobility of the DNA fragment in the presence of Cd2+ is not known but was already described for another metal-dependent repressor (42).
Zinc-dependent binding of Zur was also observed for the other palindrome-containing regions described above (data not shown).
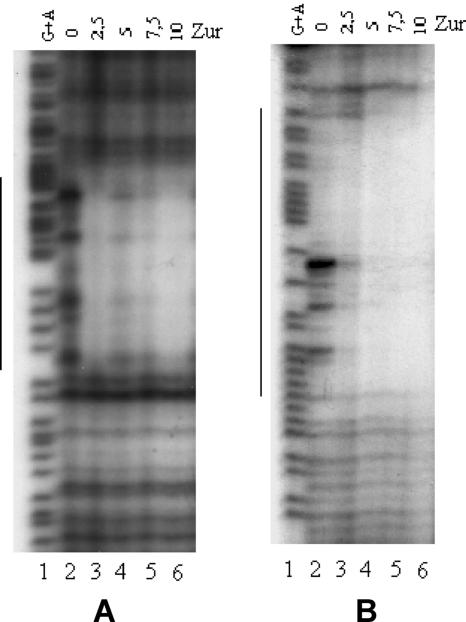
Identification of the Zur box by DNase I footprinting.
EMSA indicated that Zur is able to bind to Rv0280 and Rv0282 upstream regions as well as to rpmB2-Rv2059 and Rv0106-rpmB1 intergenic region. DNase I footprint analysis was performed on the putative promoter/operator regions of these genes (containing the palindromes showed in Fig. 1) in order to better define the consensus sequence recognized by Zur. Fragments of 250 to 300 bp, containing the putative promoter/operator region, were radiolabeled at their 5′ end, incubated with Zur, subjected to DNase I digestion, and analyzed by polacrylamide gel elctrophoresis. Under these conditions, the addition of Zur to the DNA fragment containing the Rv0280 putative promoter/operator region resulted in an area of protection spanning from −20 to −45 bp upstream of the translational start site of the gene (Fig. 4A). Analogous results were obtained with the Rv0282 putative promoter/operator region, where the protection extended from −163 to −188 bp upstream of the translational start site (data not shown). Footprinting experiments with the DNA fragment located in the rpmB1-Rv0106 and rpmB2-Rv2059 intragenic sites showed the presence of two different protected regions (data not shown). In Fig. 4B, the area of protection spanning from −80 to −106 bp upstream of the rpmB2 translational start site is shown.
FIG. 4.
DNase I protection assays. (A) Footprint analysis of the Zur binding site upstream of Rv0280. The −156/+38 DNA region, digested with BamHI and ScaI restriction enzymes, was labeled at the −156 end, incubated in the presence of 0 to 10 μg of Zur for 20 min at room temperature, and finally digested with DNase I. Maxam-Gilbert A+G sequences of the same fragment were loaded in the first lane. (B) Footprint analysis of the Zur binding site upstream of rpmB2. The −181/+78 DNA region, digested with BamHI and XbaI restriction enzymes, was labeled at the +69 end, incubated in the presence of 0 to 10 μg of Zur protein for 20 min at room temperature, and finally digested with DNase I. Maxam-Gilbert A+G sequences of the same fragment were loaded in the first lane. Vertical bars on the sides of the gels indicate the protected regions.
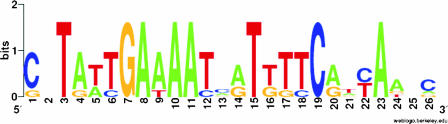
Analysis of Zur binding site and identification of consensus sequence.
The footprinting experiments allowed us to better identify the regions bound by Zur. The alignment of these sequences with ClustalW allowed us to map the conserved nucleotides and to generate a Zur consensus logo sequence (Fig. 5) (43); this sequence is highly homologous to the well known E. coli Fur box (15) which is the palindrome GATAATGATAATCATTATC.
FIG. 5.
The protected regions defined in the footprint analysis were used to define the Zur consensus sequence. WebLogo was used to obtain a consensus sequence logo in which the height of individual letters within a stack of letters represents the relative frequency of that letter at a given position, and the overall height of the stack represents the degree of conservation at that position (http://weblogo.berkeley.edu/).
Determination of the transcriptional start sites of Zur-regulated genes.
