Abstract
We have shown previously that perturbation of origin firing in chromosome replication causes DNA lesions and triggers DNA damage checkpoint control, which ensures genomic integrity by stopping cell cycle progression until the lesions are repaired or by inducing cell death if they are not properly repaired. This was based on the observation that the temperature-sensitive phenotype of orc1-4 and orc2-1 mutants required a programmed action of the RAD9-dependent DNA damage checkpoint. Here, we report that DNA lesions in the orc mutants are induced much more quickly and frequently within the rRNA gene (rDNA) locus than at other chromosomal loci upon temperature shift. orc mutant cells with greatly reduced rDNA copy numbers regained the ability to grow at restrictive temperatures, and the checkpoint response after the temperature shift became weak in these cells. In orc2-1 cells, completion of chromosomal duplication was delayed specifically on chromosome XII, where the rDNA array is located, and the delay was partially suppressed when the rDNA copy number was reduced. These results suggest that the rDNA locus primarily signals abnormalities in the initiation program to the DNA damage checkpoint and that the rDNA copy number modulates the sensitivity of this monitoring function.
Eukaryotic DNA replication initiates from multiple origins on each chromosome. These origins are activated at different time points during S phase and are utilized with different efficiencies in individual cells (16, 31, 40). The initiation program of chromosomal replication from multiple origins is crucial for the maintenance of genetic integrity in eukaryotic cells. When origin firing is perturbed, replication from fewer origins can lead to chromosome loss and rearrangement. Chromosome instability is induced in Saccharomyces cerevisiae cells having defects in components of the prereplicative complex, ORC, MCM, Cdc6, and Cdt1, which are required for origin firing and a proper initiation program (3, 8, 14, 21, 22, 38). These notions are important for understanding the causes of chromosome abnormality involved with carcinogenesis as well as the development of malignancy, because cells expressing oncogenes ectopically often display perturbed origin firing and an abnormal initiation program (1, 13).
Among the initiation proteins, ORC (origin recognition complex) plays a central role in initiating replication by associating with the replication origin and recruiting other replication factors (2, 4, 15). ORC also composes the chromatin environment near origins, which determines the timing of origin firing during chromosomal replication (28). Using yeast cells defective in ORC function, we have shown previously that chromosome loss, allelic recombination, and other chromosomal aberrations are induced in cells grown at the restrictive temperature but that this chromosomal instability is efficiently suppressed by DNA damage checkpoint control functions (38). Yeast orc mutants with temperature-sensitive (ts) growth phenotypes show either a severe defect in entering the S phase of the cell cycle or a reduced capacity to regulate the timing and efficiency of origin firing at nonpermissive temperatures (4, 15, 20, 26, 33, 34, 38). In the latter case, at the nonpermissive temperature, cells divide with a slightly prolonged S phase that correlates with mildly reduced efficiencies of origin firing at several replication origins examined, indicating that the origin firing is not abolished but is perturbed to some extent. However, following two or three rounds of cell division after the shift up to the restrictive temperature, the cell cycle is arrested at the G2/M boundary and cells start to die after prolonged incubation. These cell fate characteristics after the temperature shift were observed with orc1-4 cells at 37°C and orc2-1 cells at 26°C (38). We found that the G2/M arrest and subsequent cell death in the orc mutants were suppressed by a rad9 null mutation, thus revealing that RAD9-dependent DNA damage checkpoint control is activated in orc mutant cells when the initiation program of chromosomal DNA replication is perturbed. This implies that some DNA lesion(s) with a perturbed initiation program is produced in cells and triggers the checkpoint response which ensures genome integrity, either by blocking cell cycle progression at the G2/M boundary until the lesions are properly repaired or by inducing cell death if they are not. It was recently shown that Mec1p, which functions upstream of Rad9p in checkpoint control, is responsible for the induction of yeast apoptotic processes in orc2-1 cells that include the production of reactive oxygen species and activation of the metacaspase Yca1p (9, 39). The checkpoint-mediated apoptotic program is likely to be involved in the cell death phenotype observed with orc1-4 and orc2-1 mutants at nonpermissive temperatures.
It is largely unknown how DNA lesions are produced by perturbation of origin firing. Here, we provide evidence that the rRNA gene (rDNA) cluster (rDNA array) is the site most sensitive to such lesions in the yeast genome and primarily signals an abnormality in the initiation program to the DNA damage checkpoint control. The rDNA is a highly repetitive DNA sequence, usually in a head-to-tail consecutive configuration, and comprises one to several huge clusters in eukaryotic genomes. In budding yeast, the rDNA array resides in a single cluster of about 150 copies, occupying about 60% of chromosome XII (Chr XII). Besides the 5S and 35S rRNA genes, each unit of the rDNA contains a replication origin (rDNA autonomously replicating sequence [ARS]) and an orientation-specific replication terminus (replication fork barrier [RFB]) that functions with a replication-blocking protein, Fob1p. Yeast cells, and probably most eukaryotes, possess an elaborate mechanism for actively maintaining the high copy number of the rDNA (24). This maintenance mechanism controls and utilizes sister chromatid recombination and involves the action of Fob1p on the RFB. We initially found extremely high Chr XII instability in orc mutant cells grown at the nonpermissive temperature. Further studies revealed that DNA lesions were induced much more quickly and frequently within the rDNA locus than at other chromosomal loci when cells underwent perturbations of origin firing. During these experiments, we found that orc mutant clones surviving a prolonged incubation at the restrictive temperature included some having extensively shortened rDNA. Surprisingly, these surviving clones with truncated rDNA were cured of the ts phenotype. The reduction in the rDNA copy number also compromised the activation of the DNA damage checkpoint response upon perturbation of origin firing in the orc mutants. Furthermore, in orc2-1 cells, completion of chromosomal duplication was delayed specifically on Chr XII, and this delay was partially suppressed when the rDNA copy number was reduced. More interestingly, relatively weak rDNA ARS origin activities were significantly increased in cells with reduced rDNA copy numbers. Therefore, the copy number of the rDNA is a crucial determinant for the capacity of the rDNA array to initiate DNA replication and, thus, for the sensitivity of the rDNA array to perturbed origin firing. These results suggest that the rDNA locus plays an important role in monitoring abnormality in the initiation program of chromosomal replication for DNA damage checkpoint control.
MATERIALS AND METHODS
Saccharomyces cerevisiae strains and growth conditions.
Yeast strains used in this study were derived from three parental strains, FY23, FY838, and NOY408-1b (19, 24), and are listed in Table S1 in the supplemental material. Media for yeast strains, including complex glucose (yeast extract-peptone-dextrose [YPD]), synthetic complete (SC), and various dropout media, were prepared as previously described (19). Cells were precultured at the permissive temperature of 23°C in YPD until mid-log phase and subsequently inoculated at a density of 5 × 105 cells/ml into fresh YPD. Cultures were preincubated at the permissive temperature until cells doubled once or twice and were then shifted to nonpermissive temperatures (37°C for orc1-4/orc1-4 cells and 26°C for orc2-1cells).
Isolation of orc1-4 and orc2-1 cells surviving after incubation at nonpermissive temperatures.
Surviving orc1-4 cells (orc1-4 surviving cell [orc1-4 SvC]) were isolated from RD603 (orc1-4/orc1-4) as described previously (38). RD603 cells were incubated in YPD at 37°C for 24 h, spread on YPD plates after appropriate dilution, and incubated at 23°C until colonies of surviving cells were formed. Six independent colonies were picked and named orc1-4 SvC a to f. Surviving orc2-1 cells were isolated from YNV1 in the same way, except that the YNV1 cells were incubated in YPD at 26°C for 12 h. Eight independent colonies were picked and named orc2-1 SvC1 to SvC8.
