Abstract
Ubiquitin/26S proteasome-dependent degradation of topoisomerase I (TOP1) has been suggested to be a unique repair response to TOP1-mediated DNA damage. In the current study, we show that treatment of mammalian cells or yeast cells expressing human DNA TOP1 with camptothecin (CPT) induces covalent modification of the TOP1 by SUMO-1/Smt3p, a ubiquitin-like protein. This conclusion is based on the following observations: (i) Mammalian DNA TOP1 conjugates induced by CPT were cross-reactive with SUMO-1/Smt3p-specific antibodies both in yeast expressing human DNA TOP1 as well as mammalian cells. (ii) The formation of TOP1 conjugates was shown to be dependent on UBC9, the E2 enzyme for SUMO-1/Smt3p. (iii) TOP1 physically interacts with UBC9. (iv) Ubc9 mutant yeast cells expressing human DNA TOP1 was hypersensitive to CPT, suggesting that UBC9/SUMO-1 may be involved in the repair of TOP1-mediated DNA damage.
Keywords: UBC9, Smt3p, ubiquitin
Topoisomerase-mediated DNA damage represents an unique type of DNA damage whose importance has become increasingly appreciated. Many antibiotics, anticancer drugs, toxins, carcinogens, and physiological stresses are known to abort the catalytic cycles of topoisomerases resulting in the formation of topoisomerase-mediated DNA damage (1–7). Despite its importance, little is known about the molecular basis for the repair of this unique type of DNA damage.
Recent studies have demonstrated that topoisomerase I- (TOP1) mediated DNA damage activates a ubiquitin/proteasome pathway, resulting in degradation of TOP1 (down-regulation of TOP1) (8). It has been speculated that this pathway may represent a unique repair mechanism for TOP1-mediated DNA damage because down-regulation of human TOP1 (hTOP1) is expected to result in resistance/tolerance to TOP1 poisons (8).
Human SUMO-1 (small ubiquitin-like modifier) is an 11-kDa protein that shares 18% sequence homology with ubiquitin (9–11). Smt3p, which is 48% identical to human SUMO-1, has been identified in yeast (12, 13). SUMO-1/Smt3p is activated by a heterodimeric E1 enzyme in both the yeast and mammalian systems (13–15). In addition, UBC9, an E2 enzyme, has been shown to specifically conjugate SUMO-1/Smt3p to the target proteins (11, 16–18). Recently, an enzyme Ulp1p, which specifically cleaves proteins from SUMO-1/Smt3p conjugates and is distinct from isopeptidases for ubiquitin conjugates, also has been identified in yeast (19). These results suggest that the SUMO-1/Smt3p pathway is very similar to the ubiquitin pathway but distinct E1 and E2 enzymes as well as proteases are involved in these pathways. The biological function of the SUMO-1/Smt3p pathway appears diverse. UBC9 was originally identified to play an essential function in G2-M cell cycle progression in yeast (20). However, UBC9 is also known to be important for DNA repair (21). It has been shown that SUMO-1 can be conjugated to a number of proteins such as PML (22), RanGAP1 (23, 24), IκBα (25), FAS/apolipoprotein-1 (26), p53 (27, 28), RAD51, and RAD52 (29). A growing number of proteins also have been found to interact with UBC9, including p53 (30), adenovirus E1A oncoprotein (31), centromere DNA-binding core complex (32), c-Jun (33), FAS antigen (CD95) (34), RAD51, and RAD52 (30, 35). Whether the SUMO-1/Smt3p pathway is involved in a common function for all these proteins remains unclear. However, SUMO-1 modification of RanGAP1 is known to cause RanGAP1 to bind to the nuclear pore complex (36–38). Other studies have demonstrated that SUMO-1 conjugates are often located in the nuclear bodies (39, 40).
In this study, we have shown that camptothecin (CPT), a TOP1-specific poison, can induce rapid and extensive conjugation of SUMO-1/Smt3p to human DNA TOP1. In addition, we have shown that UBC9 mutant yeast cells expressing human DNA TOP1 is hypersensitive to CPT, suggesting that UBC9/SUMO-1 may be involved in the repair of TOP1-mediated DNA damage.
Materials and Methods
Materials.
Glutathione Sepharose 4B and Protein A Sepharose CL-4B beads were purchased from Pharmacia Biotech. The anti-FLAG antibodies were obtained from Sigma. The anti-hTOP1 antibodies were obtained from scleroderma patients. The anti-SUMO-1 antibodies were purchased from Zymed. 2RA, which is an simian virus 40 T-antigen transformed derivative of the human lung fibroblast WI 38 cell line, was obtained from American Type Culture Collection (CCL 75.1). The Tet-Off HeLa cell line was purchased from CLONTECH. URA3 and LEU2 marked two micron plasmids for pGAL10-driven expression of wild-type and mutant Smt3p were obtained from Erica S. Johnson (Howard Hughes Medical Institute, Rockefeller University, New York) (13). YCpGAL1-hTOP1 plasmid was obtained from Mary-Ann Bjornsti (St. Jude Children's Research Hospital, Memphis). Yeast strains Y0007 (MATa, his3-Δ200, leu2–3, 2–112, lys2–801, trp1–1(am), ura3–52, bar1∷HIS3) and Y0174 (MATa, his3-Δ200, leu2–3, 2–112, lys 2–801, trp1–1(am), ura3–52, ubc9-Δ1∷TRP1, LEU∷ubc9–1) with temperature sensitive ubc9 were kindly provided by Stefan Jentsch (Friedrich-Miescher-Laboratorium der Max-Planck-Gesellschaft, Tubingen, Germany) (20). Human UBC9 (hUBC9) cDNA was isolated from a yeast clone in a yeast two-hybrid screening by using hTOP2β as the bait (Y.M. and L.F.L., unpublished results).
