Abstract
Plant embryo development is regulated by a network of transcription factors that include LEAFY COTYLEDON 1 (LEC1), LEC1-LIKE (L1L), and B3 domain factors, LEAFY COTYLEDON 2 (LEC2), FUSCA3 (FUS3), and ABSCISIC ACID INSENSITIVE 3 (ABI3) of Arabidopsis (Arabidopsis thaliana). Interactions of these genes result in temporal progression of overlapping B3 gene expression culminating in maturation and desiccation of the seed. Three VP1/ABI3-LIKE (VAL) genes encode B3 proteins that include plant homeodomain-like and CW domains associated with chromatin factors. Whereas val monogenic mutants have phenotypes similar to wild type, val1 val2 double-mutant seedlings form no leaves and develop embryo-like proliferations in root and apical meristem regions. In a val1 background, val2 and val3 condition a dominant variegated leaf phenotype revealing a VAL function in vegetative development. Reminiscent of the pickle (pkl) mutant, inhibition of gibberellin biosynthesis during germination induces embryonic phenotypes in val1 seedlings. Consistent with the embryonic seedling phenotype, LEC1, L1L, ABI3, and FUS3 are up-regulated in val1 val2 seedlings in association with a global shift in gene expression to a profile resembling late-torpedo-stage embryogenesis. Hence, VAL factors function as global repressors of the LEC1/B3 gene system. The consensus binding site of the ABI3/FUS3/LEC2 B3 DNA-binding domain (Sph/RY) is strongly enriched in the promoters and first introns of VAL-repressed genes, including the early acting LEC1 and L1L genes. We suggest that VAL targets Sph/RY-containing genes in the network for chromatin-mediated repression in conjunction with the PKL-related CHD3 chromatin-remodeling factors.
Embryo development in plants is regulated by a network of transcription factors that include the LEAFY COTYLEDON 1 (LEC1) and LEC1-LIKE (L1L) genes belonging to the HAP3 family CCAAT-binding factors and a subgroup of the plant-specific B3 domain protein family composed of the LEC2, FUSCA3 (FUS3), and ABSCISIC ACID (ABA)-INSENSITIVE 3 (ABI3) genes (Giraudat et al., 1992; Lotan et al., 1998; Luerssen et al., 1998; Stone et al., 2001; Kwong et al., 2003).
A series of studies have detailed the complex sequential and regional expression patterns and mutual interactions among these regulators, which underlie the process of embryo formation (Parcy et al., 1997; Nambara et al., 2000; Raz et al., 2001; Brocard-Gifford et al., 2003; Baumbusch et al., 2004; Kagaya et al., 2005; Santos Mendoza et al., 2005; To et al., 2006). LEC1 and L1L act early in embryogenesis and initiate a complex progression of B3 transcription factor expression associated with the transition from embryo morphogenesis to embryo maturation and acquisition of desiccation tolerance. RNA interference of L1L function has been shown to cause embryo arrest during morphogenesis (Kwong et al., 2003), whereas lec1 embryos complete morphogenesis and are capable of further development into plants if rescued prior to desiccation of the seed (Meinke, 1992, 1994; West et al., 1994). Ectopic expression of LEC1 or L1L is sufficient to induce embryo formation in vegetative organs (Lotan et al., 1998; Kwong et al., 2003). The lec1, fus3, and lec2 mutations cause partial transformation of cotyledons to leaf-like organs (Meinke, 1992, 1994; Keith et al., 1994; West et al., 1994). The fus3 mutant embryos, if rescued, are also viable (Keith et al., 1994; Meinke et al., 1994). Overexpression of LEC2 or FUS3 causes ectopic expression of embryonic traits in vegetative tissues, albeit not as strongly as induced by LEC1 or L1L misexpression (Stone et al., 2001; Tsuchiya et al., 2004). Finally, ABI3 is required for maturation-related and ABA-regulated gene expression and establishment of embryo dormancy during mid-to-late embryo development (Koornneef et al., 1984). Null abi3 mutant embryos are morphologically almost normal and viable if rescued prior to desiccation (Giraudat et al., 1992; Nambara et al., 1992, 1994). Plants that ectopically express ABI3 or the maize (Zea mays) ortholog VP1 develop normally, but have dramatically altered ABA-dependent gene expression in vegetative tissues (Parcy et al., 1994; Suzuki et al., 2001).
A major function of the LEC2, FUS3, and ABI3 B3 transcription factors is activation of genes involved in accumulation of storage protein and lipid reserves in the embryo during seed maturation. Activation of downstream genes is mediated by specific binding of the B3 domain (Suzuki et al., 1997; Kroj et al., 2003; Carranco et al., 2004; Monke et al., 2004; Braybrook et al., 2006) to the Sph/RY cis-element (Hattori et al., 1992; Kao et al., 1996; Ezcurra et al., 1999; Reidt et al., 2000; Chandrasekharan et al., 2003; Nag et al., 2005). In addition, ABI3 mediates ABA-regulated gene expression in the seed through interaction with specific basic Leu-zipper transcription factors that bind ABA response elements (Finkelstein et al., 2002). FUS3 and LEC2 are implicated in repression of GA biosynthesis during seed development (Curaba et al., 2004; Gazzarrini et al., 2004). The B3 factors themselves are regulated in part through mutual interactions, as well as unidentified region-specific signals within the embryo (Kagaya et al., 2005; Santos Mendoza et al., 2005; To et al., 2006).
The LEC1/B3 network is repressed prior to germination and resumption of vegetative development. Chromatin-based repression of the pathway is implicated by pickle (pkl), a conditional recessive mutant that causes expression of embryonic characteristics in roots of seedlings treated with GA biosynthesis inhibitors (Ogas et al., 1997). The PKL gene encodes a CHD3 chromatin-remodeling factor (Ogas et al., 1999). Several of the embryonic regulatory genes, including LEC1, LEC2, and FUS3, are derepressed in roots of pkl seedlings (Dean Rider et al., 2003).
