Abstract
We describe a “protein knockout” technique that can be used to identify essential proteins in bacteria. This technique uses phage display to select peptides that bind specifically to purified target proteins. The peptides are expressed intracellularly and cause inhibition of growth when the protein is essential. In this study, peptides that each specifically bind to one of seven essential proteins were identified by phage display and then expressed as fusions to glutathione S-transferase in Escherichia coli. Expression of peptide fusions directed against E. coli DnaN, LpxA, RpoD, ProRS, SecA, GyrA, and Era each dramatically inhibited cell growth. Under the same conditions, a fusion with a randomized peptide sequence did not inhibit cell growth. In growth-inhibited cells, inhibition could be relieved by concurrent overexpression of the relevant target protein but not by coexpression of an irrelevant protein, indicating that growth inhibition was due to a specific interaction of the expressed peptide with its target. The protein knockout technique can be used to assess the essentiality of genes of unknown function emerging from the sequencing of microbial genomes. This technique can also be used to validate proteins as drug targets, and their corresponding peptides as screening tools, for discovery of new antimicrobial agents.
Despite the success of antimicrobial agents, infectious disease is now the second leading cause of death worldwide (14). This is in large part due to increased resistance of pathogens to a variety of antimicrobials and is an accelerating trend. It is evident that the dynamic and resilient nature of microbial life, coupled with the profligate use of antibiotics, has selected drug-resistant microbes across a wide spectrum of pathogenic organisms. There is a clear and urgent need for novel antibiotics to help combat this problem (8, 10, 30).
Although most antibiotics in use today are directed against a small number of bacterial targets, there is no shortage of potential targets. Analyses of bacterial genomes have allowed estimates of the “minimal genome set” of genes essential for viability. A comparison of the genomes of Haemophilus influenzae and Mycoplasma genitalium identified 256 genes considered necessary and sufficient for cell viability, while a study of the frequency of indispensable genes in the Bacillus subtilis genome arrived at 250 to 450 essential genes (25, 32). This range of genes should thus constitute the number of potential targets for antimicrobial drug discovery, a number far larger than the dozen or so sites of action for all currently existing antibiotics. Genome-sequencing efforts have increased the potential for development of new classes of antibiotics directed against novel targets (15, 39, 42). Among the potential targets revealed by sequencing are those of unknown function, as well as those of known function that have never been used as drug targets. There is a pressing need for technologies that can validate or invalidate targets for drug discovery by showing whether they are essential for microbial survival.
Here we describe a “protein knockout” method for demonstrating the essentiality of bacterial proteins that requires no prior knowledge of protein function. Proteins are expressed, purified, and used in phage display to isolate peptides that bind specifically to them. The peptides are then expressed intracellularly as glutathione S-transferase (GST) fusions, and their effect on growth is monitored. If the protein is essential, cell growth should be inhibited.
The protein knockout strategy builds on several observations reported previously. We and others have shown that many peptides selected in phage display are directed to functional sites on target proteins (reviewed in reference 26). In an extension of our previous work in isolating a peptide inhibitor of prolyl-tRNA synthetase (ProRS) (24), Tao et al. showed that intracellular expression of this peptide fused to GST could slow bacterial growth and rescue mice from an otherwise lethal dose of virulent Escherichia coli. (46). Specific inhibition of ProRS in the cell was demonstrated by a decrease in the intracellular pool of charged tRNAPro. Walker et al. (47) expressed 20,000 peptides in E. coli and found that 21 of them were growth inhibitory; however, the targets of these peptides were unknown. This study showed the importance of using peptides fused to proteins or amino acid motifs to improve stability. Blum et al. (1) induced the expression of a library of peptides fused to thioredoxin and selected for cells possessing the ThyA− phenotype (trimethoprim resistance and a thymine requirement for growth). They discovered peptides that inhibited cell growth and found that the phenotype could be suppressed by overexpression of ThyA protein. Peptide expression has also been used to knock out protein function and dissect signal transduction pathways in eukaryotic cells (5, 7, 9, 13, 17, 18, 41).