Isolation and Characterization of Smt3p-hTOP1 Conjugates in Yeast.
JN362a (MATa ade1 ura3–52 leu2 his7 trp1 tyr1 ise2) was cotransformed with YCpGAL1-hTOP1 plasmid and either wild-type HF-SMT3 (Y101) or mutant HF-Smt3 (G97) plasmid. Yeast cells harboring both plasmids were inoculated to 3 ml of Ura− Leu− synthetic dropout with 2% glucose medium. After overnight incubation, cultures were diluted 1:100 with 20 ml of Ura− Leu− synthetic dropout medium containing 3% glycerol and 2% lactic acid. When OD600 reached 0.5–0.7, 2% galactose was added to the culture. After a 12-h galactose induction, cells were treated with 100 μM CPT for 30 min. Yeast cells were then collected and lysed directly with the alkali/glass bead procedure (8). In brief, cell pellets were resuspended in 200 μl of lysis buffer (200 mM NaOH/2 mM EDTA) and mixed with ≈100 μl of acid-washed glass beads (Sigma). The mixtures were vigorously vortexed four times (20 sec vortexing and 40 sec on ice each time), and the mixtures were then neutralized by adding 40 μl of neutralizing buffer (1 M HCl/600 mM Tris, pH 8.0). After neutralization, 25 μl of a 10 × staphylococcal S7 nuclease reaction buffer (50 mM MgCl2/50 mM CaCl2/5 mM DTT/1 mM EDTA/1 mM phenylmethylsulfonyl fluoride (PMSF)/50 μg/ml of leupeptin, aprotinin, and pepstatin A) and 60 units of S7 nuclease (Boehringer Mannheim) were added to the samples. After S7 nuclease digestion (15 min at room temperature), 3 × SDS sample buffer was added to each sample, which was then analyzed by 5% SDS-PAGE. Immunoblotting was performed by using antibodies against either hTOP1 or the FLAG epitope.
Immunoprecipitation and Immunoblotting.
Yeast lysates were prepared by the alkaline lysis procedure as described above. The lysates were incubated with anti-hTOP1 antibodies-bound protein A Sepharose beads at 4°C for 2 h. After incubation, beads were washed three times with rinse buffer (20 mM Tris⋅HCl, pH 8.0/150 mM NaCl/10 mM MgCl2/1 mM DTT/1 mM PMSF) and were resuspended into 50 μl of 3 × SDS sample buffer. The bead samples were analyzed on 5% SDS gel. The immunoblotting was performed by using anti-FLAG antibodies as described (8).
Purification of Glutathione S-Transferase (GST) and GST-hUBC9 Fusion Proteins.
Human UBC9 cDNA was constructed to be in frame with GST in pGEX-2T (Pharmacia Biotech). GST and GST-hUBC9 fusion protein were overexpressed in Escherichia coli and purified by affinity glutathione (GSH) Sepharose 4B batch elution (41) with some modifications.
GST Pull Down Assay.
The GST pull down assay was performed as described (25) with some modifications.
Coimmunoprecipitation Assay.
Protein A Sepharose CL-4B beads were equilibrated with buffer N (50 mM Tris⋅HCl, pH 7.5/1 mM EDTA/1 mM EGTA/0.5% Nonidet P-40/1 mM NaF/5 mM MgCl2/1 mM PMSF/10 μg/ml aprotinin/10 μg/ml leupeptin/10 μg/ml pepstatin A). Equilibrated sepharose beads were incubated with anti-hTOP1 antibodies at room temperature for 30 min. After incubation, beads were washed three times with the rinse buffer. Nuclear extracts prepared from 2RA cells were mixed with 15 μg of purified GST-hUBC9 fusion proteins in buffer N at room temperature for 30 min. After mixing, nuclear extracts were incubated with antibody-bound Sepharose beads at room temperature for 30 min. Supernatant was collected for SDS-PAGE and immunoblotting. The bead fractions were washed three times with the rinse buffer and then mixed with 50 μl of 2 × SDS sample buffer. Twenty microliters of the supernatant fraction and 30 μl of the bead fraction were loaded onto 5% SDS gel. Immunoblotting was performed by using (1:2,000) monoclonal anti-GST antibody (from Pharmacia Biotech).
Immuno-Characterization of hTOP1 Conjugates in Human Cells.
Subconfluent HeLa cells were treated with 10 μM CPT [in 1% dimethyl sulfoxide (DMSO)] or 1% DMSO for 10 min. Cell lysates were prepared by an alkali lysis procedure (8). Lysates were analyzed on 5% SDS gel and immunoblotted with either anti-hTOP1 or anti-SUMO-1 antibodies.
Sensitivity of Yeast to CPT.
CPT sensitivity was determined as described previously (43).
Results
CPT Induces Rapid Covalent Modification of hTOP1 in Both Human and Yeast Cells.