Here we show that the VP1/ABI3-LIKE (VAL) family of B3 domain transcription factors that form a sister clade to the ABI3/FUS3/LEC2 family are required for repression of the LEC1/B3 transcription factor network in germinating seedlings. The val1 val2 double mutant exhibits strong expression of embryonic characteristics in shoot as well as root tissues. Moreover, germination of val1 monogenic mutant seeds on low doses of paclobutrazol, a GA biosynthesis inhibitor, induces embryonic phenotypes reminiscent of pkl (Ogas et al., 1997). Derepression of the LEC1/B3 network in seedlings lacking VAL function is associated with a shift in global gene expression to a profile characteristic of late-torpedo-stage embryos. The Sph/RY consensus motif is highly enriched in promoters and introns of VAL-repressed genes, including LEC1 and L1L.
RESULTS
VAL Genes and Proteins
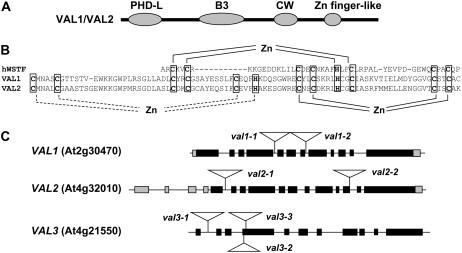
The three Arabidopsis (Arabidopsis thaliana) VAL genes encode proteins that contain B3 domains that are closely related to the ABI3/FUS3/LEC2 family of B3 transcription factors. The VAL1 gene is identical to HSI2, which was recently identified as a transcriptional repressor for a sugar-inducible gene (Tsukagoshi et al., 2005). In addition, PSI-BLAST analysis (Altschul et al., 1997; Schaffer et al., 2001) of the proteins identified two other conserved domains, a CW domain of unknown function described previously (Perry and Zhao, 2003) and a putative plant homeodomain (PHD)-like zinc (Zn)-finger domain (Fig. 1A). CW and PHD domains are frequently found in chromatin factors (Aasland et al., 1995; Perry and Zhao, 2003). An alignment of the PHD-like region with a canonical PHD domain showed that, in addition to the conserved His and Cys residues that comprise the interleaved Zn-finger structure, the VAL domain potentially includes a third, interleaved Zn finger (Fig. 1B). While there is precedent for similar expansion of the first loop in known PHD domains, to our knowledge the hypothetical third finger is novel though consistent with structural constraints of the canonical PHD fold. Except for the VAL3 sequence inferred from the Arabidopsis genome annotation that lacks an intact PHD-like domain, the domain architecture is conserved in VAL orthologs identified in rice (Oryza sativa), maize, and poplar (Populus spp.; data not shown). These structural features imply that VAL proteins are DNA-binding proteins that potentially have chromatin-related functions. The Zn-finger-like domain in the C-terminal region is highly novel and the function of this domain is unknown. Interestingly, the only closely related sequence detected by PSI-BLAST occurs in an isoform of CAAT binding factor subunit C found only in primates (data not shown).
Figure 1.
Structures of VAL1, VAL2, and VAL3. A, The four conserved domains of VAL1 and VAL2 include PHD-like (PHD-L), B3, CW (Perry and Zhao, 2003), and a putative Zn-finger-like motif. B, PHD-L sequences of VAL1 and VAL2 are aligned with the canonical PHD domain of human Williams syndrome transcription factor (hWSTF; AAC97879). The two interleaved Zn fingers, which are conserved in canonical PHD domains, are indicated with solid brackets. A potential third finger structure of VAL factors is shown by a dashed bracket. C, Gene structures of the VAL genes as well as the T-DNA insertion mutations in each gene are shown. Filled and gray boxes represent coding exons and untranslated transcribed regions, respectively. The VAL2 gene has four noncoding exons in the 5′ region. The VAL1 gene is identical to HSI2 (Tsukagoshi et al., 2005).
VAL Genes Regulate Plant Development in a Functionally Redundant Manner
To determine the functions of VAL genes in plant development, we identified multiple T-DNA insertion alleles for each of the three VAL genes (Fig. 1C). The single mutants lack discernible morphological phenotypes, albeit val1 homozygous plants tend to grow more slowly and flower slightly later than wild type.
Because genetic redundancy could mask essential functions of the VAL genes, we generated double mutants between VAL1, VAL2, and VAL3 as well as triple mutants. The val2-1 val3-3 double mutant and all pairwise combinations of val1 alleles with val2 and val3 alleles, except val1-2 val3-2, were tested. Whereas the val2 val3 double mutant did not have a discernible phenotype, the double mutants involving val1 showed altered development. In the val1 homozygous mutant background, plants with val2/+, val3/+, or val3/val3 genotypes exhibited pronounced variegation of rosette leaves (Fig. 2, A–C; Supplemental Fig. S1). Under a long-day growth regime (16-h light/8-h dark), variegation was evident in rosette leaves produced immediately prior to flowering and in cauline leaves, but was absent during early vegetative growth. The variegation phenotypes produced by val3/+ heterozygous, val2/+ heterozygous, and val3 homozygous plants exhibited a similar range of variation and were visually indistinguishable. Hence, val2 and val3 mutations had similar dominant effects in the val1 homozygous background.
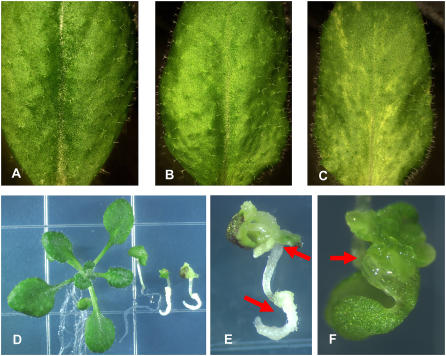
Figure 2.