Analysis of yeast chromosomes by PFGE.
Samples for pulsed-field gel electrophoresis (PFGE) were prepared as described previously (19). To determine the size of Chr XII, electrophoresis was carried out with a 0.8% agarose gel with Tris-acetate-EDTA (TAE), using a CHEF Mapper XA with a pulse time of 20 min to 22 min 53 s, at 2.0 V/cm, and at an include angle of 106° for 72 h at 14°C in block1 and with a pulse time of 25 s to 2 min 26 s, at 6.0 V/cm, and at an include angle of 120° for 7.5 h at 14°C in block2. Following electrophoresis, the gel was stained with ethidium bromide (0.5 μg/ml) for 30 min, destained in deionized water for 20 min, photographed, and then subjected to Southern hybridization analysis, using a probe for the site proximate to YLL058W on Chr XII (probe 058). To analyze Chr XII in exponentially growing cells, electrophoresis was carried out with a 1.0% agarose gel with 0.5× Tris-borate-EDTA (TBE), using a CHEF-DRIII with a pulse time of 0.2 s to 266 s, at 6.0 V/cm, and at an include angle of 120° for 15.2 h at 14°C. Southern hybridization was performed with probe 058 and a probe for the MCM2 gene on Chr II (probe MCM2).
Construction of orc2-1 and orc1-4 derivatives with reduced copy numbers of rDNA repeats.
Using hygromycin (300 μg/ml) and plasmid pRDN-hyg1, cells carrying Chr XII with extensively reduced copy numbers of rDNA repeats were constructed (10). To construct orc2-1 derivatives with reduced copy numbers of rDNA repeats, pRDN-hyg1 was introduced into NOY408-1bf. Hygromycin-resistant clones of the resulting transformant were selected. Based on the size of Chr XII determined by PFGE, a hygromycin-resistant clone whose rDNA copy number was about 30 was isolated and grown on an SC plate containing 5-fluoroorotic acid to drop the plasmid off. An orc2-1 mutation was introduced into the resulting strain (NOY408-1bf30) by the method of Foss et al. (15). For orc1-4 derivatives, YSI7 (orc1-4 MATa) cells were transformed with pRDN-hyg1 and mated with YMJ26 (orc1-4 MATα), and the resulting diploid strain (RD702) was grown in the presence of hygromycin, from which a hygromycin-resistant clone (RD703) was selected. These strains with reduced copy numbers of rDNA repeats were maintained on hygromycin until the cells were spotted on a YPD plate or precultured in YPD liquid medium at 23°C.
Determination of ARS plasmid loss rate.
Plasmids YCplac111 (ARS1) (17) and pRS414 (ARSH4) (35) were introduced separately into NOY408-1bf (fob1Δ), NOY408-1bf30 (fob1Δ rdnΔ30), YNV2 (orc2-1 fob1Δ), and YNV15 (orc2-1 fob1Δ rdnΔ30). Transformants were inoculated into nonselective medium (SC) and maintained in exponential-phase growth at 23°C for 3 days. Aliquots were withdrawn at various times, spread onto SC plates and SC-Trp or SC-Leu plates after appropriate dilution, and incubated at 23°C until colonies formed. The percentage of cells losing the plasmid per generation was determined as described previously (11).
Western blotting.
Yeast whole-cell extracts were prepared by the trichloroacetic acid method (29). Proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to a polyvinylidene difluoride membrane (Millipore), and subjected to Western blotting analysis as described previously (32). For detection of Rad53p, antihemagglutinin monoclonal antibody 12CA5 (Roche) was used. For detection of Orc2p and Mcm2p, affinity-purified anti-Orc2p antibody (a gift from Y. Kawasaki, Osaka University) and goat anti-MCM antibody (Santa Cruz Antibodies) were used, respectively.
FACS analysis.
To measure cellular DNA content, cells were stained with propidium iodide and analyzed by fluorescence-activated cell sorting (FACS) with a Becton Dickinson FACScan and Cell Quest software.
2D gel electrophoresis analysis of replication intermediates.
The origin activities of rDNA ARS and ARS1 were analyzed by two-dimensional (2D) gel electrophoresis and quantified using a Bio-Image BAS2000 analyzer (Fuji Photo Film) as described previously (5).
RESULTS
Extreme instability of Chr XII in orc1-4 cells after the shift up to the restrictive temperature.
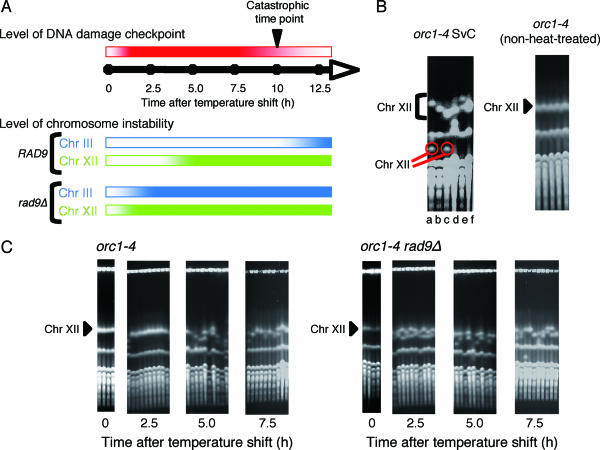
orc1-4 diploid cells showed increased chromosome instability at a nonpermissive temperature (38). However, when we analyzed the fate of Chr III, elevated chromosome instability was not seen for the first 10 h after the temperature shift to 37°C. Then, beyond this catastrophic time point, at which cells began to die, chromosome instability started to increase sharply (Fig. 1A). After incubation of the orc1-4 cells at 37°C for 24 h, about 10% survived and formed colonies when plated at 23°C. In the surviving cells, the frequency of loss of heterozygosity (LOH) for a URA3 marker placed on the right arm of Chr III, a measure of chromosome instability, was 4.5 × 10−3, about 20-fold higher than that determined with non-heat-treated cells. About 90% of the LOH events were losses of Chr III, while LOH caused by allelic recombination was not enhanced in the orc1-4 diploid cells (38). During these experiments, we found that almost all of the surviving orc1-4 clones acquired a variably abnormal-sized Chr XII (Fig. 1B). Such unusual chromosome instability was not observed in the orc1-4 non-heat-treated clones. Since two different-sized Chr XIIs were present in each surviving clone, the size alterations were occurring in both homologous chromosomes. In about one-third of the clones (Fig. 1B, clones a and c), one of the altered chromosomes was very small while the other was only slightly shortened. Thus, the instability of Chr XII was distinct from that of other chromosomes, not only because the frequency of size aberration of Chr XII was extremely high but also because both homologous chromosomes were simultaneously but independently altered in each aberrant clone.
FIG. 1.
Extreme instability of chromosome XII in orc1-4 mutant cells grown at the restrictive temperature. (A) Time profiles for the status of DNA damage checkpoint control and the induction of instability in Chr III and Chr XII in orc1-4 cells after the temperature shift. Intensities of colors (red for the DNA damage checkpoint, blue for Chr III instability, and green for Chr XII instability) indicate the strength for each category. The time profiles for the DNA damage checkpoint and the instability of Chr III are based on our previous observations (38), and the time profile for the instability of Chr XII is from data obtained in this study. (B) Size aberration of chromosome XII in orc1-4 diploid cells surviving after incubation at 37°C for 24 h. Six clones of orc1-4 SvC (a to f) were isolated, grown in YPD at 23°C, and subjected to PFGE analysis followed by ethidium bromide staining, all as described in Materials and Methods. As a control experiment, another set of non-heat-treated orc1-4 clones was similarly examined by PFGE analysis. Positions of Chr XII in the gel are indicated by a triangle for the normal size, a bracket for aberrant sizes, and red circles for extensively reduced sizes. (C) Time course for induction of Chr XII instability in orc1-4 and orc1-4 rad9Δ diploid cells after shifting the growth temperature to 37°C. RD603 (orc1-4/orc1-4) and RD613 (orc1-4/orc1-4 rad9Δ/rad9Δ) were grown as described in Materials and Methods. Aliquots were withdrawn at the indicated time points after the temperature shift to 37°C, appropriately diluted with distilled water, spread on YPD plates, and incubated at 23°C for 5 to 7 days. Five or 10 colonies were randomly chosen from cells recovered at each time point, grown in YPD at 23°C, and subjected to PFGE analysis.