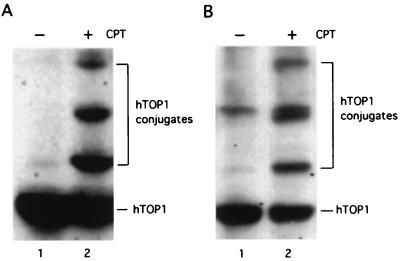
Previous studies in mammalian cells have demonstrated that CPT can induce covalent modification of TOP1 (8). These covalently modified TOP1 species were presumed to be TOP1-ubiquitin conjugates (8). Consistent with previous studies, we show that human WI38 cells treated with CPT (25 μM) resulted in the formation of multiple high molecular weight hTOP1 species (Fig. 1A, compare lanes 1 and 2). However, the spacing between these high molecular weight TOP1 species would predict a protein mass of 20 kDa, which is significantly larger than the predicted molecular weight for ubiquitin (Mr = 8 kDa). In order to characterize these putative TOP1-ubiquitin conjugates, we turned to a yeast system expressing hTOP1 (44). When the yeast strain JN362a expressing hTOP1 was treated CPT (100 μM), similar high molecular weight species were induced as evidenced by immunoblotting with anti-hTOP1 antibodies (Fig. 1B, compare lanes 1 and 2). However, overexpression of dominant negative ubiquitin mutants (UbK29R, UbK48R, UbK63R, and UbKRRR (K29,48,63R) triple mutant), which has been demonstrated to interfere with the formation of multiubiquitin chains (45), failed to abolish the high molecular weight species (data not shown). In addition, the formation of hTOP1 conjugates in cells treated with CPT was also independent of RAD6, a ubiquitin E2 enzyme (46), and RSP5, a ubiquitin E3 enzyme (47) (data not shown).
Figure 1.
CPT induces the formation of hTOP1 conjugates in human cells and yeast cells expressing hTOP1. (A) Human lung fibroblast WI38 cells were treated with 25 μM CPT in 1% DMSO (+) or 1% DMSO (−) for 15 min and then lysed using the alkali lysis procedure described in Materials and Methods. Samples were analyzed on 6% SDS-PAGE and immunoblotted with anti-hTOP1 antibodies. hTOP1 conjugates were marked by a bracket. (B) Yeast JN362a harboring YCpGAL1-hTOP1 plasmid was induced by 2% galactose for 12 h and then treated with 100 μM CPT in 1% DMSO (+) or 1% DMSO (−) for 30 min. Cells were lysed using the alkali/glass bead methods. The SDS-PAGE and immunoblotting using anti-hTOP1 antibodies were performed as described in A.
hTOP1-Mediated DNA Damage Leads to SUMO-1/Smt3p Modification of hTOP1 in Yeast.
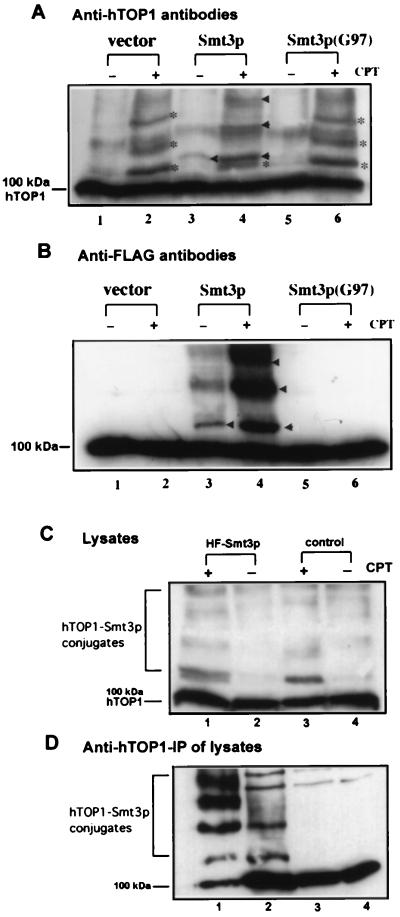
The results from the above experiments challenge the previous conclusion that TOP1 conjugates in CPT-treated cells represent TOP1-ubiquitin conjugates (8). Another possible candidate for the protein in TOP1 conjugates is the ubiquitin-related protein, SUMO-1/Smt3p. To test this possibility, we overexpressed His-6/FLAG-tagged Smt3p in JN362a and then treated the yeast with CPT. As shown in Fig. 2A (compare lanes 1 and 2), CPT treatment of JN362a expressing hTOP1 (no exogenous Smt3p) resulted in the formation of high molecular weight hTOP1 species (see bands marked with *) as revealed by immunoblotting with anti-hTOP1 antibodies. When His-6/FLAG-tagged Smt3p was overexpressed in JN362a expressing hTOP1, CPT treatment resulted in the formation of another series of high molecular hTOP1 species (marked by arrow heads) in addition to the series of high molecular weight hTOP1 species marked by * (Fig. 2A, compare lanes 3 and 4). When a mutant form of Smt3p, Smt3p(G97), was overexpressed under the same conditions, CPT treatment did not result in the formation of this new series of high molecular weight species (Fig. 2A, compare lanes 5 and 6). A duplicate gel with identical samples was immunoblotted with anti-FLAG antibodies (Fig. 2B). CPT treatment of yeast cells overexpressing His-6/FLAG-tagged Smt3p was shown to stimulate the formation of a series of high molecular weight species (see bands marked with arrow heads in Fig. 2B, lane 4) whose electrophoretic mobilities coincided with those of the new series of high molecular weight hTOP1 species mentioned above (see bands marked with arrow heads in Fig. 2A, lane 4). Expression of the vector alone (Fig. 2B, lanes 1 and 2) or mutant Smt3p (Fig. 2B, lanes 5 and 6) did not result in the formation of these high molecular weight species. These results suggest that hTOP1 conjugates formed in cells treated with CPT could be hTOP1-Smt3p conjugates. The formation of another series of high molecular weight hTOP1 species shown in lanes 4 of Fig. 2 A and B (see bands marked with arrow heads) could be due to the slight increase in the size of exogenous His-6/FLAG-tagged Smt3p relative to endogenous Smt3p.
Figure 2.