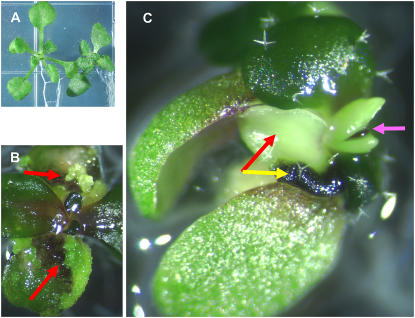
Phenotypes of val1 val3 and val1 val2 double mutants. A to C, Leaf variegation of the val1 val3 double mutant. The first cauline leaves from Col wild type (A), val1-2/val1-2 (B), val1-2/val1-2; val3-1/+ (C) plants are shown. D, Wild-type seedling and three val1-2 val2-1 double-mutant seedlings at 12 d are arranged left to right. E, Close image of the rightmost val1-2 val2-1 mutant seedling shown in D. Embryonic callous formation at shoot and root regions is highlighted by red arrows. F, Shoot apical region of the leftmost val1-2 val2-1 double-mutant seedlings shown in D. In this seedling representing the mildest expression of the double-mutant phenotype, embryo-like outgrowths are present in the shoot apical meristem region.
The val1/+ val2/val2, and val1/val1 val2/+ plants segregated seedlings that arrested in development shortly after germination (Fig. 2D). Progeny tests of normal segregants and molecular genotyping confirmed that the arrested seedling phenotype was due to the val1 val2 double homozygous mutant. Germination of double-mutant seeds was delayed by at least 2 d compared to wild type. Double-mutant seedlings survived for at least 30 d following germination on Murashige and Skoog medium, but failed to produce leaf primordia, indicating absence of a functional apical meristem. Root growth was stunted, resulting in a thick club-like root (Fig. 2E). Double-mutant seedlings developed cell proliferations that have embryonic characteristics, including callous formation from both shoot and root regions, formation of embryo-like structures at the position of the apical meristem and on cotyledon margins (Fig. 2F). Penetrance of the phenotype varied slightly among allele and ecotype combinations. The embryonic seedling phenotype was stronger in the val1-1 val2-1 and val1-1 val2-2 combinations constructed in the Wassilewskija (Ws) background (Supplemental Fig. S2) compared to the val1-2 val2-1 double mutant in Columbia (Col; Fig. 2D). In the Ws background, monogenic val1-1 homozygous seedlings formed embryonic callous at the cotyledon margins at low frequency (1% to 8%) when germinated on Murashige and Skoog medium, whereas this rare phenotype was not observed in val1-2 Col seedlings (Supplemental Fig. S2). Moreover, leaf variegation was more pronounced in val1-1 val3-3 Ws plants compared to val1-2 val3-3 Col plants (Supplemental Fig. S2).
Triple-mutant seedlings produced by self pollination of val1/+ val2/val2 val3/val3 plants had an embryonic seedling phenotype similar to the val1 val2 double mutant, although the triple-mutant seedlings germinated more slowly and typically arrested growth before emergence of the cotyledon from the seed coat (based on comparison of val1-1 val2-1 val3-3 and val1-1 val2-1 in Ws; data not shown). Hence, val1 behaved as a recessive in the val2 and val2 val3 homozygous backgrounds. Consistent with genetic evidence that VAL3 has a minor role in plant development, expression of VAL3 mRNA was low compared to VAL1 and VAL2 in seedlings (Supplemental Fig. S3) and other tissues (Arabidopsis Gene Atlas database; The Arabidopsis Information Resource [TAIR]).
Our analysis of double and triple mutants indicated that VAL genes are functionally redundant and required for development and/or maintenance of a functional apical meristem and for repression of embryonic development prior to or during seedling development.
VAL Is Required for Global Repression of Embryonic Gene Expression
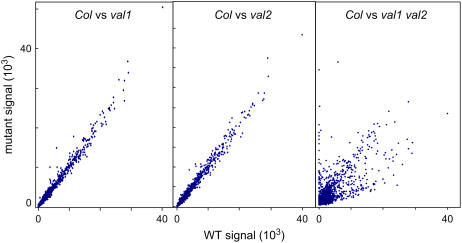
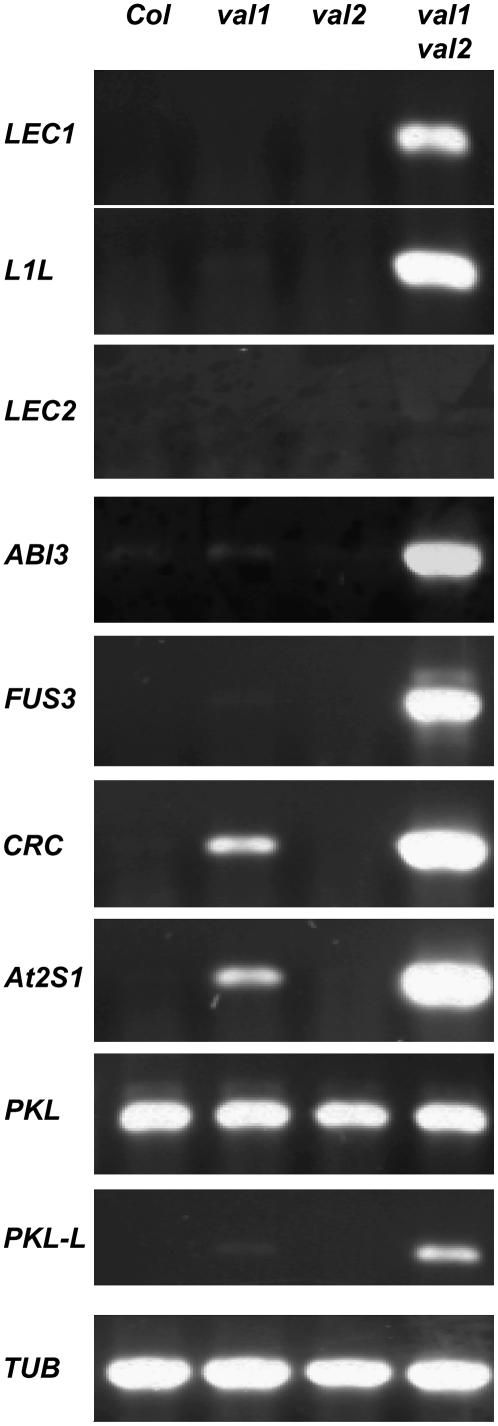
To obtain a global picture of gene expression changes conditioned by val mutants and to detect subtle gene expression differences in the val1 and val2 single mutants, we performed microarray analysis of mutant and wild-type seedlings using the Affymetrix 22K chip (Supplemental Table S1). Pairwise comparisons detected a limited number of gene expression differences between the wild type and either of the single mutants, whereas global gene expression patterns were profoundly altered in the val1 val2 double mutant relative to wild-type and single mutants (Fig. 3). Sets of 837 genes and 656 genes were more than 4-fold up-regulated and down-regulated, respectively, in the double mutant compared to wild type (Supplemental Tables S2 and S3, respectively). Genes implicated in regulation of seed development, including LEC1, L1L, and the ABI3 and FUS3 B3 transcription factors, were strongly up-regulated in the double mutant (Fig. 4; Supplemental Table S4). LEC2 expression was