In this study, we showed that protein-specific peptides can first be selected in vitro and then used to assess the essentiality of a wide variety of proteins in bacteria. We demonstrated this method with proteins known to be essential for E. coli, including those involved in nearly every basic cellular process: DNA replication (DnaN), lipid A biosynthesis (LpxA), transcription initiation (RpoD or σ70), translation (ProRS), protein secretion (SecA), and DNA supercoiling (GyrA), as well as one essential protein of unknown function (Era). We also assessed the specificity of the intracellular protein-peptide interaction by performing protein overexpression rescue experiments.
MATERIALS AND METHODS
Bacterial strains and media.
For all work, the base medium was 2×YT supplemented when appropriate with the antibiotics chloramphenicol (to 30 μg/ml), spectinomycin (to 100 μg/ml), and/or kanamycin (to 30 μg/ml). E. coli DH5αPro was purchased from BD Biosciences/Clontech (Palo Alto, Calif.). E. coli BL21(DE3) cells and electrocompetent JS5 cells were purchased from Bio-Rad (Hercules, Calif.). Other electrocompetent cells, when needed, were prepared as follows. The cells were grown overnight at 37°C in 20 ml of the appropriate medium, diluted into 1 liter, and grown to an optical density of 0.5 at a wavelength of 600 nm. The cells were pelleted and washed in ice-cold sterile 10% glycerol in volumes of 1 liter, 500 ml, 250 ml, 125 ml, and finally 5 ml. They were frozen at −80°C.
DNA manipulations and synthesis of oligonucleotide and peptides.
PCR was carried out using the Expand High-Fidelity PCR system (Roche, Indianapolis, Ind.) in conjunction with the Techne Genius thermal cycler (Techne, Princeton, N.J.). Restriction enzymes and T4 DNA ligase were from New England Biolabs (NEB; Beverly, Mass.). DNA was purified using kits purchased from Qiagen (Valencia, Calif.). Electroporation was carried out in 0.1-cm cuvettes in a Bio-Rad E. coli Pulser device at 1.8 kV. DNA sequence analysis was carried out by Sequetech Corp. (Mountain View, Calif.). Oligonucleotides were synthesized by The Midland Certified Reagent Co., Inc. (Midland, Tex.). Peptides were synthesized by AnaSpec, Inc. (San Jose, Calif.).
Oligonucleotides.
Two oligonucleotides were annealed and used to make a cassette to introduce SalI and NheI sites into pPROTet.E332-lacZ (BD Biosciences/Clontech; see below). They are ProtetSal/Nhe5 and ProtetSal/Nhe3, respectively (5′-GAGAAAGGTACCCATGTCGACAGGTTAGGCTAGCACAAGCTTGGATACT and 5′-AGTATCCAAGCTTGTGCTAGCCTAACCTGTCGACATGGGTACCTTTCTC). Two oligonuclcotides were annealed and used to make a cassette to introduce a new multicloning site into pPROLar.A222. They are pKBU-Pro-Lar5 and pKBU-Pro-Lar3, respectively (5′-CATATGCTCATGAGGTAATAACCCGGGCGTTAAT and 5′-TAACGCCCGGGTTATTACCTCATGAGCATATGGTAC). The PCR primers used to clone GST (and a hinge) into pPROTet.E332 (Hind3-PstI) are primers tGST5 and tGST3, respectively (5′-GATCGATCAAGCTTGGGATCAGGATCCATGTCCCCTATACTAGGTTATTGG and 5′-GATCGATCCTGCAGCCTCACAGATCCGATTTTGGAGGATGGTCGCC).