CPT induces Smt3p modification of hTOP1 in yeast. (A) Modification of hTOP1 by HF-Smt3p in yeast. JN362a was cotransformed with YCpGAL1-hTOP1 and HF-Smt3 plasmids. After 2% galactose induction, cells were treated with either 100 μM CPT in 1% DMSO (+) or 1% DMSO (−) for 30 min. Cells were lysed using the alkali lysis procedure described in Materials and Methods. After SDS-PAGE, immunoblotting was performed using antibodies against hTOP1. Lanes 1 and 2: vector control, no HF-Smt3p expression. The high molecular weight bands induced by CPT were marked with *. Lanes 3 and 4: overexpression of HF-Smt3p from HF-Smt3p expressing plasmid. The new series of high molecular weight bands were indicated by arrow heads. Lanes 5 and 6: overexpression of HF-Smt3p (G97) from plasmid expressing mutant HF-Smt3p(Smt3p (G97)). (B) The same six samples shown in A were also analyzed in parallel by SDS-PAGE and immunoblotted with anti-FLAG antibodies instead of anti-hTOP1 antibodies. (C) Yeast cell lysates from JN362a cells harboring HF-Smt3p (lanes 1 and 2) or cells harboring HF-Smt3p(G97) (lanes 3 and 4) were prepared as described in A. Lysates were immunoblotted with anti-hTOP1 antibodies. (D) The lysates from C were immunoprecipated with anti-hTOP1 antibodies as described in Materials and Methods. The beads fractions were analyzed on 5% SDS-PAGE and immunoblotted with anti-FLAG antibodies. Lanes 1 and 2: IP beads fractions from JN362a expressing hTOP1 and HF-Smt3p with (+) (lane 1) or without (−) (lane 2) CPT treatment. Lanes 3 and 4: beads fractions from JN362a expressing hTOP1 and HF-Smt3p(G97) with (+) (lane 3) or without (−) (lane 4) CPT treatment. The 100-kDa bands indicated in B and D are protein(s) in yeast lysates which are cross-reactive to anti-FLAG antibodies.
To positively identify these hTOP1 conjugates as hTOP1-Smt3p conjugates, yeast cell lysates were immunoprecipitated with anti-hTOP1 antibodies. The immunoprecipiates were then analyzed by immunoblotting with anti-FLAG antibodies (Fig. 2D). Again, CPT was shown to stimulate the formation of a series of high molecular weight species in JN362a expressing both hTOP1 and His-6/FLAG-tagged Smt3p (Fig. 2D, lane 1). These high molecular weight species had electrophoretic mobilities similar to those of the high molecular weight species detected by anti-hTOP1 antibodies (see lane 1 in Fig. 2C). These results suggest strongly that the high molecular weight species induced by CPT contain both hTOP1 and Smt3p.
To further support our conclusion that the high molecular weight species induced by CPT is hTOP1-Smt3p conjugates, we examined the role of UBC9 in the formation of these conjugates. UBC9 is known to be the E2 enzyme for Smt3p (16, 18, 38). Consequently, inactivation of UBC9 is expected to abolish the formation of hTOP1-Smt3p conjugates. An UBC9 temperature sensitive mutant Y0174 was transformed with a hTOP1 expression plasmid and then treated with CPT at both the permissive and nonpermissive temperatures. As shown in Fig. 3A, at the permissive temperature, CPT treatment resulted in the formation of the expected high molecular weight species (compare lanes 1 and 2). However, at the nonpermissive temperature, CPT treatment did not induce the formation of these high molecular weight species (Fig. 3A, compare lanes 3 and 4). The inability of CPT to induce the formation of these high molecular weight species was not due to the temperature effect since CPT was able to induce their formation in wild-type UBC9 yeast (Y0007) at both temperatures (Fig. 3B, lanes 1 and 3).
Figure 3.
CPT -induced formation of hTOP1-Smt3p conjugates is UBC9 dependent in yeast. (A) Yeast ubc9 ts mutant strain Y0174 was transformed with YCpGAL-hTOP1 plasmid. Cells were treated with CPT (+) (lanes 1 and 3) or DMSO (−) (lanes 2 and 4) at both permissive (25°C) (lanes 1 and 2) and nonpermissive (36°C) (lanes 3 and 4) temperatures. Cells were lysed and analyzed on SDS-PAGE and immunoblotted with anti-hTOP1 antibodies as described in Materials and Methods. (B) The formation of hTOP1-Smt3p conjugates was examined in the UBC9 wild-type strain Y0007 as described in A.
CPT Induces the Formation of hTOP1-SUMO-1 Conjugates in Human Cells.
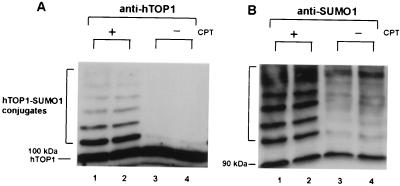
As presented above, our studies in the yeast model system have suggested that CPT can stimulate the formation of hTOP1-Smt3p conjugates. One of the homologs of Smt3p in human cells is SUMO-1. SUMO-1 is known to be conjugated to target proteins by hUBC9 (16, 38). To test whether the hTOP1 conjugates stimulated by CPT in human cells is hTOP1-SUMO-1 conjugates, two sets of experiments were performed. First, lysates from HeLa cells treated with CPT (10 μM) were immunoblotted with both anti-hTOP1 (Fig. 4A) and anti-SUMO-1 (Fig. 4B) antibodies. In both cases, CPT treatment resulted in the formation of a series of high molecular weight species (see the bracketed region marked hTOP1-SUMO-1 conjugates in Fig. 4). The mobilities of these high molecular weight species detected by either anti-hTOP1 antibodies or anti-SUMO-1 antibodies were similar if not identical. This result suggests that these high molecular weight species induced by CPT are most likely hTOP1-SUMO-1 conjugates. The gradual increase in the intensities of these high molecular weight species detected by anti-SUMO-1 antibodies (Fig. 4B, lanes 1 and 2) as opposed to the gradual decrease in the intensities of these high molecular weight species detected by anti-hTOP1 antibodies (Fig. 4A, lanes 1 and 2) can be explained by the presence of multiple SUMO-1 molecules on each hTOP1 molecule.