below the threshold for detection in all microarray treatments and in quantitative reverse transcription (RT)-PCR assays (Fig. 4). Consistent with the deregulation of FUS3 and ABI3, downstream targets of the B3 factors, such as the 2S albumin and cruciferin genes (Baümlein et al., 1994; Parcy et al., 1994, 1997; Nambara et al., 1995, 2000; Vicient et al., 2000; Suzuki et al., 2003; Tsuchiya et al., 2004; Braybrook et al., 2006), were also expressed at very high levels in the double mutant compared to levels below detection in wild type (Fig. 4; Supplemental Table S2). These two seed storage protein genes are among 61 of the 837 up-regulated genes that were also at least 2-fold up-regulated in the val1 single mutant. In contrast, only 11 of the 837 up-regulated genes showed >2-fold response in the val2 single mutant. Recently, the HSI2 gene, which is identical to VAL1, was identified as a transcriptional repressor for sugar-inducible genes (Tsukagoshi et al., 2005). In the hsi2 mutant, activation of a sugar-inducible seed storage protein gene was observed under noninducing conditions. A similar weak derepression of several seed storage protein genes was observed in the val1 single mutant.
Figure 3.
Altered gene expression in seedlings of wild type and val mutants. Global alteration of gene expression in val1 val2 double-mutant seedlings. Means for all elements, excluding controls from two biological replicate hybridizations, are plotted for Col wild type versus val1-2, val2-1, and the val1-2 val2-1 double mutant, respectively. [See online article for color version of this figure.]
Figure 4.
Confirmation of gene expression differences in wild type and val mutants. Expression of seed-expressed genes in the seedlings of wild type and val mutants. RT-PCR analysis was performed to verify expression of seed-specific genes as well as PKL and PKL-like (PKL-L) genes. Tubulin (TUB) was used as a control.
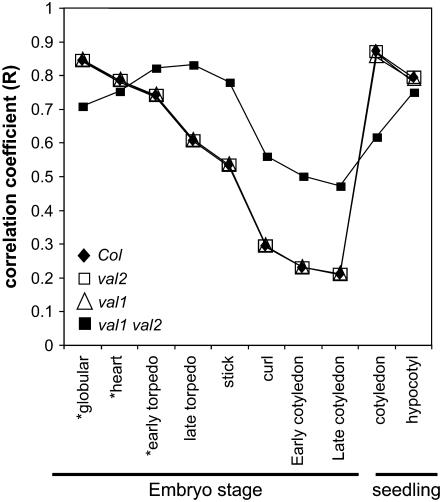
Overall, our results suggested that failure to achieve or maintain repression of the LEC1-related and ABI3/FUS3 B3 transcription factors during seed germination is likely responsible for the embryonic seedling phenotype of the val1 val2 double mutant. A correlation analysis of our microarray data in comparison to the Arabidopsis Gene Atlas dataset (described in “Materials and Methods”) indicated that the global gene expression profile of the double mutant was strongly correlated (r2 = 0.9) with late-torpedo-stage embryo development (Fig. 5). Based on the Gene Atlas dataset, the late torpedo stage corresponds to the peak in expression of VAL1 during embryo development and marks a temporal transition from peak expression of early embryonic regulators, LEC1, L1L, and LEC2, to the induction of the maturation phase B3 transcription factors, FUS3 and ABI3 (Supplemental Fig. S4).
Figure 5.
Correlation analysis of gene expression profiles of wild-type and val mutant seedlings with wild-type embryo development. Correlations of microarray data with Affymetrix 22K microarray data for staged embryos contained in the Arabidopsis Gene Atlas dataset (TAIR). Globular, heart, and early torpedo stages (shown by asterisks) of the Gene Atlas include silique tissue.
LEC2, FUS3, and ABI3 activation of downstream maturation genes is mediated in part by specific binding of the B3 domain to Sph/RY cis-elements (Suzuki et al., 1997; Kroj et al., 2003; Carranco et al., 2004; Monke et al., 2004; Braybrook et al., 2006). Because the VAL B3 domain is closely related, there is potential for direct interaction with downstream genes as well as indirect regulation via derepression of the B3 activators. We analyzed the frequency of consensus Sph/RY motifs (CATGCA; Suzuki et al., 2003) in the promoter regions (500 and 1,000 bp upstream of the annotated coding sequence) and introns of VAL-regulated genes. Genes derepressed 4-fold or greater in the val1 val2 double mutant showed significant enrichment of the Sph/RY motif in promoters as well as first introns (Table I). The presence of the Sph/RY consensus in the first intron was the strongest single correlate (P < 3.6 × 10−56) of VAL-repressed genes. This set of Sph/RY-containing genes included the non-B3 LEC1 and L1L genes, as well as FUS3, ABI3, VAL1, and VAL2 (Table II). LEC2, on the other hand, which did not respond to loss of VAL function, lacks a CATGCA consensus motif. Overall, about 54% (353/655 = 0.539) of genes >4-fold up-regulated in the double mutant contained at least one consensus motif in the 500-bp promoter region or first intron.