The instability of Chr XII in orc1-4 cells showed another unique characteristic. We found that after the orc1-4 diploid cells were transferred to the restrictive temperature, the size aberration of Chr XII began to occur much earlier than the catastrophic time point, at which the instability of other chromosomes started (Fig. 1C). Aliquots of orc1-4 cells grown in YPD liquid medium were taken every 2.5 h after the temperature shift and plated on YPD solid medium to form colonies at the permissive temperature. The chromosomes of each heat-treated clone were extracted and subjected to PFGE analysis. The size aberration of Chr XII was not observed at 2.5 h but was clearly visible at 5 h after the temperature shift, indicating that the hyperinstability of Chr XII was induced nearly simultaneously in all the orc1-4 cells at some time between the 2.5-h and 5-h time points. We previously demonstrated that the instability of Chr III (and probably of other chromosomes), caused by a defect in Orc1p, was strongly suppressed by the RAD9-dependent checkpoint control until 10 h after the temperature shift. Thus, unlike the Chr III instability, the hyperinstability of Chr XII induced in orc1-4 cells at 37°C seemed to be free from suppression by checkpoint control. However, we obtained evidence that the RAD9-dependent checkpoint control was operative for the suppression of strictly the hyperinstability of Chr XII until at least the 2.5-h time point (Fig. 1C). In orc1-4 rad9Δ cells, the hyperinstability of Chr XII was seen as early as the 2.5-h time point and, thus, was probably induced immediately after the temperature shift. This also clearly indicated that the stability of Chr XII is maintained by checkpoint control until 2.5 h after the temperature shift, as for other chromosomes, but that Chr XII became unstable at some time between the 2.5-h and 5-h time points in those cells where checkpoint control was still functioning to suppress rigorously the instability of other chromosomes. In orc1-4 rad9Δ diploid cells where DNA damage checkpoint control was not functioning, the instability of Chr III began to increase sharply immediately after the temperature shift (38). The frequency of LOH, mostly chromosome loss events, for the URA3 marker on Chr III was 2.7 × 10−3 at 5 h after the temperature shift (38), whereas almost all of the orc1-4 rad9Δ cells suffered size aberration of Chr XII at the same time point (Fig. 1C). From these data, we speculated that very soon after the temperature shift, DNA lesions would be induced within every Chr XII at an extremely high frequency and within other chromosomes at much lower frequencies and that all the DNA lesions could be properly corrected by functions of DNA damage checkpoint control before the 2.5-h time point. However, these functions would become insufficient to correct completely the DNA lesions within Chr XII after the 2.5-h time point, whereas lesions within other chromosomes were still efficiently corrected until the catastrophic time point (Fig. 1A). The extent of DNA lesions afflicting Chr XII may surpass the capacities of repair functions that suppress chromosome aberration.
orc mutant cells carrying extensively shortened Chr XIIs are cured of the ts phenotype.
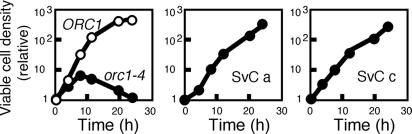
As shown in Fig. 1B, about one-third of orc1-4 clones that survived heat treatment at 37°C for 24 h carried a very small Chr XII. We found that the surviving orc1-4 clones with extensively shortened Chr XIIs (SvC a and c) were able to grow at 37°C (Fig. 2). Such very small Chr XIIs, with deletions exceeding 500 kb, probably reflect a severe reduction in the copy number of rDNA repeats. Therefore, we reasoned that the reduced copy numbers of the rDNA repeats might lead to the suppression of the ts phenotype in orc1-4 diploid cells.
FIG. 2.
Growth of surviving orc1-4 clones at the restrictive temperature. Two surviving orc1-4 clones (SvC a and SvC c) were grown in YPD at 37°C as described in Materials and Methods. At the indicated time points, aliquots were withdrawn, appropriately diluted with distilled water, spread on YPD plates, and incubated at 23°C for 5 to 7 days. From the numbers of colonies formed, the viable density at each time point was calculated. As controls, the growth of wild-type and orc1-4 diploid cells at 37°C is also shown.
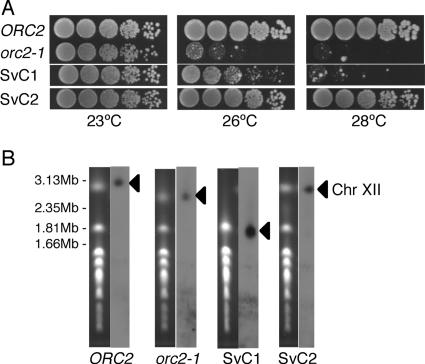
To ascertain the relation between ts phenotype and size of Chr XII, we examined orc2-1 cells surviving after a prolonged incubation at 26°C. We previously reported that orc2-1 diploid cells also trigger RAD9-dependent checkpoint control and subsequently induce cell death at 26°C (38). In the orc2-1 strain, however, the instability of Chr III was never induced, even after a prolonged incubation at 26°C, suggesting that DNA lesions produced in orc1-4 and orc2-1 strains differ in structure or in distribution within the genome. Nevertheless, almost all of the orc2-1 diploid cells surviving treatment at 26°C for 24 h acquired a variably abnormal-sized Chr XII (data not shown). This clearly indicates that the extreme instability of Chr XII is another characteristic common to orc1-4 and orc2-1 strains. Although the underlying mechanisms are unknown, the checkpoint-dependent induction of cell death was observed in orc2-1 haploid cells but not in orc1-4 haploid cells (K. Watanabe and H. Maki, unpublished results). Since haploid strains are more versatile for genetic analyses than diploid strains, we decided to use orc2-1 haploid strains for further studies. Similar to that of orc1-4 diploid cells, the survival rate of orc2-1 haploid cells after heat treatment was about 10%. We picked eight surviving orc2-1 clones (SvC1 to SvC8) and examined whether they were cured of the ts phenotype. Five clones were able to grow at the nonpermissive temperature, among which we found two distinct types. Type 1 (SvC1) grew at 26°C but not at 28°C, while type 2 (SvC2, SvC6, SvC7, and Svc8) could grow at temperatures higher than 28°C (Fig. 3A). As shown in Fig. 3B, the size of Chr XII was greatly reduced in the type 1 surviving clone, suggesting a strong correlation between the cure of the ts phenotype and the extensive reduction in the rDNA copy number. On the other hand, the type 2 clones had a normal-sized Chr XII. We found that the intracellular levels of Orc2p in the type 2 clones were much higher than those in the parental orc2-1 cells (data not shown). As reported previously (33), a genetic alteration other than shortened Chr XII probably stabilized the unstable mutant Orc2p in the surviving type 2 clones. Since the surviving type 1 orc2-1 clone showed a ts phenotype at 28°C and produced unstable Orc2p indistinguishably from non-heat-treated orc2-1 cells (data not shown), it seemed likely that the defect in origin-firing capacity remained unchanged in this clone. The ts phenotype of orc2-1 as well as orc1-4 mutants was suppressed when the RAD9-dependent checkpoint control was inactivated (38). Therefore, we hypothesized that an extensive reduction in the copy number of the rDNA repeats would make the RAD9-dependent checkpoint response insensitive to the perturbation of origin firing in orc mutant cells.