Figure 4.
CPT induces SUMO-1 conjugation to hTOP1 in HeLa cells. HeLa cells were treated with 10 μM CPT (+) or DMSO (−) for 15 min. Cells were lysed and analyzed as described in Materials and Methods. (A) Samples were analyzed on 5% SDS-PAGE and immunoblotted with anti-hTOP1 antibodies. Duplicate samples were loaded. Lanes 1 and 2 were samples from cells treated with CPT. Lanes 3 and 4 were samples from cells without CPT treatment. (B) The same membrane filter was stripped and reblotted with anti-SUMO-1 antibodies.
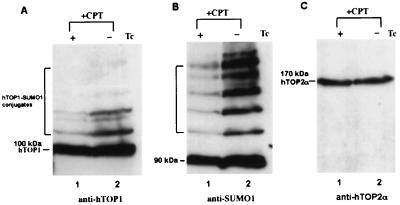
We also have tested whether human UBC9 is involved in the formation of these hTOP1 conjugates in HeLa cells. We established a stable hUBC9 expression Tet-Off HeLa cell line, HeLa-hUBC9. Northern analysis confirmed an elevated expression of hUBC9 in this stable transfectant upon removal of tetracycline (data not shown). As shown in Fig. 5A (compare lanes 1 and 2), overexpression of hUBC9 (removal of tetracycline, −Tc) in this cell line greatly enhanced the formation of the CPT-induced high molecular weight hTOP1 species as revealed by immunoblotting with anti-hTOP1 antibodies. The same membrane filter was stripped and reblotted with anti-SUMO-1 antibodies (Fig. 5B). Again, the CPT-induced SUMO-1-containing high molecular weight species were significantly enhanced upon removal of tetracycline (compare lanes 1 and 2 in Fig. 5B). To rule out the possibility that the difference is due to different loading of samples, the same membrane filter was stripped and reblotted with anti-hTOP2α antibodies. hTOP2α levels were shown to be the same in these two samples (Fig. 5C). These results support the notion that CPT-induced hTOP1 conjugates in HeLa cells are hTOP1-SUMO-1 conjugates, and that hUBC9 is the E2 enzyme responsible for SUMO-1 conjugation to hTOP1. Additionally, we have immunoprecipitated hTOP1 from HeLa cells treated with CPT and immunoblotted the immunoprecipitates with anti-SUMO-1 antibodies (data not shown). The results again showed that CPT induced SUMO-1 modification on hTOP1.
Figure 5.
Overexpression of hUBC9 enhances SUMO-1 conjugation to hTOP1 in HeLa cells treated with CPT. (A) Tet-Off HeLa cells were transfected with pTRE-hUBC9 and the stable transfectant (clone 2) was selected by Northern blot screening. Clone 2 was cultured in the presence (+) or absence (−) of tetracycline for 48 h before CPT treatment (10 μM for 10 min). The same amount of samples (based on protein concentrations) were analyzed on 5% SDS-PAGE and immunoblotted with anti-hTOP1 antibodies. (B) The same membrane filter from (A) was stripped and reblotted with anti-SUMO-1 antibodies. (C) The same membrane filter from (B) was stripped and reblotted with anti-hTOP2α antibodies.
Binding of Human DNA TOP1 to hUBC9.
We had also performed GST pull down and coimmunoprecipitation assays to demonstrate physical interaction between hTOP1 and hUBC9. As shown in Fig. 6A, both purified hTOP1 and hTOP1 in a nuclear extract prepared from human 2RA cells (T-antigen-transformed WI38 cells) were retained in beads with bound GST-hUBC9 fusion protein (lanes 3 and 4). By contrast, no hTOP1 was detectable in beads with bound GST (lanes 1 and 2). In a coimmunoprecipitation experiment, we also have demonstrated that GST-hUBC9 fusion protein can bind hTOP1 in the nuclear extract prepared from 2RA cells. As shown in Fig. 6B, GST-hUBC9 was coprecipitated with hTOP1 by using beads with bound hTOP1 antibodies. As a control, GST-hUBC9 was shown to be absent in the immunoprecipitate using beads with bound HA antibodies (Fig. 6B).
Figure 6.
Binding of hTOP1 to GST-hUBC9. (A) hTOP1 binds to GST-hUBC9 in a GST pull down assay. The GST pull down assay was performed as described in Materials and Methods. Lanes 1 and 2 were lysates from GST bead fractions. Lanes 3 and 4 were lysates from GST-hUBC9 bead fractions. NE indicates that nuclear extracts were used in the pull down assay (lanes 1 and 3). “TOP1” indicates that purified recombinant hTOP1 was used in the pull down assay (lanes 2 and 4). (B) hTOP1 binds to GST-hUBC9 in a coimmunoprecipitation assay. The coimmunoprecipitation assay was performed as descried in Materials and Methods. Purified GST-hUBC9 is shown in lane 1. Lanes 2 (bead) and 4 (supernatant): anti-HA antibody (control antibody) was used in coimmunoprecipitation. Lanes 3 (bead) and 5 (supernatant): anti-hTOP1 antibodies were used in coimmunoprecipitation.
UBC9/Smt3p Affects Sensitivity of Yeast to TOP1-Mediated DNA Damage.