To clone target genes into pTYB2AT or pTYB4AT (see below), the following pairs of PCR primers were used, each set named after the protein of interest and the end of the gene to which they hybridize (5 = 5′; 3 = 3′): LpxA5 (5′-ATGATCGACATATGATTGATAAATCCGCCTTTGTGC) and LpxA3 (5′-GATCGACCCGGGACGAATCAGACCGCGCGTTGAGCG), DnaN5 (5′-TCGTCATCCATATGAAATTTACCGTAGAACGTGAGCA) and DnaN3 (5′-TCGTCATCCCCGGGCAGTCTCATTGGCATGACAACATAA), RpoD5 (5′-ATGATCGACATATGGAGCAAAACCCGCAGTCACAGC) and RpoD3 (5′-GATCGACCCGGGATCGTCCAGGAAGCTACGCAGC), Era5 (5′-GATCGTACCATGGGAAGCATCGATAAAAGTTACTGCGG) and Era3 (5′-GATCGACCCGGGAAGATCGTCAACGTAACCGAGACTGCG), GyrA5 (5′-TCGTCATCCATATGAGCGACCTTGCGAGAGAAATTAC) and GyrA3 (5′-TCGTCATCGATATCTTCTTCTTCTGGCTCGTCGTCAACG), and SecA5 (5′-TCGTCATCCATATGCTAATCAAATTGTTAACTAAAG) and SecA3 (5′-TCGTCATCCCCGGGTTGCAGGCGGCCATGGCACTGCTTG).
The sequencing primer used to confirm cloning of peptides was 5′ProTet (5′-CATCAGCAGGACGCACTGAC). The sequencing primer used to confirm cloning of target genes into pKBU-Pro-Lar was 5′ProLar (5′-GTGAGCGCTCACAATTATGATAG).
Cloning of target genes and protein purification.
Genes encoding target proteins were amplified by PCR with specific oligonucleotides. The amplified products were cloned into pTYB2AT or pTYB4AT, vectors based on TYB2 and TYB4 (NEB), respectively, altered to include a 15-amino-acid in vivo biotinylation signal at the C terminus of the purified protein. Each construct was transformed into BL21(DE3) cells (Stratagene, La Jolla, Calif.) carrying the pBirAcm plasmid (Avidity, Denver, Colo.) that encodes biotin ligase. Plasmid DNA was isolated, and the inserts were confirmed by DNA sequencing. Protein was produced and purified by the NEB IMPACT method.
Phage display.
Immulon 4 96-well plates (Thermo Labsystems, Franklin, Mass.) were coated with streptavidin (1 μg/well) in 0.1 M sodium bicarbonate and blocked with 1% bovine serum albumin. Biotinylated protein (4 pmol/well) diluted into TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) was allowed to bind to the plate for 1 h. The plates were washed with TBST, and phage from each of 20 recombinant M13 libraries (total complexity of 2 × 1010) were allowed to bind for 1 h. The plates were washed again, and the bound phage were eluted, amplified, and used for the next round of phage selection as described previously (44). After three or four rounds of selection, phage pools were plated for single plaques and purified and the DNA sequence of individual isolates was determined. Peptides were synthesized to include the 12- to 15-amino-acid sequence displayed by the phage followed by a short linker and a biotin at the C terminus to allow labeling for analysis of peptide binding. Isolation and use of ProRS-specific phage has been described previously (24, 46); peptide Pro-3 in those publications is referred to as peptide 72 in this work.
Cloning vectors for intracellular expression of peptide fusion and protein.
Two commercially available plasmids were altered for cloning purposes.
(i) Vector for peptide expression.