FIG. 3.
A surviving orc2-1 clone with extensively shortened Chr XII shows a partially suppressed ts phenotype. (A) ts phenotypes of surviving orc2-1 clones. Cells of wild-type (NOY408-1b), orc2-1 (YNV1), and two surviving orc2-1 clones (SvC1 and SvC2) were grown in YPD at 23°C. Cells were serially fivefold diluted with distilled water, spotted onto YPD plates, and incubated at 23°C, 26°C, and 28°C for 2.5 days. (B) Sizes of Chr XII in surviving orc2-1 clones. Chromosomes in wild-type, orc2-1, SvC1, and SvC2 cells were analyzed by PFGE as described in Materials and Methods. The left panel for each strain shows the ethidium bromide-stained chromosome profile, and the right panel is an autoradiogram obtained after Southern hybridization with probe 058 for Chr XII. Positions of Chr XII are indicated by triangles.
The copy number of rDNA repeats modulates activation of DNA damage checkpoint response by perturbing origin firing in orc mutant cells.
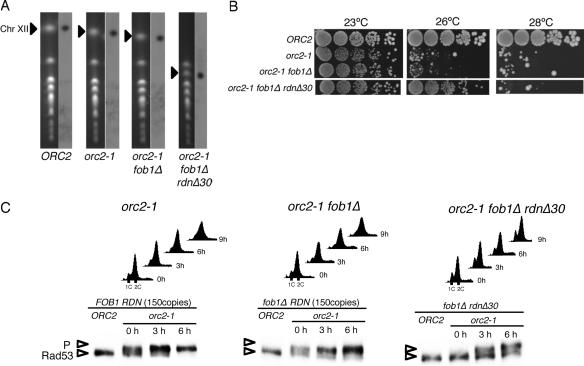
The genetic method of Chernoff et al. (10) makes it possible to construct yeast strains with reduced copy numbers of rDNA repeats. It has been shown that such a reduced copy number of rDNA recovers to 150 copies, the normal copy number of rDNA in wild-type cells, during many cycles of mitotic growth and that recovery depends on the FOB1 gene (24). By combining the Chernoff method with fob1Δ mutation, a series of strains stably inheriting 30 copies of the rDNA repeats (rdnΔ30) was constructed and used to determine whether the reduction of the rDNA copy number alone is the reason for the suppression of the ts phenotype observed with the heat-surviving orc mutants. The orc2-1 point mutation was introduced into a fob1Δ rdnΔ30 strain as well as a fob1Δ strain with the normal copy number of rDNA. The copy number in each strain was examined by measuring the size of Chr XII by PFGE (Fig. 4A) and by Southern hybridization with probes specific for rDNA after digestion of the chromosomal DNA with BglII (see Fig. S1 in the supplemental material).
FIG. 4.
Reduction of rDNA copy numbers suppresses the temperature sensitivities of orc2-1 cells and compromises the activation of the DNA damage checkpoint in orc2-1 cells after the temperature shift. (A) Sizes of Chr XIIs in orc2-1 fob1Δ rdnΔ30 cells. NOY408-1b (wild type), YNV1 (orc2-1), YNV2 (orc2-1 fob1Δ), and YNV15 (orc2-1 fob1Δ rdnΔ30) cells were separated by PFGE as described in Materials and Methods. The left panel for each strain shows an ethidium bromide-stained chromosome profile, and the right is an autoradiogram obtained after Southern hybridization with probe 058 for Chr XII. Positions of Chr XII are indicated by triangles. (B) Suppression of the temperature sensitivity of orc2-1 by reduction of the rDNA copy number. NOY408-1b (wild type), YNV1 (orc2-1), YNV2 (orc2-1 fob1Δ), and YNV15 (orc2-1 fob1Δ rdnΔ30) cells grown in YPD at 23°C were serially fivefold diluted with distilled water, spotted onto YPD plates, and incubated at 23°C, 26°C, and 28°C for 2.5 days. (C) Cell cycle progression and activation of DNA damage checkpoint control after the temperature shift. YNV1 (orc2-1), YNV2 (orc2-1 fob1Δ), and YNV15 (orc2-1 fob1Δ rdnΔ30) cells were grown in YPD at 23°C, and the growth temperature was shifted to 26°C at the 0-hour time point. At the indicated time points, aliquots were withdrawn and subjected to FACS analysis for cell cycle progression or to Western blot analysis for phosphorylation of checkpoint kinase Rad53p, as described in Materials and Methods. P indicates the phosphorylated form of Rad53p. As controls, the status of Rad53p in the corresponding ORC2 strain (NOY408-1b, NOY408-1bf, or NOY408-1bf30) for each treated strain is shown in the first lane of each panel.
As shown in Fig. 4B, the orc2-1 fob1Δ rdnΔ30 strain was able to form colonies at the nonpermissive temperature, 26°C, but not at 28°C. The growth pattern of the ts phenotype is the same as that observed with orc2-1 SvC1. In FACS profiles of orc2-1 and orc2-1 fob1Δ strains, the proportions of cells in G1 phase were significantly reduced even at 23°C, and cell cycle arrest at G2/M was clearly seen at 9 h after the shift to 26°C (Fig. 4C). On the other hand, orc2-1 fob1Δ rdnΔ30 cells grown at 23°C contained more G1-phase cells, and G2/M arrest was not observed at the 9-h time point after the temperature shift. Consistent with these observations, Rad53p was mostly phosphorylated in orc2-1 and orc2-1 fob1Δ cells grown at 23°C (Fig. 4C, 0 h) and the amount of phosphorylated Rad53p increased in these cells after the temperature shift. In orc2-1 fob1Δ rdnΔ30 cells grown at 23°C (0 h) and treated at 26°C for 3 h, however, a significant proportion of Rad53p remained unphosphorylated. The accumulation of Rad53p after the temperature shift was mitigated in the orc2-1 fob1Δ rdnΔ30 cells. Similar results were obtained with orc1-4 diploid cells with reduced copy numbers of rDNA (see Fig. S2 in the supplemental material). These data clearly indicate that the reduction of the rDNA copy number leads to the suppression of the ts phenotype of orc mutant strains by a weakening of the checkpoint response in the mutant cells at the restrictive temperature. It appeared, therefore, that the generation of DNA lesions in orc mutant cells, and/or the subsequent transmission of the checkpoint signal to Rad53p, would be affected by the reduction of the rDNA copy number.
Reduced rDNA copy numbers affect neither the impaired origin-firing capacities in orc2-1 cells nor the cellular function of DNA damage checkpoint control.