Previous studies have suggested that a ubiquitin/proteasome pathway is involved in the repair of hTOP1-mediated DNA damage (8). To test whether the SUMO-1/UBC9 pathway is also involved in repair of hTOP1-mediated DNA damage, we have performed two types of experiments in yeast cells expressing hTOP1. Yeast cells expressing hTOP1 is known to be sensitive to killing by hTOP1-mediated DNA damage induced by CPT (43, 44). First, we tested the sensitivity of a ubc9 ts mutant (Y0174 expressing hTOP1) to CPT. As shown in Fig. 7A, Y0174 expressing hTOP1 was hypersensitive to CPT (50 μM) compared with its isogenic wild-type strain (Y0007 expressing hTOP1) at the semipermissive temperature (30°C). At the permissive temperature (25°C), mutant (Y0174) yeast, like wild-type yeast, was less sensitive to CPT. Second, we tested the effect of Smt3p overexpression on CPT sensitivity. As shown in Fig. 7B, overexpression of wild-type Smt3p from a two micron-based multicopy plasmid Y101 in JN362a expressing hTOP1 resulted in a slight but reproducible increase in survival in response to CPT treatment as compared with the vector-transformed control cells or mutant Smt3p (G97)-transformed cells. The G97 mutant Smt3p is known to be completely defective in Smt3p conjugation to target proteins (13). These two experiments suggest possible involvement of UBC9/Smt3p in repair of TOP1-mediated DNA damage.
Figure 7.
UBC9 and Smt3p affect CPT sensitivity in yeast expressing hTOP1. (A) An ubc9 ts mutant is hypersensitive to CPT treatment. Yeast strains Y0007 (wild-type) and Y0174 (ubc9ts) were transformed with YCpGAL1-hTOP1. The CPT sensitivity were determined at 30°C as described in Materials and Methods. (B) The effect of Smt3p overexpression on CPT sensitivity. hTOP1 and Smt3p (wild-type or G97 mutant) were coinduced in JN362a cells by 2% galactose. CPT sensitivity was determined at 30°C as described in Materials and Methods.
Discussion
Topoisomerase-mediated DNA damage is unique due to the properties of the reversible topoisomerase cleavable complexes (1, 2). The bulkiness of the protein-DNA adducts makes it difficult to conceive of their repair by conventional means. Previous studies have shown that TOP1-mediated DNA damage can trigger a ubiquitin/26S proteasome pathway resulting in the degradation of TOP1. Reduction of the intracellular level of TOP1 by this ubiquitin/proteasome pathway could be considered as a potential repair mechanism for TOP1-mediated DNA damage since the amount of DNA damage is presumably proportional to the amount of cellular TOP1 (48).
Previous studies have demonstrated that TOP1 down-regulation induced by CPT is dependent on E1 and 26S proteasome (8). Consequently, the TOP1 conjugates in cells treated with CPT were suggested to be TOP1-ubiquitin conjugates. To study these putative TOP1-ubiquitin conjugates, we have developed a yeast system expressing hTOP1 which upon CPT treatment can produce hTOP1 conjugates with similar properties as those produced in mammalian cells. Our studies in yeast have suggested that these hTOP1 conjugates are primarily hTOP1-SUMO-1 conjugates rather than hTOP1-ubiquitin conjugates. This conclusion is based on a number of observations; (1) Dominant negative mutant ubiquitins with various critical lysines mutated to arginines, which are known to block the formation of ubiquitin multichains, were unable to affect the formation of hTOP1 conjugates. In addition, RAD6, an E2 enzyme, and RSP5, an E3 enzyme, had no effect on the formation of these conjugates. (2) Overexpression of tagged Smt3p in yeast resulted in the formation of a series of new hTOP1 conjugates with a slight increase in molecular weights. The formation of these new hTOP1 conjugates is CPT-stimulated and UBC9-dependent. (3) Tagged Smt3p is associated with immunoprecipiated hTOP1 conjugates in yeast treated with CPT.
Studies in HeLa cells also have demonstrated that hTOP1 conjugates induced by CPT are primarily hTOP1-SUMO-1 conjugates. First, both anti-hTOP1 and anti-SUMO-1 antibodies detected a group of high molecular weight proteins which were induced by CPT. The electrophoretic mobilities of these high molecular weight proteins were similar if not identical. Second, overexpression of human UBC9 significantly enhanced the level of these high molecular weight proteins. Lastly, hTOP1 can physically interact with UBC9 as evidenced by GST pull-down and coimmunoprecipitation assays. Based on our studies both in yeast and HeLa cells, we conclude that CPT can induce SUMO-1/Smt3p modification of hTOP1. Previous studies have demonstrated that inactivation of E1 (for ubiquitin) can abolish the formation of “TOP1-ubiquitin” conjugates (8). Our current identification of these TOP1-ubiquitin conjugates as TOP1-SUMO-1 conjugates would suggest that inactivation of E1 may somehow interfere with SUMO-1/UBC9 function.
The signal that triggers SUMO-1 conjugation of hTOP1 is most likely the hTOP1-CPT-DNA cleavable complex. Two types of experiments support this view. First, majority of SUMO-1 conjugated hTOP1 was covalently linked to DNA (8). Second, mutant cell lines (e.g., CPT-K5 and U-937/CR; refs. 49 and 50) are defective in CPT induced SUMO-1 conjugation to hTOP1 (S.D.D. and L.F.L., unpublished results).
The possible role of the UBC9 pathway in DNA repair has been suggested from studies of ubc9 mutants (21). Indeed, our results have also demonstrated that a ubc9 ts mutant is hypersensitive to hTOP1-mediated DNA damage induced by CPT (Fig. 7A). In addition, overexpression of SUMO-1/Smt3p results in increased resistance to CPT (Fig. 7B). These results suggest that the SUMO-1/UBC9 pathway could be involved in the repair of hTOP1-mediated DNA damage. However, whether SUMO-1 modification of hTOP1 plays a critical role for repair of hTOP1-mediated DNA damage remains unclear. It should be noted that CPT-induced SUMO-1 modification of hTOP1 is independent of ongoing DNA synthesis since aphidicolin, which substantially abolishes CPT cytotoxicity (48), does not affect the formation of hTOP1-SUMO-1 conjugates (data not shown).