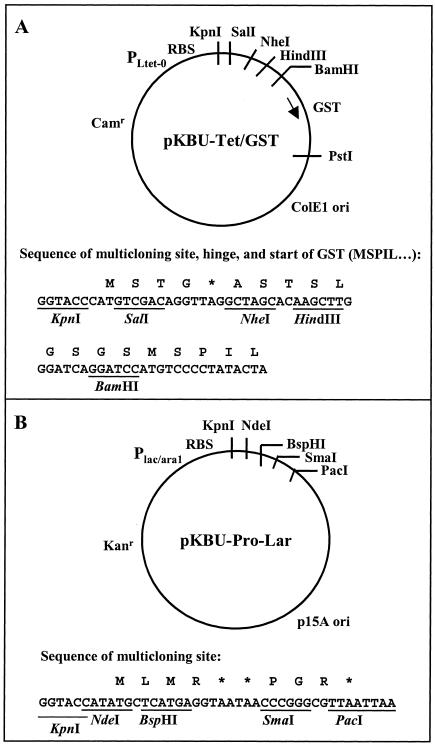
Plasmid pPROTet.E332-lacZ was modified for cloning of peptide sequences as GST fusions by first introducing a cassette with SalI and NheI sites for peptide sequence cloning and then replacing the lacZ gene with the gst gene. Oligonucleotides ProtetSal/Nhe5 and ProtetSal/Nhe3 were annealed at 95°C for 2 min in 1× SURE/CUT buffer B (Roche) and then allowed to cool to room temperature. The DNA was then cut with HindIII and KpnI and ligated into the pPROTet.E332-lacZ vector previously cut with HindIII and KpnI. Transformants were screened for the presence of the NheI site, and DNA sequences of the plasmids were confirmed. The gst gene was amplified by PCR from pGEX2T (Amersham BioSciences, Piscataway, N.J.) with primers tGST5 and tGST3. This set of primers also introduced a linker region between the multiple-cloning site and gst. The PCR product was cut with HindIII and PstI and ligated into the modified pPROTet.E332-lacZ vector cut with the same enzymes. Transformants were screened for presence of an introduced BamHI site, and plasmids were confirmed by sequence analysis. The resulting plasmid is pKBU-Tet/GST (Fig. 1A).
FIG. 1.
Expression vectors. (A) Peptide intracellular expression vector pKBU-Tet/GST is based on the Clontech pPROTet.E332-lacZ vector. SalI and NheI sites were introduced on a cassette, and then the gene for GST and a 5′ hinge region were inserted into the HindIII and PstI sites. Subsequently, peptides were cloned into the SalI and NheI sites. (B) Protein intracellular expression vector pKBU-Pro-Lar is based on the Clontech pPROLar.A222 vector. NdeI, BspHI, and SmaI sites were introduced on a cassette cloned into the KpnI and PacI sites. Subsequently, target genes of interest were cloned into the NdeI or BspHI and SmaI sites. RBS, ribosome binding site.
(ii) Vector for target protein expression.
Plasmid pProLar.A222 (Clontech) was modified by introducing a cassette with NdeI, BspHI, and SmaI cloning sites between the KpnI and PacI sites to create pKBU-Pro-Lar (Fig. 1B).
Construction of plasmids for peptide expression.
Each peptide expression plasmid (with the exception of the ProRS peptide-encoding plasmid) was made with a specific pair of 5′-phosphorylated oligonucleotides for insertion into the SalI and NheI cloning sites of pKBU-Tet/GST (Fig. 1A). Each oligonucleotide was resuspended to 1 μg/μl in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA (TE), and 9.5 μl of the sense oligonucleotide and 9.5 μl of the antisense oligonucleotide were mixed with 1 μl of 5M NaCl in TE and placed in a heat block at 80°C. After 30 min, the block was removed from its heater and allowed to cool to 30°C. Each annealed cassette was diluted with TE to 200 μl and then ligated into pKBU-Tet/GST; 2 μl of each ligation mixture was transformed into 20 μl of electrocompetent DH5αPro cells. Cells were plated onto medium containing chloramphenicol and grown overnight at 37°C. Several isolates from each ligation were picked and cultured, and insert-positive isolates were identified by PCR amplification with primers outside the insert region. Positive isolates were then sequenced with the oligonucleotide 5′ProTet to confirm correct sequences of the peptide-coding inserts.
Construction of plasmids for protein expression.