Table I.
Occurrence of Sph (CATGCA) motif in the noncoding regions of VAL-regulated genes
| Gene Region | Totala | >4-Fold Up | >4-Fold Down | % Upb | P Value |
|---|---|---|---|---|---|
| Sph/RY promoterc (500 bp) | 1,563 | 228 | 61 | 14.7 | 1.7 × 10−30 |
| Sph/RY first intron | 888 | 184 | 22 | 20.7 | 3.6 × 10−56 |
| Sph/RY promoter and first intron | 167 | 59 | 5 | 35.3 | 1.1 × 10−43 |
| Sph/RY promoterc (1,000 bp) | 3,424 | 350 | 149 | 10.2 | 2.5 × 10−13 |
| All genesd | 9,333 | 655 | 473 | 7.0 | – |
Total number of genes that have at least one consensus motif in either orientation in the indicated regions.
Percent of Sph-containing genes that were up-regulated.
500 and 1,000 bp upstream of annotated coding sequence extracted from the TAIR promoter database. Intron sequences were extracted from the TAIR intron database.
Elements on the chip selected for analysis had unambiguous gene assignments and absolute signal values of >200 in either the wild-type or val1 val2 double-mutant treatment.
Table II.
Occurrence of the Sph (CATGCA) motif in the noncoding regions of embryonic regulators
Promoter (1,000 bp) and first intron regions are obtained from the TAIR database.
| LEC1 | L1L | LEC2 | FUS3 | ABI3 | VAL1 | VAL2 | VAL3 | PKL | PKL-L | |
|---|---|---|---|---|---|---|---|---|---|---|
| Promoter | 2 | 0 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 3 |
| First intron | 2 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 |
Inhibition of GA Synthesis Induces an Embryonic Seedling Phenotype in the val1 Single Mutant
The embryonic seedling phenotype of the val1 val2 double mutant is reminiscent of the pkl mutant (Ogas et al., 1997). To test whether VAL function, like PKL, is also conditioned by GA biosynthesis, we tested germination of val1 single-mutant seeds on medium containing paclobutrazol, an inhibitor for GA biosynthesis (Fig. 6). On 5 nm paclobutrazol, a dosage below the threshold for inhibition of germination (Ogas et al., 1997), 23% of the seedlings exhibited strong embryonic transformations, including embryonic callous on cotyledon and leaf margins, ectopic embryo formation on leaves, and partial or complete transformation of leaves to cotyledon-like organs (Fig. 6, B and C). Whereas VAL repression of embryonic development was also affected by GA signaling, PKL expression was not affected in the double mutant, indicating that VAL genes do not act upstream of PKL (Fig. 4). However, a related CHD3 remodeling gene, PKL-like, was up-regulated in the val1 val2 double mutant.
Figure 6.
Phenotype of the val1 mutant seedling treated with an inhibitor of GA biosynthesis. A, 14-d-old seedlings of control (left) and 5 nm paclobutrazol-treated val1-2 mutant (right). The mild dwarf phenotype of the GA inhibitor-treated seedling was typical of the majority of seedlings (77%). B, Embryonic characteristics observed in 23% of the paclobutrazol-treated val1-2 seedlings are highlighted by a red arrow. C, Paclobutrazol-treated val1 seedling showing an emerging leaf with normal (yellow arrow) and embryonic (red arrow) sectors as well as a pair of transformed cotyledon-like leaves (pink arrow) is shown.
DISCUSSION
Our results show that VAL genes are essential for repression of embryonic pathways, as well as maintenance and/or differentiation of functional apical meristem function during seedling development. The three VAL genes are expressed throughout the plant life cycle (Arabidopsis Gene Atlas) and the seedling and variegated leaf phenotypes of double-mutant genotypes indicate that VAL genes have redundant functions in vegetative as well as seed development.
Among the three genes, VAL1 alone is sufficient for normal development. A nonredundant function of VAL1 is revealed by the variegated leaf phenotype that is manifest in val1 val3/+ and val1 val2/+ plants, but not in the val2 val3 double mutant. Dominant leaf variegation could arise from a variety of mechanisms, including haploinsufficiency, interference by defective VAL2 or VAL3 proteins, or transmission of an epigenetic state established in the haploid gametophyte phase. The similarity of val3 and val2 heterozygotes to the val3 homozygote in the val1 background does not suggest dosage sensitivity consistent with haploinsufficiency. The lack of allele-specific differences among the val3 and val2 insertion mutations argues against expression of a dominant inhibitory product, although this possibility is not ruled out. The possibility of epigenetic origin is intriguing in light of circumstantial evidence linking VAL to chromatin regulation and the classical relationship between variegation and epigenetic phenomena (Wakimoto, 1998; Schotta et al., 2003).
In addition to the functional relationship with PKL, the domain architecture of the VAL factors is consistent with a possible role in chromatin regulation. Whereas the VAL PHD-like domain bears the most resemblance to PHD domains of chromatin factors, the structure is quite divergent from canonical PHD domains known to bind chromatin. Four aromatic residues that are conserved in VAL orthologs, for example, evidently do not align with conserved Tyr and Trp residues implicated in the binding of specific methylated histones by the ING2 PHD domain (Peña et al., 2006). The CW domain is also frequently associated with chromatin factors (Perry and Zhao, 2003). For example, a histone methyltransferase equipped with a CW domain has recently been shown to regulate flowering in Arabidopsis (Zhao et al., 2006). Intriguingly, chromatin remodeling has been implicated in gene regulation mediated by other B3 domain factors, including ABI3 (Li et al., 1999; Levy et al., 2002; Fukaki et al., 2006; Ng et al., 2006), with association of PKL function (Fukaki et al., 2006). Hence, VAL B3 factors may associate with general chromatin factors, perhaps as part of the protein complex, to repress expression of a specific set of genes.