It is interesting to speculate the possible role(s) of SUMO-1/Smt3p in the repair of hTOP1-mediated DNA damage. Currently, the SUMO-1 function is still unclear (51–53). However, there are suggestions that SUMO-1 may be involved in targeting proteins to either the nuclear pore complexes in the nuclear envelope or nuclear bodies in the nucleus (36–38). The notion that SUMO-1 may be involved in targeting proteins to the nuclear pore complex came from studies of SUMO-1 conjugated RanGAP1 which is known to bind to the nuclear pore complex (24, 37, 39, 40). SUMO-1 conjugates also have been shown to be primarily located in the nucleus (54) and often targeted to the nuclear bodies as most evidently demonstrated in the case of PML (52). One could speculate that SUMO-1 conjugated hTOP1 molecules in cells treated with CPT are either targeted to the nuclear pore complex for nuclear export or to the nuclear bodies. Relocation of hTOP1 to different cellular compartments is expected to effectively reduce hTOP1-mediated DNA damage due to the absence of DNA bound hTOP1. Indeed, relocation of hTOP1 has been demonstrated to occur in cells treated with CPT (55, 56). In one case, CPT treatment results in relocation of TOP1 from nucleoli to nucleoplasm (55). In the other, CPT treatment results in relocation of TOP1 from the nucleus to the cytoplasm (56). Whether targeting of SUMO-1-conjugated hTOP1 to other cellular compartments is associated with protein degradation and/or refolding remains unclear. It is also possible that SUMO-1 conjugation to TOP1 may signal binding of the repair proteins to the site of TOP1-mediated DNA damage (e.g., recruitment of tyrosyl-DNA phosphodiesterase; ref. 57).
Studies of IκBα have demonstrated that SUMO-1 modification of IκBα antagonizes ubiquitin modification of IκBα (58). Apparently, the antagonism is due to competition of the two proteins on the same lysine residue on IκBα, which is used for conjugation (58, 59). It remains to be seen whether SUMO-1 and ubiquitin may similarly compete on the same lysine residue(s) on hTOP1 for conjugation.
The SUMO-1/UBC9 pathway is probably involved in the repair of both TOP1- and TOP2-mediated DNA damage. Our preliminary studies have demonstrated that both topoisomerase IIα (TOP2α) and topoisomerase IIβ (TOP2β) are modified by SUMO-1 in cells treated with topoisomerase II poison, VM-26. In addition, both TOP2α and TOP2β can physically interact with UBC9 (Y.M. and L.F.L., unpublished results). Whether SUMO-1/UBC9 is generally involved in the repair of protein-linked DNA breaks remains to be determined. However, recent studies have demonstrated that SUMO-1/UBC9 is activated by a number of stress conditions known to damage proteins (Y.M. and L.F.L., unpublished results). It seems possible that the SUMO-1/UBC9 pathway is evolved to process nuclear proteins which are either damaged (e.g., by oxidative stress or heat shock) or have undergone specific conformational changes (e.g., topoisomerase cleavable complexes induced by topoisomerase poisons). Our demonstration that CPT can induce rapid and extensive SUMO-1 conjugation to TOP1 could provide a new system for studying both the repair mechanism of protein-linked DNA damage and more generally the function of SUMO-1.
Acknowledgments
We are grateful to Dr. Erica S. Johnson for providing us with wild-type and mutant HF-SMT3 plasmid DNAs and Dr. Stefan Jentsch for providing us with Y0007 and Y0174 yeast strains. We also thank Dr. R. Haguenauer-Tsapis for providing us ubiquitin dominant negative mutants. This work was supported by National Institutes of Health Grants CA39662 and GM27731.
Abbreviations
- TOP1
topoisomerase I
- hTOP1
human TOP1
- SUMO-1
small ubiquitin-like modifier
- CPT
camptothecin
- PMSF
phenylmethylsulfonyl fluoride
- GST
glutathione S-transferase
- GSH
glutathione
- DMSO
dimethyl sulfoxide
- hUBC9
human UBC9
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080536597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080536597
References
- 1.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Liu L F, Duann P, Lin C T, D'Arpa P, Wu J. Ann NY Acad Sci. 1996;803:44–49. doi: 10.1111/j.1749-6632.1996.tb26375.x. [DOI] [PubMed] [Google Scholar]
- 3.Kingma P S, Osheroff N. Biochim Biophys Acta. 1998;1400:223–232. doi: 10.1016/s0167-4781(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 4.Nambi P, Mattern M, Bartus J O, Aiyar N, Crooke S T. Biochem J. 1989;262:485–489. doi: 10.1042/bj2620485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palitti F. Environ Mol Mutagen. 1993;22:275–277. doi: 10.1002/em.2850220416. [DOI] [PubMed] [Google Scholar]
- 6.Pommier Y, Tanizawa A, Kohn K W. Adv. Pharmacol. 1994. 73–92. [DOI] [PubMed] [Google Scholar]
- 7.Li T K, Chen A Y, Yu C, Mao Y, Wang H, Liu L F. Genes Dev. 1999;13:1553–1560. doi: 10.1101/gad.13.12.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desai S D, Liu L F, Vazquez-Abad D, D'Arpa P. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- 9.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 10.Saitoh H, Pu R T, Dasso M. Trends Biochem Sci. 1997;22:374–376. doi: 10.1016/s0968-0004(97)01102-x. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meluh P B a K, D. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson E S, Schwienhorst I, Dohmen R J, Blobel G. EMBO J. 1997;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. Biochem Biophys Res Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 15.Desterro J M, Rodriguez M S, Kemp G D, Hay R T. J Biol Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 16.Desterro J M, Thomson J, Hay R T. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee G W, Melchior F, Matunis M J, Mahajan R, Tian Q, Anderson P. J Biol Chem. 1998;273:6503–6507. doi: 10.1074/jbc.273.11.6503. [DOI] [PubMed] [Google Scholar]