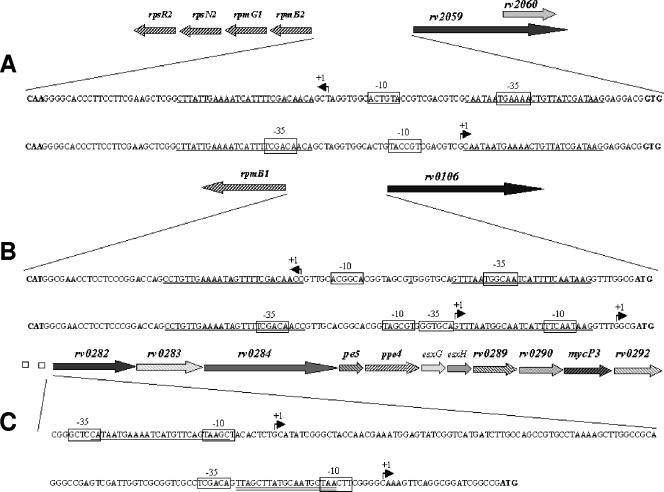
In order to determine the positions of Zur binding sites relatively to the promoter sequences, transcriptional start sites of rpmB1, Rv0106, Rv0282, rpmB2, and Rv2059 were identified by 5′ RACE PCR. The determination of transcriptional start sites provided the basis for the identification of potential −35 and −10 promoter regions, according to the known features of mycobacterial promoter sequences (30). In all cases, at least three nucleotides of both the potential −35 and −10 hexamers were found to be identical to those of the consensus sequences TTGCCA (−35) and TA(C/T)AAT (−10).
Consistent with the physiological role of Zur as a repressor of transcription, all Zur binding sites were shown to overlap the promoter sequences (Fig. 6). Interestingly, we found that both Rv0106 and Rv0282 are transcribed from two different promoters. In the case of Rv0106, both of them overlap a Zur operator (Fig. 6B). However, in the case of Rv0282, one of the promoters overlaps a Zur binding box, as hypothesized for a Zur-repressed promoter, while the other one overlaps an IdeR binding site (39) (Fig. 6C), supporting the previously reported IdeR dependency of these gene clusters.
FIG. 6.
Detailed genetic maps of the upstream region of selected genes belonging to the Zur regulon. (A) rpmB2-Rv2059 intergenic region. (B) rpmB1-Rv0106 intergenic region. (C) Rv0282 upstream. In panels A and B, the intergenic region is reported twice: the regulatory regions of the left-oriented genes are indicated in the upper lane, while the regulatory regions of the right-oriented genes are indicated in the lower lane. Boldface nucleotides indicate putative translational start codons, Zur binding sites are underlined, the IdeR binding site upstream of Rv0282 is double underlined, the identified transcriptional start sites (+1) are shown, and the deduced −35 and −10 promoter regions are boxed.
Effect of zinc concentration on Zur-regulated genes.
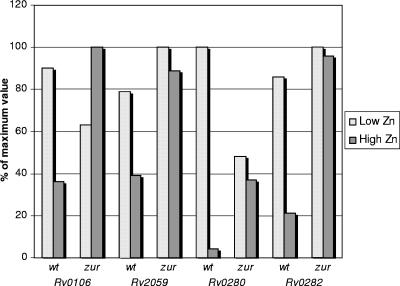
β-Galactosidase reporter assays were used to determine the role of zinc in Zur regulatory activity. A previously described M. smegmatis zur mutant (mcJF3) (8) and its wild-type parental strain (mc2155) were transformed with plasmids in which the promoters of Rv0106, Rv2059, Rv0280, and Rv0282 were cloned upstream of the promotorless reporter gene lacZ. After metal starvation, each culture was divided into two subcultures, which were supplemented with zinc up to concentrations of 100 μM (high zinc) and 1.4 nM (low zinc), according to a previously developed protocol (27). After protein extraction, β-galactosidase activity was measured to characterize the promoter activity. As shown in Fig. 7, the addition of zinc was able to repress the activity of all of the tested promoters in the wild-type strain but not in the zur mutant, favoring the hypothesis of Zur being a Zn-sensing metalloregulatory protein.
FIG. 7.
β-Galactosidase assays. The activities of four Zur-regulated genes were measured in a zur mutant of M. smegmatis and in its wild-type parental strain under conditions of low (1.4 nM) and high (100 μM) zinc concentrations. Values are indicated as percentages of maximal activity.