At a minimum, VAL function is required for maintenance of repression of embryonic gene expression during germination. The conditional phenotype of val1 mutant seedlings treated with the GA synthesis inhibitor, paclobutrazol, strongly implies that VAL function during germination is necessary. Whereas we suggest that VAL function is also required for establishing repression of the embryo pathway during seed development, the evidence for VAL action during embryo development is indirect. The global profile of gene expression in val1 val2 double-mutant seedlings resembles late-torpedo-stage embryogenesis, albeit combined with aspects of seed maturation and normal seedling development. This correlation is consistent with down-regulation of the early regulators of embryogenesis, LEC1 and L1L, and the relative peak in VAL1 expression at the late torpedo stage, suggesting that LEC1 and L1L may be direct targets of VAL.
The pkl mutant is the only other recessive mutant known to cause derepression of the embryonic program in vegetative organs. PKL is a CHD3 homolog implicated in establishing chromatin-based silencing of the embryo pathway during germination. Like pkl (Ogas et al., 1997), the val1 embryonic seedling phenotype is conditioned by inhibition of GA synthesis, suggesting these genes function in a common pathway. However, the val1 val2 double-mutant phenotype differs from pkl in at least three respects: (1) the embryonic seedling phenotype of the val double mutant is not dependent on GA synthesis inhibitor treatment; (2) whereas in pkl, embryonic characteristics are predominantly expressed in the distal region of the root (Ogas et al., 1997; Dean Rider et al., 2003), the val1 val2 phenotype is strongly expressed in shoot as well as root tissues; and (3) pkl and the val loss-of-function phenotypes affect different sets of embryo pathway genes. For instance, in contrast to the val double mutant, ABI3 is not derepressed in pkl mutant seedlings, whereas LEC2 is activated in the pkl, but not in the val double mutant (Dean Rider et al., 2003). These differences may be partly due to differential expression of PKL and VAL genes in root and shoot regions of the embryo. Although we have not determined the spatial distribution of VAL gene expression during development, PKL is predominantly expressed in the distal region of the root where the embryonic phenotype is manifest, as well as at the shoot apical meristem (Li et al., 2005). In contrast, the LEC2 gene is strongly expressed in the root axis, but not in the shoot apical meristem during seed development (To et al., 2006). Consistent with these expression patterns, the paclobutrazol-induced phenotype of val1 is most prominent in the shoot apical region. Hence, the LEC2 gene, which also lacks of a consensus Sph/RY, may be regulated independently of VAL1 and VAL2.
The conditional embryonic seedling phenotypes of the val1 and pkl mutants implicate GA signaling in repression of embryonic pathways. One interpretation is that, on normal medium, GA signaling enhances the function of VAL2 in the val1 single mutant sufficiently to allow normal development, whereas VAL2 activity is insufficient under GA-deficient conditions. The VAL genes may, in turn, at least indirectly regulate GA synthesis during seed development. AtGA3ox1, one of the key 3-oxidase genes expressed during seed germination (Yamaguchi et al., 1998; Mitchum et al., 2006), is down-regulated more than 10-fold in the val double mutant compared to wild type (Supplemental Table S4). Simultaneous strong up-regulation in the double mutant ( approximately 1,700-fold for AtGA20ox3) of genes encoding earlier steps in GA biosynthesis is consistent with loss of negative feedback regulation of the pathway by biologically active GA (Mitchum et al., 2006). VAL regulation of the GA pathway may occur indirectly through regulation of other B3 factors. Synthesis of active GA in the developing seed is enhanced by the lec2 and fus3 mutants (Curaba et al., 2004). FUS3 is proposed to repress GA biosynthesis (Curaba et al., 2004; Gazzarrini et al., 2004) through direct binding to Sph/RY elements in the promoter of a biosynthesis gene, AtGA3ox2 (Curaba et al., 2004). Hence, the delayed germination of val double- and triple-mutant seeds compared to wild type may be due to prolonged expression of FUS3 in the mutant seed.
Because many of the VAL-repressed genes are transcription factors, including representatives of most major families in the Arabidopsis genome, the prospects for distinguishing primary and secondary targets among the large number of affected genes detected by microarray analysis are seemingly limited. In spite of this complexity, we find that a majority of VAL-regulated genes contain a consensus Sph/RY element (CATGCA) either upstream of transcription initiation or within the first intron, and this enrichment is highly significant. This result suggests two limiting cases: (1) VAL and ABI3/FUS3/LEC2 families of B3 proteins may recognize the same or overlapping sets of downstream targets, the former functioning as repressors and the latter primarily as activators; and (2) VAL genes may primarily effect global repression of the pathway indirectly through repression of the B3 factors, or one step further removed, through repression of LEC1 and L1L. An intermediate view that we find attractive is that there is not necessarily a clear distinction between upstream and downstream targets within the B3 network. Initially proposed by Parcy et al. (1997), there is mounting evidence of mutual interactions among LEC1, FUS3, LEC2, and ABI3 during embryo development (Brocard-Gifford et al., 2003; Kagaya et al., 2005; Santos Mendoza et al., 2005; To et al., 2006). Moreover, our findings imply that the set of Sph/RY-regulated genes includes the early acting LEC1 and L1L genes in the embryo pathway. Recently, deletion of the 5′ region of LEC1 in the tnp mutant is shown to induce ectopic expression of this gene (Casson and Lindsey, 2006). The deleted region of the LEC1 promoter in this mutant contains a consensus Sph (ACATGCAT) at 666 bp upstream of the translation initiation, suggesting that this motif may mediate VAL repression function.