Since the rDNA array contains about 150 ORC-binding sites, one-fourth of the total number of ORC-binding sites in the genome (3), the ratio of ORC molecules to ARS-containing DNA segments may increase in cells with reduced copy numbers of rDNA. It was also shown that the ts phenotype of orc2-1 is suppressed by increasing the level of mutated Orc2p (33). Thus, it seemed probable that the reduction in the total number of ORC-binding sites per cell could restore the impaired ORC function in the orc mutants, thereby decreasing the checkpoint response at the restrictive temperature. To clarify this possibility, we examined the origin-firing capacities in orc2-1 cells with reduced rDNA copy numbers by measuring the loss rates for plasmids carrying either ARS1 or ARSH4 in the orc2-1 fob1Δ rdnΔ30 strain grown at a permissive temperature, 23°C. Yeast mutants defective in the origin-firing process, such as orc and mcm mutants, often exhibit a high index of ARS plasmid loss at the permissive temperature because they frequently fail to initiate DNA replication from the plasmid-borne origin in every cell cycle (15). ORC2 wild-type strains (ORC2 fob1Δ and ORC2 fob1Δ rdnΔ30) lost each plasmid at rates of less than 5% per generation under nonselective conditions for the plasmids. On the other hand, the orc2-1 fob1Δ mutant showed highly elevated plasmid loss rates of 15% for ARS1 and 21% for ARSH4. The high plasmid loss rate, indicative of the impaired origin-firing capacities in orc2-1 cells, was unchanged when the rDNA copy number was reduced to 30 in the orc2-1 mutant (see Fig. S3A in the supplemental material). In addition, we confirmed that intracellular concentrations of mutant Orc2p in orc2-1 fob1Δ and orc2-1 fob1Δ rdnΔ30 strains were the same at 23°C and decreased in similar manners after the temperature shift (see Fig. S3B in the supplemental material). These results clearly indicate that the defective ORC function in the orc2-1 cells was unchanged by the reduction of the rDNA copy number and that the suppression of the ts phenotype of orc2-1 was not due to a recovery of ORC function.
The conclusion described above strongly suggested that the reduction of the rDNA copy number made cells ineffective for activation of the DNA damage checkpoint control against the perturbation of origin firing. This suggested in turn that the DNA damage checkpoint might malfunction when the rDNA copy number is greatly decreased. We therefore examined the DNA damage checkpoint response after treatment of the fob1Δ rdnΔ30 cells with a DNA alkylating agent, methyl methanesulfonate (MMS). The progression of the cell cycle and the phosphorylation of Rad53p were analyzed for wild-type, fob1Δ, and fob1Δ rdnΔ30 cells grown in liquid medium containing MMS. The cell cycle in fob1Δ rdnΔ30 cells was completely arrested at the G2/M boundary by 180 min after the addition of MMS to the medium, with the same pattern of DNA damage checkpoint response as those observed with the wild type (FOB1) and fob1Δ cells. The time courses and the extents of Rad53p phosphorylation after the MMS treatment were also the same in all three strains (see Fig. S4 in the supplemental material). From these data, we concluded that the sensing, transmitting, and executing functions of the DNA damage checkpoint control were normal in cells with reduced rDNA copy numbers.
Completion of chromosomal duplication is delayed specifically for Chr XII in orc2-1 cells and is relieved by reducing the rDNA copy number.
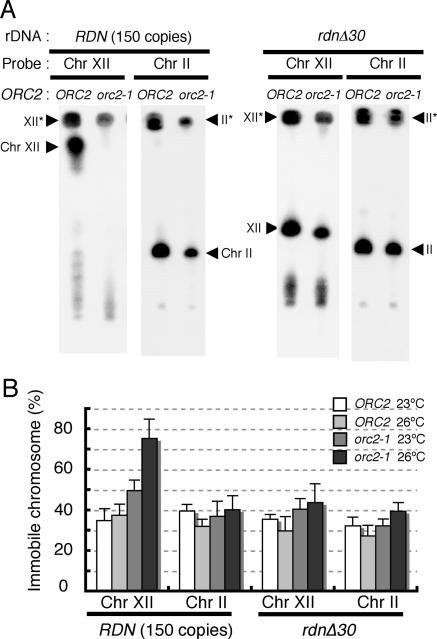
Considering that the function of DNA damage checkpoint control was not affected in cells with extensively reduced copy numbers of rDNA, we speculated that the reduction of rDNA repeats to 30 copies would significantly decrease the total number of DNA lesions per cell when origin firing was perturbed in orc mutants at restrictive temperatures. If the DNA lesions induced within the rDNA array accounted for the vast majority of all DNA lesions within the whole genome of orc mutant cells, the reduction of the rDNA copy number would compromise the activation of the checkpoint response and suppress the ts phenotype of the orc mutants. To ascertain that DNA lesions are induced more frequently within the rDNA locus than at other chromosomal loci and that the DNA lesions are reduced by the extensive reduction of the rDNA copy number, we compared the status of Chr XII with that of Chr II in the orc2-1 cells with and without rdnΔ30 after incubation for 6 h at 26°C, the nonpermissive temperature for the mutant.
Using PFGE, in which chromosomes with branched DNA structures, such as replication forks and recombination intermediates, cannot migrate (18, 23), we measured the proportion of chromosomes undergoing DNA replication or recombination for Chr II and Chr XII (Fig. 5). In exponentially growing wild-type cells at 23°C, 35 to 40% of both chromosomes did not migrate in PFGE, reflecting chromosomes with branched DNA structures. The percentages of immobile Chr II and Chr XII were unchanged when the wild-type cells were grown at 26°C. In orc2-1 cells grown at 23°C, the PFGE-immobile fraction of Chr II was also 35 to 40%. When the orc2-1 cells were incubated at 26°C for 6 h, the percentage of immobile Chr II was unchanged. This clearly indicated that the duplication of Chr II in orc2-1 cells was completed in almost the same time at 23°C and 26°C. On the other hand, the PFGE-immobile fraction of Chr XII increased visibly in the orc2-1 cells; 50% of Chr XII in orc2-1 cells grown at 23°C did not enter the gel, and this proportion increased to 75% when the cells were grown at 26°C. Furthermore, using α-factor-mediated synchronization of the cell cycle, we examined the fate of Chr XII during S phase in wild-type and orc2-1 cells at 23°C (see Fig. S5 in the supplemental material). Even at the permissive temperature, the PFGE-immobile fraction of Chr XII in orc2-1, but not wild-type, cells remained beyond the G2/M boundary, whereas duplication of other chromosomes was fully completed by this stage. From these observations, we concluded that completion of chromosomal replication and/or sister chromatid recombination was delayed specifically for Chr XII in orc2-1 cells. This Chr XII-specific delay strongly suggested that DNA lesions were induced more frequently within Chr XII than within other chromosomes in orc2-1 mutant cells. The reduction of the rDNA copy number relieved the delay in Chr XII duplication (Fig. 5), whereas deletion of the FOB1 gene had little effect on the delay (data not shown). The proportion of PFGE-immobile Chr XII was reduced from 75% in orc2-1 cells to 43% in orc2-1 rdnΔ30 cells grown at 26°C. It appears that the copy number of rDNA affects the delay in the completion of Chr XII duplication and thus determines the extent of DNA lesions occurring within the rDNA locus.
FIG. 5.
Completion of Chr XII duplication is specifically delayed in orc2-1 cells but restored by reduction of the rDNA copy numbers. (A) The status of Chr XII and Chr II in exponentially growing NOY408-1b (ORC2 FOB1 RDN150), YNV1 (orc2-1 FOB1 RDN150), NOY408-1bf30 (ORC2 fob1Δ rdnΔ30), and YNV15 (orc2-1 fob1Δ rdnΔ30) was examined by PFGE as described in Materials and Methods. Each strain was incubated in YPD at 26°C for 6 h after the temperature shift from 23°C. Cells were collected and prepared in agarose plugs for PFGE. Chromosome patterns were detected and quantified by Southern hybridization using Chr XII- and Chr II-specific probes, as described in Materials and Methods. (B) The percentages of chromosomes which remained in the plug after electrophoresis (XII* and II*) were quantified for NOY408-1b (ORC2 FOB1 RDN150), YNV1 (orc2-1 FOB1 RDN150), NOY408-1bf30 (ORC2 fob1Δ rdnΔ30), and YNV15 (orc2-1 fob1Δ rdnΔ30) grown at 23°C and 26°C for 6 h. Average values from three independent experiments are shown. Error bars represent standard deviations.
The origin activities of rDNA ARSs are affected by orc2-1 mutation to an extent similar to that associated with the effect on ARS1.