DISCUSSION
In this study, we have investigated the regulatory role of the global transcriptional regulator Zur in M. tuberculosis. We constructed an M. tuberculosis zur mutant strain, examined its phenotypes, and compared its transcriptional profile to that of the parental strain (H37Rv).
When the zur mutant strain was exposed to stressing agents or conditions, it did not show any difference from the wild-type strain and did not behave differently from the wild type in a mouse model of infection. It is possible that Zur is not involved in virulence, but we cannot rule out the possibility that we could find a phenotype by using a different model of infection (e.g., different mouse strains, routes of infection, or mortality assays instead of bacterial growth, etc.). It should also be noted that Zur is a repressor, so the genes under its transcriptional control are constitutively expressed in the mutant: if some of them are required for virulence or for stress responses, no phenotype is necessarily expected in the mutant.
We identified 32 genes, included in 16 putative transcriptional units, up-regulated in the mutant strain. However, no down-regulated genes were observed, suggesting that Zur acts specifically as a repressor in M. tuberculosis. The sequence upstream of all of the identified putative transcriptional units were searched for conserved sequences, and a conserved AT-rich palindrome was found upstream of eight up-regulated putative transcriptional units (including 24 genes). EMSA showed that Zur was able to specifically bind to DNAs containing this conserved palindrome and that this binding was zinc dependent. Zur binding was further investigated by DNase protection assay, which confirmed that the previously identified palindrome was indeed its binding target. Finally, the transcriptional start sites of five of these genes were identified, allowing the localization of the promoter sequences of these Zur-regulated genes. Interestingly, mapping of these promoters indicated that Zur operator sequences overlap either the −35 region (rpmB2, Rv2059, rpmB1, and Rv0106 upstream promoters) (Fig. 6) or the entire −10/−35 region (Rv0106 downstream promoter and the Zur-dependent promoter of Rv0282) (Fig. 6), suggesting that Zur binding will prevent transcriptional initiation. This conclusion is consistent with the results of the DNA array analyses (Table 2).
No conserved sequence was found upstream of the remaining eight induced genes, suggesting that their regulation by Zur is indirect. For example, the zur mutant strain could accumulate more zinc than the wild type and this could cause the variation of their transcription as a side-effect. Another possibility is that another gene(s) encoding a regulatory protein(s) is induced and that this postulated regulator up-regulates these genes not directly controlled by Zur.
Using an M. smegmatis zur mutant as a model, we showed that the Rv0106, Rv0280, Rv0282 and Rv2059 promoters are repressed by zinc in a Zur-dependent manner. We did not use M. tuberculosis in these experiments because of the difficulty of obtaining zinc starvation in this bacterium (data not shown). We also showed that Zur binds its consensus sequence in a zinc-dependent manner and that several genes under its transcriptional control have putative functions related to zinc uptake or are homologous to genes belonging to the Zur regulon in other bacteria (see below). Taken together, these findings strongly suggest that Zur has a function in regulating zinc uptake. Further evidence supporting this idea is the finding that the transcription of its structural gene is repressed by Rv2358 in a zinc-dependent manner (8).
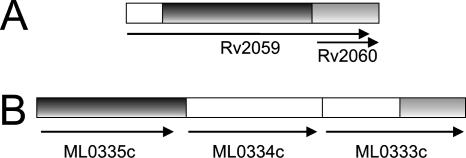
Among the genes directly regulated by Zur, Rv0106 encodes a protein similar to the Zur-regulated low-affinity zinc transporter YciC characterized in B. subtilis (16). Rv2059 and Rv2060 encode two components of an incomplete ABC-transporter system: the Rv2059 product belongs to the TroA superfamily (21) proteins functioning as initial receptors in ABC transport of Zn2+ and Mn2+ in many eubacterial species. However, Rv2060 encodes a truncated membrane permease component of an ABC-type Zn2+ transport system. Interestingly, Rv2060 almost totally overlaps the second half of Rv2059, even if it uses a different reading frame. It is possible that this peculiar structure is derived from a deletion leading to a frameshift which caused the fusion of the terminal part of Rv2059 with a sequence internal to Rv2060. This hypothesis is supported by the fact that in the Mycobacterium leprae genome sequence (10), this region contains a putative operon encoding a complete ABC transporter system: the product of the first gene is similar to the N terminus of Rv2059, and the product of the second is not encoded by M. tuberculosis, while the C terminus of the third gene product is similar to Rv2060 (Fig. 8). Since the overlap between Rv2059 and Rv2060 is perfectly conserved in Mycobacterium bovis, it is possible that the recombination event is specific for bacteria belonging to the M. tuberculosis complex. Whether Rv2060 is translated into a functional protein and whether the products of Rv2059 and Rv2060 retain some functionality remain to be determined.