For these reasons, we suggest that B3-regulated genes comprise a gene system that includes two classes of B3 factors that recognize the Sph/RY element leading to activation and repression, respectively. Many, but not all, of these genes are involved in seed development. We further suggest that, by enabling recognition of Sph/RY in an active chromatin context, VAL proteins target genes in the system for repression by recruiting a chromatin-remodeling complex that includes PKL and related CHD3 proteins. Identification of cis-elements recognized by the VAL B3 domains is critical to understanding the mechanism of VAL-mediated repression. Our preliminary experiments aimed at testing specific binding of VAL1 and VAL2 B3 domains to the Sph motif in vitro are so far inconclusive (M. Suzuki and D.R. McCarty, unpublished data). Conceivably, the DNA-binding activity of the VAL B3 domain may depend on a chromatin context.
MATERIALS AND METHODS
Plant Growth Conditions
For phenotypic characterization of seedlings of val mutants, seeds were sterilized and sown on plates containing 1× Murashige and Skoog salt, 0.05% MES, 1% Suc, and 0.15% of Phytagel (Sigma). Seedlings were grown for 5 to 18 d at 22°C under continuous light. For phenotypic characterization of mature plants, seeds were germinated on sterile plates and transferred to soil or sown directly on soil after sterilization and cold stratification treatment and grown at 23°C to 25°C in a 16-h light/8-h dark regime.
Genetic Analysis and Construction of val Mutant Lines
We obtained T-DNA insertion alleles, val1-1, val2-1, val2-2, and val3-3 in Ws ecotype from the University of Wisconsin Arabidopsis Knockout facility (Sussman et al., 2000). The val3-3 allele, which has a T-DNA insertion within a coding exon of the VAL3 gene, was derived from an activation tag population. Alleles derived from the SALK Institute population (Alonso et al., 2003), val1-2 (SALK_088606), val3-1 (SALK_015582), and val3-2 (SALK_013113) in Col-0 ecotype, were obtained from the Arabidopsis Biological Resource Center. The mutant lines were backcrossed at least twice prior to genetic and phenotype analysis. The double and triple mutants were constructed by PCR-based genotyping. Because val2 and val3 loci are weakly linked, we screened recombinants to establish a val2 val3 double homozygous mutant. The primers used for genotyping the val alleles were listed in Supplemental Table S5.
Microarray Analysis
For Affymetrix ATH1 GeneChip microarray analysis, total RNA was prepared from two independent biological replicates using the RNeasy Plant mini kit (Qiagen). cDNA synthesis, in vitro transcription reactions, and hybridization were performed at the University of Florida Interdisiplinary Center for Biotechnology Research Core facility. Five-day-old whole seedlings were sampled from Col-0, val1-2, and val2-1, and 7.5-d-old whole seedlings were sampled for the val1-2 val2-1 double mutants. Due to their delayed germination relative to wild type, the double-mutant seedlings were grown 2.5 d longer to obtain comparable postgermination development. Because val2-1 was originally generated in Ws ecotype background, we backcrossed this allele with Col-0 five times. The backcrossed val2-1 mutant was used to generate val2 and val1 val2 mutants for microarray analysis. The val1 val2 double mutants were sampled based on the phenotypic differences among the seedlings segregating val2/+ in a val1 homozygous background. The genotypes of RNA samples were confirmed by RT-PCR before microarray hybridization (Supplemental Fig. S5).
RT-PCR Analysis
A 100-ng sample of total RNA was used for RT-PCR reactions (28 cycles) in a total volume of 12 μL with the One-Step RT-PCR kit (Qiagen). The primers used for RT-PCR are listed in Supplemental Table S5.
Statistical Analysis of cis-Elements and Microarray Data
Statistical analysis of Affymetrix microarray data derived experimentally or obtained from the Gene Atlas dataset of TAIR was performed using Excel spreadsheet functions. Motif frequencies in promoters and introns of coregulated genes were determined using a simple word search implemented in a custom java program and analyzed using chi-square (Suzuki et al., 2005). Gene sequence subregions were extracted from TAIR promoter (500 bp and 1,000 bp) and intron sequence compilations, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Leaf variegation.
Supplemental Figure S2. val double mutants.
Supplemental Figure S3. VAL gene expression.
Supplemental Figure S4. LEC1/B3 gene expression.
Supplemental Figure S5. Sample profiles for ATH1 microarray analysis.
Supplemental Table S1. Affymetrix ATH1 all genes.
Supplemental Table S2. VAL-repressed genes.
Supplemental Table S3. VAL-up-regulated genes.
Supplemental Table S4. Selected set of genes.
Supplemental Table S5. Primers in this study.
Supplementary Material
Acknowledgments
We acknowledge Dr. Michael Popp and Joint Shands Cancer Center-Interdisciplinary Center for Biotechnology Research at the University of Florida for assistance in analysis of microarray data. We also thank the Arabidopsis Biological Resource Center and the Arabidopsis research community for providing seeds and data.