For intracellular expression experiments, target genes were removed from the pTYB2AT or pTYB4AT constructs used for protein production with the restriction sites used to clone them (or with flanking sites) and recloned into compatible sites in the vector pKBU-Pro-Lar (Fig. 1B). The resulting constructs were transformed into JS5 cells, sequences were confirmed, and the plasmids were repurified and transformed into DH5αPro cells.
Induction of peptide fusions.
Overnight cultures of sequence-confirmed constructs were diluted 1:100 (10 μl into 1 ml) into a 96-well block with fresh selective medium and the addition of the inducer anhydrotetracycline (BD Biosciences/Clontech) to 100 ng/ml. These cultures were grown at 22°C with shaking, and optical densities (absorbance at 600 nm) were read in a SpectraMax 190 spectrophotometric plate reader (Molecular Devices, Sunnyvale, Calif.) at 12, 24, 36, and 48 h.
Coinduction of peptide fusions and target proteins.
Cells bearing pKBU-Tet-peptide-GST constructs were made electrocompetent and were transformed with the pKBU-Pro-Lar-target constructs, plated onto 2× YT plus kanamycin, and grown overnight at 37°C. Isolates were picked and grown in medium with chloramphenicol, spectinomycin, and kanamycin, and the presence of both constructs in the cells was confirmed by sequencing of the DNA with individual sets of plasmid-specific primers. Overnight cultures of cells bearing these plasmids were diluted 1:100 (10 μl into 1 ml) into a 96-well block, with fresh selective medium containing the inducer of peptide expression, anhydrotetracyline (100 ng/ml), as well as the inducers of protein expression, l-(+)-arabinose (0.2 %) and isopropyl-1-thio-β-d-galactopyranoside (1 mM). Induced cultures were grown at 22°C, and optical densities were read after 48 h.
Cultures were also diluted 1:1,000 at 3 h after induction; plated onto 2× YT solid medium containing chloramphenicol, spectinomycin, kanamycin, and inducers; and grown overnight at 37°C.
Western blotting.
Equal volumes of overnight cultures of cells carrying pKBU-Tet-peptide-GST constructs were diluted into fresh selective medium containing inducer and grown at 37°C. At 0, 1, 2, 3, and 4 h, optical densities were measured and samples were taken. Cell samples were resuspended at a concentration of 1 absorbance unit at 600 nm per ml in SDS sample buffer (3% sodium dodecyl sulfate, 1 mM dithiothreitol, 10 % glycerol, 63 mM Tris-HCl [pH 6.8]), boiled for 2 min, and applied to 4 to 20% gradient polyacrylamide gels. The resolved proteins were electroblotted to polyvinylidene difluoride membranes and probed with anti-GST antibody covalently linked to alkaline phosphatase (Santa Cruz Biotechnology, Santa Cruz, Calif.). The blots were developed with WesternBlue substrate (Promega Corp., Madison, Wis.).
RESULTS
Isolation of peptides that bind specifically to essential E. coli proteins.
The genes encoding DnaN, LpxA, RpoD, ProRS, SecA, GyrA, and Era are all essential in E. coli (2, 16, 29, 33, 35, 36, 40). These genes were amplified by PCR from E. coli strain K-12 genomic DNA and cloned into expression vectors, their DNA sequences were confirmed, and the proteins were expressed and purified. Each biotinylated target protein was immobilized in individual wells of a streptavidin-coated plastic 96-well plate. Recombinant M13 phage from libraries with a total expected diversity of 2 × 1010 peptides were added to the plate to isolate binding phage. The selections carried out on all of the bacterial proteins yielded phage particles that bound specifically to the protein of interest and not to several irrelevant control proteins. Replicative intermediate DNA from these phage was sequenced to determine the amino acid sequences of the peptides displayed by the phage. Correlations were then drawn between amino acid sequences and strong binding to target (as assessed by relative phage affinity experiments [data not shown]), and representative peptides were chosen for synthesis. These synthetic peptides were characterized for relative binding and cross-competition (data not shown) to confirm binding and specificity. Peptides identified as being strong, specific binders were expressed intracellularly as GST fusions. Sequences of the peptides described in this report are shown in Table 1.