FIG. 8.
Map of Rv2059 and Rv2060 in M. tuberculosis and M. bovis (A) and of the corresponding region in M. leprae (B). Arrows indicate open reading frames. Shadowed regions indicate regions of homology.
The Zur regulon also includes five genes encoding ribosomal proteins, four of which form a single putative transcriptional unit (rpmB2, rpsR2, rpsN2, and rpmG1). It has been previously hypothesized that zinc-binding ribosomal proteins could be involved in zinc storage (29, 36), and it was recently shown that in B. subtilis following zinc starvation, this metal is mobilized from the ribosome by the replacement of a zinc-associated ribosomal protein with a paralog unable to bind zinc. Interestingly, transcription of the latter protein was shown to be under Zur control (1, 31). Three of the ribosomal proteins encoded by Zur-regulated genes (RpsR2, RpsN2, and RpmG1) in M. tuberculosis have paralogs containing a Zn ribbon in their sequences (RpsR1, RpsN1, and RpmG2, respectively), suggesting that also in this bacterium the ribosome represents a zinc storage compartment and that in the absence of zinc, Zur-regulated ribosomal proteins (unable to bind zinc) replace their zinc binding paralogs to mobilize the stored metal.
Another group of Zur-regulated genes of particular interest is represented by those encoding five proteins of the ESAT-6/CFP-10 family (esxG, esxH, esxQ, esxR, and esxS). The M. tuberculosis genome encodes 23 members of this family (esxA to esxW), whose genes are located in 11 loci where they are usually present in pairs (9). These genes encode small secreted proteins present in the M. tuberculosis cell culture supernatant as ESAT-6/CFP-10 heterodimers (5). In five cases, the esx gene couple is flanked by blocks of conserved genes indicated as clusters of ESAT-6 and numbered from 1 to 5 (17). Functional analysis performed on the ESAT-6-like cluster 1 of M. tuberculosis and on a similar cluster from M. smegmatis suggested that they encode a secretion apparatus specific for the CFP-10/ESAT-6 heterodimer encoded by the cluster (4, 11).
Within the Zur regulon, esxG and esxH are part of ESAT-6 cluster 3, while esxQ, esxR, and esxS are physically associated but do not belong to any of the five clusters. Interestingly, esxR and esxS were reported to be lost in several clinical isolates (25). All 11 genes of ESAT-6 cluster 3 (Rv0282 to Rv0292) were induced in the Zur mutant, suggesting their organization in an operon. Interestingly, the same gene cluster is induced by iron starvation and is repressed by iron and IdeR (39). Consistent with dual regulation of this gene cluster by Zur and by IdeR, we identified two different promoters upstream of its first gene (Rv0282); one overlaps the Zur binding site, while the other overlaps the IdeR binding site (Fig. 6C). Additional studies will further characterize the interactions of Zur and IdeR at these promoters.
Practically nothing is known regarding the physiological role of the proteins secreted by ESAT-6 clusters. It is known that cluster 1 is essential for M. tuberculosis virulence, and it was shown that its deletion from the vaccine strain Mycobacterium bovis BCG genome is one of the main causes of its attenuation (19). However, no clear mechanism of their role in pathogenesis was ever shown, even if a role in infected cell lysis and M. tuberculosis dissemination has been suggested (19). The finding that ESAT-6 cluster 3 genes are induced under conditions of zinc and iron starvation could be clues to their physiological function. The concentration of available iron and zinc in the body is low. In particular, the zinc concentration is low in lung alveoli (36). This suggests that cluster 3-secreted proteins could be involved in zinc and/or iron scavenging and/or uptake. It is worthwhile to mention that the proteins of the ESAT-6/CFP-10 family are known to be among the most potent T-cell antigens (5), suggesting that the modulation of their expression in response to changing iron or zinc concentrations could strongly modify the M. tuberculosis antigenic profile during the course of infection.