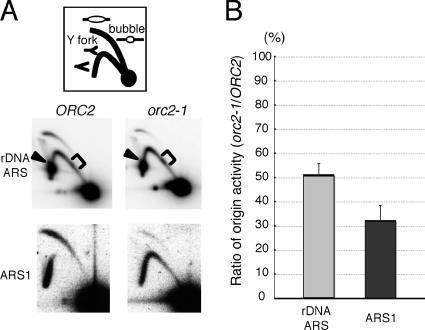
A plausible explanation for the delay in the completion of chromosomal duplication specific for Chr XII is that the origin activities of rDNA ARSs are affected by orc2-1 mutation more severely than those of other ARSs and that such a rigorous reduction of origin activity also leads to generation of DNA lesions specifically on Chr XII. Using 2D gel electrophoresis to examine this possibility, we measured the origin activities of rDNA ARSs on Chr XII as well as ARS1 on Chr IV in wild-type and orc2-1 cells growing exponentially at 23°C (Fig. 6). In orc2-1 cells, the origin activity of ARS1 was reduced to about one-third of that in wild-type cells. Contrary to our expectation, the origin activities of rDNA ARSs appeared to be affected by orc2-1 mutation to an extent similar to that associated with the effect on ARS1 (Fig. 6B). From the 2D gel patterns of replication intermediates around rDNA ARSs, we obtained no obvious indication that progression of DNA synthesis is inhibited or that the amount of recombination intermediates increases within the rDNA segment in the orc2-1 cells. Interestingly, the intensity of a spot corresponding to the Y-fork intermediate pausing at the 5S rRNA gene increased slightly in the 2D-gel pattern for orc2-1 cells. From these data, we concluded that the rDNA ARSs are not exceptionally sensitive to the defective origin-firing capacities of orc2-1 mutant cells. Therefore, it seemed unlikely that the delay in the completion of Chr XII duplication in orc2-1 cells is simply due to decreased origin activities of rDNA ARSs in the cells.
FIG. 6.
Effects of orc2-1 on the origin activities of rDNA ARSs in Chr XII. (A) Two-dimensional gel electrophoresis of replication intermediates of chromosomal fragments containing rDNA ARSs and ARS1 from wild-type and orc2-1 strains. DNA was prepared from NOY408-1b (wild type) and YNV1 (orc2-1) cells growing at 23°C and digested with NheI for mapping of rDNA ARSs or with NcoI for mapping of ARS1. The DNA preparations were subjected to 2D gel electrophoresis followed by Southern hybridization using an rDNA-specific probe or a probe specific for the site around ARS1, as described in Materials and Methods. Bubble arc and Y fork correspond to replication intermediates as depicted in the top panel. Arrowheads show Y forks paused at the RFB site in the rDNA unit, and brackets show Y-fork intermediates paused at a region encoding 5S rRNA. (B) Reduced origin activities of rDNA ARSs and ARS1 in orc2-1 cells. Origin activities in NOY408-1b (wild type) and YNV1 (orc2-1) cells were determined for each ARS by dividing the intensities of bubble intermediates by those of Y-fork intermediates, and the ratio of origin activity in YNV1 (orc2-1) to that in NOY408-1b (the wild type) was calculated for each ARS. The thick bars indicates the average ratios (%) of origin activity from two independent experiments. Error bars represent standard deviations.
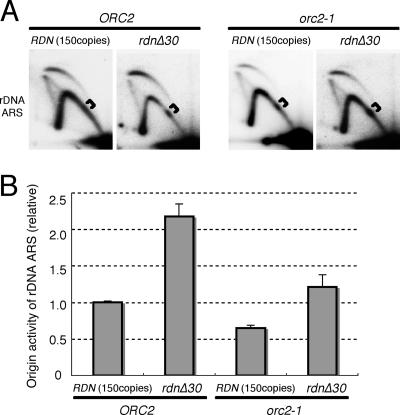
The origin activities of rDNA ARSs are enhanced when the rDNA copy numbers are reduced.
We observed reproducibly that the origin activities of rDNA ARSs in orc2-1 cells at 23°C were reduced to about 50% of those in wild-type cells. If this decreased origin activity involves the activation of the DNA damage checkpoint and the specific delay in the completion of Chr XII in orc2-1 cells, the effect of orc2-1 mutation on rDNA ARSs should be counteracted in cells having reduced copy numbers of rDNA units. As shown in Fig. 7, this was the case. The origin activities of rDNA ARSs in the orc2-1 strain increased twofold when rdnΔ30 was introduced and thus became comparable to those in the wild-type strain. However, this enhancement of the origin activities of rDNA ARSs was not confined to the orc2-1 strain: the origin activities of rDNA ARSs were also enhanced twofold when the copy number of rDNA was reduced in an ORC2-proficient strain (Fig. 7). Therefore, in the genetic background with rdnΔ30, the origin activities of rDNA ARSs were affected by orc2-1 mutation, being reduced to about 50% of that in the ORC2-proficient strain (see Fig. S6 in the supplemental material). This implies that the defect in origin-firing capacity in the orc2-1 strain was not restored per se but was relieved by an increased competence of rDNA ARSs for initiating DNA replication when the copy number of rDNA was reduced to 30. The pausing of replication at 5S rRNA genes was apparently increased in the ORC2 rdnΔ30 strain as well as in the orc2-1 rdnΔ30 strain. From these results, we concluded that the level of origin activity for rDNA ARSs or the efficiency of origin firing at rDNA ARSs is an important determinant for activation of DNA damage checkpoint control.
FIG. 7.
Effects of rdnΔ30 on the origin activities of rDNA ARSs in Chr XII. (A) Two-dimensional gel electrophoresis of replication intermediates of chromosomal fragments containing rDNA ARSs from wild-type (150 copies of the rDNA) and rdnΔ30 (30 copies) strains. DNA was prepared from NOY408-1bf (fob1Δ), NOY408-1bf30 (fob1Δ rdnΔ30), YNV2 (fob1Δ orc2-1), and YNV15 (fob1Δ orc2-1 rdnΔ30) cells growing at 23°C, digested with NheI, and subjected to 2D gel electrophoresis followed by Southern hybridization using an rDNA-specific probe, as described in Materials and Methods. Brackets show Y-fork intermediates paused at a region encoding 5S rRNA. (B) Increased origin activities of rDNA ARSs in wild-type and orc2-1 cells with reduced copy numbers of rDNA. Origin activities in NOY408-1bf (fob1Δ), NOY408-1bf30 (fob1Δ rdnΔ30), YNV2 (fob1Δ orc2-1), and YNV15 (fob1Δ orc2-1 rdnΔ30) cells were determined for rDNA ARSs as described in the legend for Fig. 6. The thick bars indicate the average origin activities from two independent experiments, and the values expressed are relative to that obtained with NOY408-1bf. Error bars represent standard deviations.
DISCUSSION
Several groups have found that when origin firing is perturbed, the subsequent chromosomal replication with fewer origins leads to loss of chromosomes and chromosomal rearrangement (3, 8, 14, 21, 22, 38). Our previous study showed that such chromosome instability is mainly caused by perturbation of initiation program (PIP)-dependent DNA lesions, which trigger DNA damage checkpoint control (38). We now report that such DNA lesions are induced much more frequently within the rDNA locus than at other chromosomal loci when orc mutants are transferred to the restrictive temperature. Furthermore, we reveal that the copy number of rDNA repeats modulates cellular capacity to monitor an abnormal initiation program of chromosomal DNA replication through the checkpoint response. Finally, we deduce that the rDNA array plays an important role in the maintenance of the yeast genome.
The rDNA locus becomes extremely fragile when the initiation program is perturbed.