TABLE 1.
Amino acid sequences of peptides described in this report
| Peptide ID no. | E. coli target | Amino acid sequence |
|---|---|---|
| 72a | ProRS | SREWHFWRDYNPTSR |
| 729 | RpoD | SRSFSEWREEWLRSR |
| 920 | LpxA | SSGWMLDPIAGKWSR |
| 1330 | SecA | SSFSFDGHVWYPVLPAQSR |
| 1337 | DnaN | SSATRGWHQLDLFSR |
| 1396 | GyrA | SSGWPQWFPGVGWSR |
| 1706 | Era | SSEAWRKWLSADFSR |
| RND | Control | SREWDRLWHPWLQSR |
Intracellular expression of peptide-GST fusions inhibits bacterial growth.
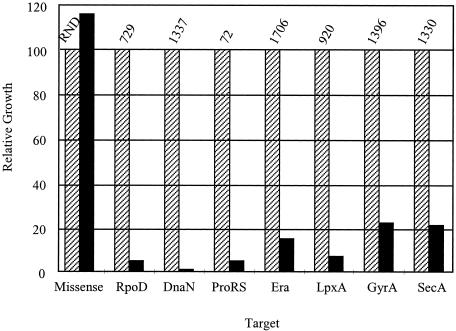
Oligonucleotide cassettes were designed encoding the amino acid sequences of the protein-specific peptides isolated from phage display. A random control peptide was also designed with the same general amino acid composition of the growth-inhibitory peptides but different in primary sequence. Oligonucleotide cassettes were cloned into the expression vector pKBU-Tet/GST. Cells containing each construct were grown overnight at 37°C and then diluted 1:100 in fresh liquid media with or without the inducer anhydrotetracycline. These cultures were grown at 22°C, and the optical densities were tracked for 48 hours. Expression of a target-specific peptide-GST fusion should inhibit cell growth (Fig. 2). The expression of protein-specific peptide-GST fusions resulted in various degrees of inhibition; representative data for peptides with the most inhibitory effects are shown in Fig. 3. These included peptides 729 (RpoD), 1337 (DnaN), 72 (ProRS), 1706 (Era), 920 (LpxA), 1396 (GyrA), and 1330 (SecA). The random peptide did not inhibit growth, as also shown in Fig. 3. Western blotting of cells very soon after the induction of peptide expression using anti-GST antibodies showed that all fusions were expressed and that there was no correlation between growth inhibition and level of peptide expression (data not shown).
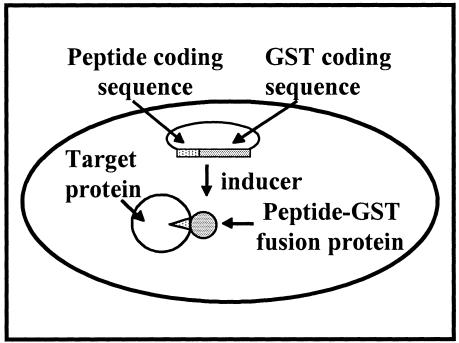
FIG. 2.
Intracellular peptide expression. In the presence of the inducer anhydrotetracycline, peptide specific for an essential cellular protein is produced inside the cell as an N-terminal fusion to GST. Specific peptide binding to the target protein inhibits the essential function and causes growth inhibition.
FIG. 3.
Intracellular peptide expression inhibits cell growth. Cells with the plasmid carrying an inducible target-specific peptide fused to GST were grown in the absence (striped bars) or presence (solid bars) of the inducer; the absorbance at 600 nm was read 44 to 48 h after induction. Data are expressed relative to the uninduced control for a given peptide, with the absorbance of the uninduced control at 600 nm representing 100%. Peptide identification numbers are given above each data set; the peptide target is given below each data set. The peptide RND is a noninhibitory peptide, using amino acids generally like that of inhibitory peptides but with a randomized primary sequence. Data shown are representative of several such experiments.