Taken together, these data strongly suggest that M. tuberculosis Zur should be considered a zinc uptake regulator involved in the derepression of genes involved in zinc uptake and mobilization from the storage compartment. The presence in the Zur regulon of an ESAT-6 cluster is an intriguing finding and could help in our understanding of the role of these proteins in M. tuberculosis physiology.
The absence of discernible phenotypes in the zur mutant suggests that the deregulation of the Zur regulon is not detrimental to any basic physiologic function in M. tuberculosis (possibly due to redundant mechanisms for controlling zinc uptake), but this does not mean that the induction of Zur-regulated genes in the presence of low zinc concentrations is not important in M. tuberculosis physiology. To study this aspect, it should be possible to obtain a dominant positive mutant of Zur that can repress target genes in the absence of zinc, as already described for DtxR and IdeR in Corynebacterium diphtheriae and M. tuberculosis (22, 44).
Acknowledgments
This work was supported by the Istituto Superiore di Sanità, Progetto Nazionale AIDS, grant no. 50F.24 (awarded to R.M.); by MIUR-COFIN 2003, grant no. 2003059340 (awarded to R.M.) and MIUR-COFIN 2003 (awarded to G.R.); by the University of Padova, Progetti di Ateneo, grant no. CPDA047993 (awarded to R.M.); by EC-VI Framework Contract no. LSHP-CT-2005-018923 (awarded to G.R.); and by National Institutes of Health grant RO1 AI-44856 (awarded to I.S.).
We are grateful to John Chan (Albert Einstein College of Medicine, New York, NY) for performing mouse infections.
Footnotes
Published ahead of print on 10 November 2006.
REFERENCES
- 1.Akanuma, G., H. Nanamiya, Y. Natori, N. Nomura, and F. Kawamura. 2006. Liberation of zinc-containing L31 (RpmE) from ribosomes by its paralogous gene product, YtiA, in Bacillus subtilis. J. Bacteriol. 188:2715-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beisel, W. R. 1977. Magnitude of the host nutritional responses to infection. Am. J. Clin. Nutr. 30:1236-1247. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe, D. K., and A. P. Morby. 2003. Zn(II) metabolism in prokaryotes. FEMS Microbiol. Rev. 27:291-311. [DOI] [PubMed] [Google Scholar]
- 4.Brodin, P., L. Majlessi, L. Marsollier, M. I. de Jonge, D. Bottai, C. Demangel, J. Hinds, O. Neyrolles, P. D. Butcher, C. Leclerc, S. T. Cole, and R. Brosch. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodin, P., I. Rosenkrands, P. Andersen, S. T. Cole, and R. Brosch. 2004. ESAT-6 proteins: protective antigens and virulence factors? Trends Microbiol. 12:500-508. [DOI] [PubMed] [Google Scholar]
- 6.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 7.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 8.Canneva, F., M. Branzoni, G. Riccardi, R. Provvedi, and A. Milano. 2005. Rv2358 and FurB: two transcriptional regulators from Mycobacterium tuberculosis which respond to zinc. J. Bacteriol. 187:5837-5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 10.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 11.Converse, S. E., and J. S. Cox. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J. Bacteriol. 187:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Pina, K., V. Desjardin, M. A. Mandrand-Berthelot, G. Giordano, and L. F. Wu. 1999. Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli. J. Bacteriol. 181:670-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubrac, S., and D. Touati. 2000. Fur-positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, Jr., and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gey Van Pittius, N. C., J. Gamieldien, W. Hide, G. D. Brown, R. J. Siezen, and A. D. Beyers. 2001. The ESAT-6 gene cluster of Mycobacterium tuberculosis and other high G+C Gram-positive bacteria. Genome Biol. 2:RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall, H. K., and J. W. Foster. 1996. The role of fur in the acid tolerance response of Salmonella typhimurium is physiologically and genetically separable from its role in iron acquisition. J. Bacteriol. 178:5683-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu, T., S. M. Hingley-Wilson, B. Chen, M. Chen, A. Z. Dai, P. M. Morin, C. B. Marks, J. Padiyar, C. Goulding, M. Gingery, D. Eisenberg, R. G. Russell, S. C. Derrick, F. M. Collins, S. L. Morris, C. H. King, and W. R. Jacobs, Jr. 2003. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc. Natl. Acad. Sci. USA 100:12420-12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusner, D. J. 2005. Mechanisms of mycobacterial persistence in tuberculosis. Clin. Immunol. 