- 18.Johnson E S, Blobel G. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 19.Li S J, Hochstrasser M. Nature (London) 1999;398:246–251. doi: 10.1038/18457. [DOI] [PubMed] [Google Scholar]
- 20.Seufert W, Futcher B, Jentsch S. Nature (London) 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- 21.Spence J, Sadis S, Haas A L, Finley D. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 23.Matunis M J, Coutavas E, Blobel G. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 25.Tashiro K, Pando M P, Kanegae Y, Wamsley P M, Inoue S, Verma I M. Proc Natl Acad Sci USA. 1997;94:7862–7867. doi: 10.1073/pnas.94.15.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okura T, Gong L, Kamitani T, Wada T, Okura I, Wei C F, Chang H M, Yeh E T. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 27.Gostissa M, Hengstermann V F, Sandy P, Schwarz S E, Scheffner M, Del Sal G. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez M S, Desterro J M P, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 30.Shen Z, Pardington-Purtymun P E, Comeaux J C, Moyzis R K, Chen D J. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 31.Hateboer G, Hijmans E M, Nooij J B, Schlenker S, Jentsch S, Bernards R. J Biol Chem. 1996;271:25906–25911. doi: 10.1074/jbc.271.42.25906. [DOI] [PubMed] [Google Scholar]
- 32.Tarsounas M, Pearlman R E, Gasser P J, Park M S, Moens P B. Mol Biol Cell. 1997;8:1405–1414. doi: 10.1091/mbc.8.8.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottlicher M, Heck S, Doucas V, Wade E, Kullmann M, Cato A C, Evans R M, Herrlich P. Steroids. 1996;61:257–262. doi: 10.1016/0039-128x(96)00032-3. [DOI] [PubMed] [Google Scholar]
- 34.Wright D A, Futcher B, Ghosh P, Geha R S. J Biol Chem. 1996;271:31037–31043. doi: 10.1074/jbc.271.49.31037. [DOI] [PubMed] [Google Scholar]
- 35.Kovalenko O V, Plug A W, Haaf T, Gonda D K, Ashley T, Ward D C, Radding C M, Golub E I. Proc Natl Acad Sci USA. 1996;93:2958–2963. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahajan R, Gerace L, Melchior F. J Cell Biol. 1998;140:259–270. doi: 10.1083/jcb.140.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matunis M J, Wu J, Blobel G. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Curr Biol. 1998;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 39.Kamitani T, Kito K, Nguyen H P, Wada H, Fukuda-Kamitani T, Yeh E T. J Biol Chem. 1998;273:26675–26682. doi: 10.1074/jbc.273.41.26675. [DOI] [PubMed] [Google Scholar]
- 40.Muller S, Dejean A. J Virol. 1999;73:5137–5143. doi: 10.1128/jvi.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bharti A K, Olson M O J, Kufe D W, Rubin E H. J Biol Chem. 1996;271:1993–1997. doi: 10.1074/jbc.271.4.1993. [DOI] [PubMed] [Google Scholar]
- 42.Gatto B, Sanders M M, Yu C, Wu H, Makhey D, Lavoie E J, Liu L F. Cancer Res. 1996;56:2795–2800. [PubMed] [Google Scholar]
- 43.Nitiss J, Wang J C. Proc Natl Acad Sci USA. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bjornsti M-A, Benedetti P, Viglianti G A, Wang J C. Cancer Res. 1989. 49. [PubMed] [Google Scholar]
- 45.Galan J-M, Haguenauer-Tsapis R. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jentsch S, McGrath J P, Varshavsky A. Nature (London) 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 47.Huibregtse J M, Yang J C, Beaudenon S L. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L F, D'Arpa P. Important Adv. Oncol. 1992. 79–89. [PubMed] [Google Scholar]
- 49.Andoh T, Ishii K, Suzuki Y, Ikegami Y, Kusunoki Y, Takemoto T, Okada K. Proc Natl Acad Sci USA. 1987;84:5565–5569. doi: 10.1073/pnas.84.16.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pantazis P, Mendoza J T, DeJesus A, Rubin E, Kufe D, Giovanella B C. Eur J Haematol. 1994;53:135–144. doi: 10.1111/j.1600-0609.1994.tb00661.x. [DOI] [PubMed] [Google Scholar]
- 51.Muller S, Miller W H, Jr, Dejean A. Blood. 1998;92:4308–4316. [PubMed] [Google Scholar]
- 52.Duprez E, Saurin A J, Desterro J M, Lallemand-Breitenbach V, Howe K, Boddy M N, Solomon E, de Th H, Hay R T, Freemont P S. J Cell Sci. 1999;112:381–393. doi: 10.1242/jcs.112.3.381. [DOI] [PubMed] [Google Scholar]
- 53.Everett R, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kamitani T, Nguyen H P, Yeh E T. J Biol Chem. 1997;272:14001–14004. doi: 10.1074/jbc.272.22.14001. [DOI] [PubMed] [Google Scholar]
- 55.Buckwalter C A, Lin A H, Tanizawa A, Pommier Y G, Cheng Y C, Kaufmann S H. Cancer Res. 1996;56:1674–1681. [PubMed] [Google Scholar]
- 56.Dank M K, Garrett K E, Marion R C, Whipple D O. Cancer Res. 1996;56:1664–1673. [PubMed] [Google Scholar]
- 57.Pouliot J J, Yao K C, Robertsob C A, Nash H A. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 58.Desterro J M, Rodriguez M S, Hay R T. Mol Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 59.Scherer D C, Brockman J A, Chen Z, Maniatis T, Ballard D W. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]