This work was supported by the National Science Foundation (grant nos. 0080175 and 0322005 to D.R.M. and M.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masaharu Suzuki (masaharu@ufl.edu).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Aasland R, Gibson TJ, Stewart AF (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci 20 56–59 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbusch LO, Hughes DW, Galau GA, Jakobsen KS (2004) LEC1, FUS3, ABI3 and Em expression reveals no correlation with dormancy in Arabidopsis. J Exp Bot 55 77–87 [DOI] [PubMed] [Google Scholar]
- Baümlein H, Miséra S, Luerssen H, Kölle K, Horstmann C, Wobus U, Müller AJ (1994) The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J 6 379–387 [Google Scholar]
- Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ (2006) Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc Natl Acad Sci USA 103 3468–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard-Gifford IM, Lynch TJ, Finkelstein RR (2003) Regulatory networks in seeds integrating developmental, abscisic acid, sugar and light signaling. Plant Physiol 131 78–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carranco R, Chandrasekharan MB, Townsend JC, Hall TC (2004) Interaction of PvALF and VP1 B3 domains with the beta-phaseolin promoter. Plant Mol Biol 55 221–237 [DOI] [PubMed] [Google Scholar]
- Casson SA, Lindsey K (2006) The turnip mutant of Arabidopsis reveals that LEAFY COTYLEDON1 expression mediates the effects of auxin and sugars to promote embryonic cell identity. Plant Physiol 142 526–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC (2003) Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J 33 853–866 [DOI] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean Rider S Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezcurra I, Ellerstrom M, Wycliffe P, Stalberg K, Rask L (1999) Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol Biol 40 699–709 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SS, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14S 15–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Taniguchi N, Tasaka M (2006) PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J 48 380–389 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P (2004) The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell 7 373–385 [DOI] [PubMed] [Google Scholar]
- Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4 1251–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori T, Vasil V, Rosenkrans L, Hannah LC, McCarty DR, Vasil IK (1992) The viviparous-1 gene and abscisic acid activate the C1 regulatory gene for anthocyanin biosynthesis during seed maturation in maize. Genes Dev 6 609–618 [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T (2005) LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol 46 399–406 [DOI] [PubMed] [Google Scholar]
- Kao CY, Cocciolone SM, Vasil IK, McCarty DR (1996) Localization and interaction of the cis-acting elements for abscisic acid, VIVIPAROUS1, and light activation of the C1 gene of maize. Plant Cell 8 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P (1994) FUSCA3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol Plant 61 377–383 [Google Scholar]
- Kroj T, Savino G, Valon C, Giraudat J, Parcy F (2003) Regulation of storage protein gene expression in Arabidopsis. Development 130 6065–6073 [DOI] [PubMed] [Google Scholar]
- Kwong RW, Bui AQ, Lee H, Kwong LW, Fischer RL, Goldberg RB, Harada JJ (2003) LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 15 5–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297 243–246 [DOI] [PubMed] [Google Scholar]
- Li G, Bishop KJ, Chandrasekharan MB, Hall TC (1999) Beta-phaseolin gene activation is a two-step process: PvALF-facilitated chromatin modification followed by abscisic acid-mediated gene activation. Proc Natl Acad Sci USA 96 7104–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HC, Chuang K, Henderson JT, Rider SD Jr, Bai Y, Zhang H, Fountain M, Gerber J, Ogas J (2005) PICKLE acts during germination to repress expression of embryonic traits. Plant J 44 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Misera S (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15 755–764 [DOI] [PubMed] [Google Scholar]
- Meinke DW (1992) A homoeotic mutant of Arabidopsis thaliana with leafy cotyledons. Science 258 1647–1650 [DOI] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum MG, Yamaguchi S, Hanada A, Kuwahara A, Yoshioka Y, Kato T, Tabata S, Kamiya Y, Sun TP (2006) Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J 45 804–818 [DOI] [PubMed] [Google Scholar]
- Monke G, Altschmied L, Tewes A, Reidt W, Mock HP, Baumlein H, Conrad U (2004) Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta 219 158–166 [DOI] [PubMed] [Google Scholar]
- Nag R, Maity MK, Dasgupta M (2005) Dual DNA binding property of ABA insensitive 3 like factors targeted to promoters responsive to ABA and auxin. Plant Mol Biol 59 821–838 [DOI] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S (2000) The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol 220 412–423 [DOI] [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1994) Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol 35 509–513 [PubMed] [Google Scholar]
- Nambara E, Keith K, McCourt P, Naito S (1995) A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121 629–636 [Google Scholar]
- Nambara E, Naito S, McCourt P (1992) A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J 2 435–441 [Google Scholar]
- Ng DW, Chandrasekharan MB, Hall TC (2006) Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell 18 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng JC, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Kohara A, Misera S, Giraudat J (1997) The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6 1567–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442 100–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Zhao Y (2003) The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem Sci 28 576–580 [DOI] [PubMed] [Google Scholar]
- Raz V, Bergervoet JH, Koornneef M (2001) Sequential steps for developmental arrest in Arabidopsis seeds. Development 128 243–252 [DOI] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerstrom M, Czihal A, Tewes A, Ezcurra I, Rask L, Baumlein H (2000) Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J 21 401–408 [DOI] [PubMed] [Google Scholar]
- Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L (2005) LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 579 4666–4670 [DOI] [PubMed] [Google Scholar]
- Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29 2994–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Dorn R, Reuter G (2003) Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 14 67–75 [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman MR, Amasino RM, Young JC, Krysan PJ, Austin-Phillips S (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol 124 1465–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, Cocciolone S, McCarty DR (2001) Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J 28 409–418 [DOI] [PubMed] [Google Scholar]
- Suzuki M, Kao CY, McCarty DR (1997) The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9 799–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, Li QB, McCarty DR (2003) Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol 132 1664–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Ketterling MG, McCarty DR (2005) Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol 139 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nambara E, Naito S, McCourt P (2004) The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J 37 73–81 [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H, Saijo T, Shibata D, Morikami A, Nakamura K (2005) Analysis of a sugar response mutant of Arabidopsis identified a novel B3 domain protein that functions as an active transcriptional repressor. Plant Physiol 138 675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient CM, Bies-Etheve N, Delseny M (2000) Changes in gene expression in the leafy cotyledon1 (lec1) and fusca3 (fus3) mutants of Arabidopsis thaliana L. J Exp Bot 51 995–1003 [DOI] [PubMed] [Google Scholar]
- Wakimoto BT (1998) Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell 93 321–324 [DOI] [PubMed] [Google Scholar]
- West M, Yee KM, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ (1994) LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6 1731–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Smith MW, Brown RG, Kamiya Y, Sun T (1998) Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2006) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7 1256–1260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.