In orc1-4 and orc2-1 mutant cells incubated at the restrictive temperature for 24 h, size aberrations in Chr XII, attributable to a reduction in the copy number of rDNA repeats, occurred at an extremely high frequency and nearly all the cells suffered. However, this extreme instability of the rDNA array was not seen in orc1-4 cells for the first several hours after the temperature shift. This is because RAD9-dependent DNA damage checkpoint control is activated immediately after the temperature shift and promotes proper repair of the lesions produced in the rDNA locus. In orc1-4 rad9Δ cells, where the checkpoint control was compromised, hyperinstability of Chr XII was induced immediately in all the cells after the temperature shift. On the other hand, the frequency of chromosomal aberration in Chr III, mostly chromosome loss, increased greatly in orc1-4 rad9Δ diploid cells grown at the restrictive temperature for 24 h but was detected only in less than 0.1% of the cells (38). Furthermore, in contrast to what was found for the orc1-4 rad9Δ cells, Chr III was maintained stably in orc2-1 rad9Δ cells grown at 26°C, the restrictive temperature for the orc2-1 mutant cells, for 24 h (38). This is consistent with the finding that the delay in the completion of chromosomal duplication was confined to Chr XII in orc2-1 cells grown at 26°C. These observations indicate that the rDNA locus is the site most sensitive to PIP DNA lesions within the yeast genome.
PIP DNA lesions induced within the rDNA locus.
What is the nature of the lesion induced within the rDNA locus when the initiation program of DNA replication is perturbed? The most likely candidate for the PIP DNA lesion is a stalled replication fork, because completion of chromosomal replication was delayed specifically on Chr XII in orc2-1 cells. It seems probable that a reduction in the efficiency of origin firing would lead to an enlargement in the average size of the replicons. In such a case, replication forks would travel a longer distance and sometimes move in the wrong direction; consequently, there would be more chance for the replication fork to encounter obstacles on the DNA, such as spontaneous DNA damage, proteins bound to DNA for various DNA transactions, and particular chromatin structures unsuitable for DNA replication. Indeed, a yeast artificial chromosome (YAC) that contains a 170-kb region lacking efficient replication origins triggers RAD9-dependent checkpoint control, and the YAC becomes unstable in the absence of Rad9p, indicating chronic induction of DNA lesions in cells carrying the origin-deficient YAC (37). Since each rDNA unit (9.1 kb) possesses a replication origin (ARS), the rDNA locus is a unique chromosomal region, having the highest density of ARSs in the yeast genome (7). One in five ARSs in the rDNA array initiates bidirectional DNA replication, and several adjacent ARSs simultaneously fire to replicate a subcluster of the rDNA array (6, 27, 30). Therefore, the size of a replicon involved in rDNA replication seems to be tightly regulated by the initiation program for this specialized chromosomal region, and the replicons in the subcluster would be maintained as the smallest in the yeast genome. If the efficiency of origin firing were reduced in the rDNA array, such small replicons would be substantially enlarged, and the probability for each replication fork to stall would be increased. On the other hand, the gap between each cluster is larger than 60 kb. Although it is not known whether the DNA segment corresponding to the gap is replicated as a single replicon or as clusters of many replicons, the rDNA locus may contain replicons larger than the average size for replicons on other chromosomes, about 46 kb (25, 30). If the gap segment is a single and very large replicon, perturbation of origin firing would result in the emergence of further larger replicons within the rDNA array.
However, the extremely high instability of the rDNA array may not be attributable only to an enlarged replicon in the rDNA array. We reasoned that the stalled replication fork may involve the transcription of 5S rRNA, of which one gene is located in each rDNA segment. In an rDNA unit which initiates bidirectional DNA replication from the origin, one replication fork proceeds in the same direction as the transcription of 5S rRNA and the other proceeds in the same direction as the transcription of 35S rRNA. When the former replication fork reaches the RFB site, it stops and never goes beyond the RFB site. In contrast, the latter proceeds over the RFB site and enters the adjacent rDNA unit if it has not been replicated. This is because RFB is highly orientation dependent (6, 24). Thus, head-on collisions between replication and transcription machineries within the 35S rRNA gene region are always prevented by the RFB, whereas such collisions are unavoidable within the 5S rRNA gene region in passively replicated rDNA units (12). Probably, in wild-type cells, one replication fork moving in the same direction as the 35S rRNA needs to pass through a 5S rRNA gene region several times in a direction opposite to that of the 5S rRNA transcription. When origin firing is perturbed within the rDNA array, each of a reduced number of replication forks would have to traverse a greater number of rDNA segments, leading to a heightened replication stress that arises on passing through the 5S rRNA gene.
The model described above is supported by the results of 2D gel analysis, which indicated that collisions between replication fork and transcription machineries within the 5S rRNA gene were more frequent in orc2-1 cells (Fig. 6) (see Fig. S6 in the supplemental material). However, an increased frequency of such collisions was also observed in cells with shorter rDNA arrays, even though the function of the ORC was normal. This is probably because, like the increased level of 35S rRNA transcription in cells with reduced copy numbers of rDNA (36), each 5S rRNA gene in the rDNA array would be much more actively transcribed. The reduction of rDNA copy numbers simultaneously enhanced the origin activities of rDNA ARSs, resulting in a reduction of replicon size in the rDNA array. Therefore, in cells with shorter rDNA arrays, collision between replication and transcription machineries may occur in a chromatin context distinct from that in cells with normal-sized rDNA arrays and be processed properly to avoid harmful consequences from the collision.
Interplay between PIP DNA lesions and DNA damage checkpoint control within the rDNA locus.
Another unique feature of the rDNA locus is the interplay occurring between the hyperinstability of the rDNA array and DNA damage checkpoint control. In RAD9-proficient orc1-4 cells, checkpoint control was activated by the PIP DNA lesions produced within the rDNA array immediately after the temperature shift and functioned to maintain the integrity of all chromosomes, including Chr XII. However, at some time between 2.5 and 5 h after the temperature shift, Chr XII became unstable, even though checkpoint control continued to suppress rigorously the instability of other chromosomes. This indicates that the PIP DNA lesions within the rDNA locus increased to a level beyond the capacities of repair functions powered by checkpoint control and remained unrepaired thereafter. It is of interest that PIP DNA lesions on chromosomes other than Chr XII were adequately repaired until the catastrophic (10-h) time point, beyond which the cells started to cancel the G2/M cell cycle arrest and to undergo cell death (Fig. 1A). As a result, it seems likely that DNA damage checkpoint control is continuously activated by the unrepaired PIP DNA lesions within the rDNA array in cells with perturbed initiation programs. Thus, the fragility of the rDNA may greatly help cells to monitor abnormalities in the initiation program of DNA replication.
rDNA copy number modulates the sensitivity of DNA damage checkpoint control in monitoring the perturbation of origin firing.
When the rDNA copy numbers were extensively reduced in orc1-4 and orc2-1 cells, the activation of checkpoint control after the temperature shift was weakened. It is clear that the reduction of the rDNA copy number affects neither the deficiency of orc mutants in origin-firing capacity nor the cellular capacity of DNA damage checkpoint control. In cells with extensively reduced rDNA copy numbers, the amounts of DNA lesions produced within the shortened rDNA array are probably greatly reduced and more readily repaired, resulting in feeble activation of checkpoint control. The balance between the extent of DNA lesions produced within the rDNA locus and the cell's capacity for DNA repair would determine the duration of the DNA damage checkpoint response: the lower the rDNA copy number, the fewer the PIP DNA lesions produced within the rDNA locus and the shorter the duration of the checkpoint response. Therefore, the rDNA copy number is an important factor that modulates the sensitivity of DNA damage checkpoint control in responding to a reduced capacity for initiation of DNA replication.