114:239-247. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y.-H., M. R. Dorwart, K. R. O. Hazlett, R. K. Deka, M. V. Norgard, J. D. Radolf, and C. A. Hasemann. 2002. The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation. J. Bacteriol. 184:2300-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manabe, Y. C., B. J. Saviola, L. Sun, J. R. Murphy, and W. R. Bishai. 1999. Attenuation of virulence in Mycobacterium tuberculosis expressing a constitutively active iron repressor. Proc. Natl. Acad. Sci. USA 96:12844-12848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 24.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor SigE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 25.Marmiesse, M., P. Brodin, C. Buchrieser, C. Gutierrez, N. Simoes, V. Vincent, P. Glaser, S. T. Cole, and R. Brosch. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483-496. [DOI] [PubMed] [Google Scholar]
- 26.Maxam, A. M., and W. Gilbert. 1977. A new method for sequencing DNA. Proc. Natl. Acad. Sci. USA 74:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milano, A., M. Branzoni, F. Canneva, A. Profumo, and G. Riccardi. 2004. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155:192-200. [DOI] [PubMed] [Google Scholar]
- 28.Milano, A., F. Forti, C. Sala, G. Riccardi, and D. Ghisotti. 2001. Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J. Bacteriol. 183:6801-6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, C. M., and J. D. Helmann. 2005. Metal ion homeostasis in Bacillus subtilis. Curr. Opin. Microbiol. 8:188-195. [DOI] [PubMed] [Google Scholar]
- 30.Mulder, M. A., H. Zappe, and L. M. Steyn. 1997. Mycobacterial promoters. Tuberc. Lung Dis. 78:211-223. [DOI] [PubMed] [Google Scholar]
- 31.Nanamiya, H., G. Akanuma, Y. Natori, R. Murayama, S. Kosono, T. Kudo, K. Kobayashi, N. Ogasawara, S. M. Park, K. Ochi, and F. Kawamura. 2004. Zinc is a key factor in controlling alternation of two types of L31 protein in the Bacillus subtilis ribosome. Mol. Microbiol. 52:273-283. [DOI] [PubMed] [Google Scholar]
- 32.Ochsner, U. A., and M. L. Vasil. 1996. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc. Natl. Acad. Sci. USA 93:4409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Halloran, T. V. 1993. Transition metals in control of gene expression. Science 261:715-725. [DOI] [PubMed] [Google Scholar]
- 34.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2004. Analysis of genes that encode DtxR-like transcriptional regulators in pathogenic and saprophytic corynebacterial species. Infect. Immun. 72:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 36.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc. Natl. Acad. Sci. USA 100:9912-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patzer, S. I., and K. Hantke. 2000. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275:24321-24332. [DOI] [PubMed] [Google Scholar]
- 38.Pym, A. S., P. Domenech, N. Honore, J. Song, V. Deretic, and S. T. Cole. 2001. Regulation of catalase-peroxidase (KatG) expression, isoniazid sensitivity and virulence by furA of Mycobacterium tuberculosis. Mol. Microbiol. 40:879-889. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Schaible, U. E., and S. H. Kaufmann. 2005. A nutritive view on the host-pathogen interplay. Trends Microbiol. 13:373-380. [DOI] [PubMed] [Google Scholar]
- 42.Schmitt, M. P. 2002. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn2+-dependent mechanism. J. Bacteriol. 184:6882-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider, T. D., and R. M. Stephens. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097-6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, L., J. vanderSpek, and J. R. Murphy. 1998. Isolation and characterization of iron-independent positive dominant mutants of the diphtheria toxin repressor DtxR. Proc. Natl. Acad. Sci. USA 95:14985-14990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timm, J., E. M. Lim, and B. Gicquel. 1994. Escherichia coli-mycobacteria shuttle vectors for operon and gene fusions to lacZ: the pJEM series. J. Bacteriol. 176:6749-6753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaquerizas, J. M., L. Conde, P. Yankilevich, A. Cabezon, P. Minguez, R. Diaz-Uriarte, F. Al-Shahrour, J. Herrero, and J. Dopazo. 2005. GEPAS, an experiment-oriented pipeline for the analysis of microarray gene expression data. Nucleic Acids Res. 33:W616-W620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahrt, T. C., J. Song, J. Siple, and V. Deretic. 2001. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39:1174-1185. [DOI] [PubMed] [Google Scholar]