The rDNA array plays an important role in genome maintenance.
From the above-described considerations, we propose that the rDNA array functions as a sensor to monitor perturbation in the initiation program of chromosomal replication and counteracts unfavorable fluctuations of initiation potential during the course of S-phase progression. Although little is known about the programmed origin firing for the multiple origins within the eukaryotic genome, the initiation potential is expected to be maintained at a certain level throughout S phase, perhaps fluctuating slightly as is usually found in biological phenomena. As we observed with the orc1-4 rad9Δ strain, cells could tolerate under initiation as well as overinitiation if the extent of such variation were small. However, even to a small degree, fluctuation of initiation potential could lead to genetic instability. Cells are able to avoid such a fatal risk if they are equipped with a feedback loop for the controlling mechanism. The rDNA array seems to be ideal for such a feedback loop: it is highly sensitive to perturbation of origin firing, is readily reparable by sister chromatid recombination, and is highly plastic because of its repetitive nature. A certain minimum level of repetitiveness is required for the rDNA array to act properly as a monitor for perturbation of the initiation program. We postulate that yeast cells maintain the copy numbers of rDNA at around 150 to attune the sensitivity of this monitoring function to fluctuations of initiation potential.
Supplementary Material
Acknowledgments
We are grateful to Hiroyuki Araki and Hideyuki Saya for discussion and advice. We thank Ian Smith for helpful comments on the manuscript for this paper. We also thank Yasuo Kawasaki for providing affinity-purified anti-Orc2p antibody.
We acknowledge the financial support of Grants-in-Aid for Scientific Research on Priority Areas (17013060 to H.M.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. S.I. was supported by the 21st Century Center of Excellence Program (Graduate School of Biological Sciences, NAIST) from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 13 November 2006.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bartkova, J., Z. Horejsi, K. Koed, A. Kramer, F. Tort, K. Zieger, P. Guldberg, M. Sehested, J. M. Nesland, C. Lukas, T. Orntoft, J. Lukas, and J. Bartek. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864-870. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. [DOI] [PubMed] [Google Scholar]
- 3.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 4.Bell, S. P., R. Kobayashi, and B. Stillman. 1993. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science 262:1844-1849. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 6.Brewer, B. J., and W. L. Fangman. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55:637-643. [DOI] [PubMed] [Google Scholar]
- 7.Brewer, B. J., and W. L. Fangman. 1991. Mapping replication origins in yeast chromosomes. BioEssays 13:317-322. [DOI] [PubMed] [Google Scholar]
- 8.Bruschi, C. V., J. N. McMillan, M. Coglievina, and M. S. Esposito. 1995. The genomic instability of yeast cdc6-1/cdc6-1 mutants involves chromosome structure and recombination. Mol. Gen. Genet. 249:8-18. [DOI] [PubMed] [Google Scholar]
- 9.Burhans, W. C., M. Weinberger, M. A. Marchetti, L. Ramachandran, G. D'Urso, and J. A. Huberman. 2003. Apoptosis-like yeast cell death in response to DNA damage and replication defects. Mutat. Res. 532:227-243. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff, Y. O., A. Vincent, and S. W. Liebman. 1994. Mutations in eukaryotic 18S ribosomal RNA affect translational fidelity and resistance to aminoglycoside antibiotics. EMBO J. 13:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dani, G. M., and V. A. Zakian. 1983. Mitotic and meiotic stability of linear plasmids in yeast. Proc. Natl. Acad. Sci. USA 80:3406-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshpande, A. M., and C. S. Newlon. 1996. DNA replication fork pause sites dependent on transcription. Science 272:1030-1033. [DOI] [PubMed] [Google Scholar]
- 13.Ekholm-Reed, S., J. Mendez, D. Tedesco, A. Zetterberg, B. Stillman, and S. I. Reed. 2004. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell Biol. 165:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elble, R., and B. K. Tye. 1992. Chromosome loss, hyperrecombination, and cell cycle arrest in a yeast mcm1 mutant. Mol. Biol. Cell 3:971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foss, M., F. J. McNally, P. Laurenson, and J. Rine. 1993. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science 262:1838-1844. [DOI] [PubMed] [Google Scholar]
- 16.Friedman, K. L., B. J. Brewer, and W. L. Fangman. 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2:667-678. [DOI] [PubMed] [Google Scholar]
- 17.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 18.Hennessy, K. M., A. Lee, E. Chen, and D. Botstein. 1991. A group of interacting yeast DNA replication genes. Genes Dev. 5:958-969. [DOI] [PubMed] [Google Scholar]
- 19.Hiraoka, M., K. Watanabe, K. Umezu, and H. Maki. 2000. Spontaneous loss of heterozygosity in diploid Saccharomyces cerevisiae cells. Genetics 156:1531-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hori, Y., K. Shirahige, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1996. Characterization of a novel CDC gene (ORC1) partly homologous to CDC6 of Saccharomyces cerevisiae. Mol. Biol. Cell 7:409-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, D., and D. Koshland. 2003. Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev. 17:1741-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson, M. D., C. X. Munoz, K. S. Knox, B. E. Williams, L. L. Lu, F. R. Cross, and E. A. Vallen. 2001. Mutations in SID2, a novel gene in Saccharomyces cerevisiae, cause synthetic lethality with sic1 deletion and may cause a defect during S phase. Genetics 159:17-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaliraman, V., and S. J. Brill. 2002. Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 41:389-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi, T., D. J. Heck, M. Nomura, and T. Horiuchi. 1998. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12:3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lengronne, A., P. Pasero, A. Bensimon, and E. Schwob. 2001. Monitoring S phase progression globally and locally using BrdU incorporation in TK(+) yeast strains. Nucleic Acids Res. 29:1433-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, C., M. Weinreich, and B. Stillman. 1995. ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81:667-676. [DOI] [PubMed] [Google Scholar]
- 27.Linskens, M. H., and J. A. Huberman. 1988. Organization of replication of ribosomal DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 8:4927-4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipford, J. R., and S. P. Bell. 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol. Cell 7:21-30. [DOI] [PubMed] [Google Scholar]
- 29.Paciotti, V., G. Lucchini, P. Plevani, and M. P. Longhese. 1998. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 17:4199-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pasero, P., A. Bensimon, and E. Schwob. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev. 16:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294:115-121. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Shimada, K., P. Pasero, and S. M. Gasser. 2002. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 16:3236-3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395:618-621. [DOI] [PubMed] [Google Scholar]
- 35.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi, Y., T. Horiuchi, and T. Kobayashi. 2003. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 17:1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Brabant, A. J., C. D. Buchanan, E. Charboneau, W. L. Fangman, and B. J. Brewer. 2001. An origin-deficient yeast artificial chromosome triggers a cell cycle checkpoint. Mol. Cell 7:705-713. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, K., J. Morishita, K. Umezu, K. Shirahige, and H. Maki. 2002. Involvement of RAD9-dependent damage checkpoint control in arrest of cell cycle, induction of cell death, and chromosome instability caused by defects in origin recognition complex in Saccharomyces cerevisiae. Eukaryot. Cell 1:200-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weinberger, M., L. Ramachandran, L. Feng, K. Sharma, X. Sun, M. Marchetti, J. A. Huberman, and W. C. Burhans. 2005. Apoptosis in budding yeast caused by defects in initiation of DNA replication. J. Cell Sci. 118:3543-3553. [DOI] [PubMed] [Google Scholar]
- 40.Yamashita, M., Y. Hori, T. Shinomiya, C. Obuse, T. Tsurimoto, H. Yoshikawa, and K. Shirahige. 1997. The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. Genes Cells 2:655-665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.