Expression of specific target protein rescues peptide-dependent growth inhibition.
DNAs encoding the target proteins LpxA and DnaN were subcloned into the expression vector pKBU/ProLar. Each new construct was transformed into cells containing either of the peptide-GST fusion constructs 920-GST or 1337-GST. Plasmid DNAs isolated from the transformants were sequenced, which confirmed the presence of both constructs (a peptide-GST fusion plasmid and a target plasmid) in each cell strain generated.
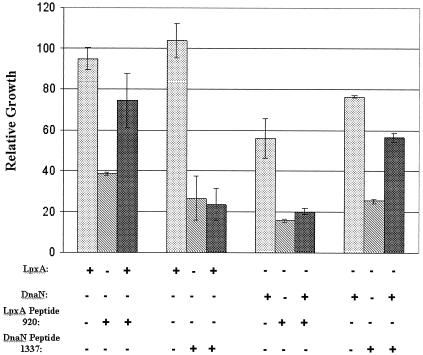
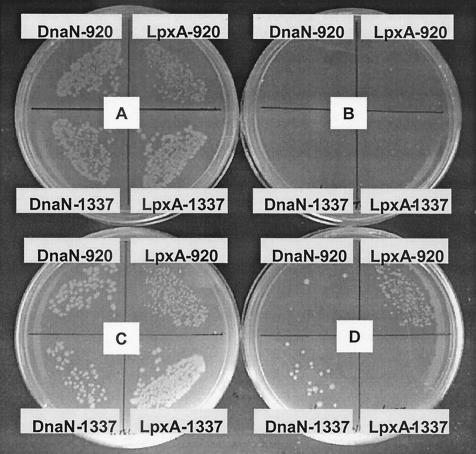
The peptide-GST fusion and the target protein were induced, either individually or simultaneously, and growth in liquid culture at 22°C was tracked for 48 h. Induction of LpxA alone had little effect on cell growth, and induction of DnaN alone had a slight inhibitory effect on cell growth. Overexpression of peptide fusions 920-GST and 1337-GST each resulted in significant growth inhibition. However, in cells expressing both a target protein and a peptide fusion, peptide-dependent growth inhibition could be fully or partially relieved, but only in a target-specific manner (Fig. 4). These experiments were repeated several times with similar results. The same effects were seen with plated cultures (Fig. 5). Plate B shows growth inhibition of cells due to induction of peptides. Plate D shows induction of peptides and targets and reveals the specificity of rescue: overexpression of LpxA rescues growth inhibition caused by its cognate peptide 920 but not that caused by the DnaN cognate peptide 1337. Similarly, overexpression of DnaN specifically rescues growth inhibition caused by its cognate peptide 1337 but not that caused by peptide 920. (Plate A shows uninduced cells, and plate C shows induction of target alone). Overexpression of SecA rescued growth inhibition caused by SecA cognate peptide 1330; however, overexpression of RpoD did not rescue growth inhibition caused by RpoD cognate peptide 729 (data not shown).
FIG. 4.
Specific rescue of peptide-dependent growth inhibition. Dotted bars, protein (LpxA or DnaN) induction; striped bars, peptide (920 or 1337) induction; cross-hatched bars: coinduction. Peptide 920 was originally selected in vitro on LpxA; peptide 1337 was selected on DnaN. The absorbance of cultures at 600 nm was read 48 h after induction of protein and/or peptide in liquid medium. Data are expressed relative to those obtained with uninduced control cultures grown in parallel; error bars show standard deviations.
FIG. 5.
Specific rescue of peptide-dependent growth inhibition. Cultures were plated on selective medium containing appropriate inducers 3 h after induction of protein and/or peptide in liquid medium and then grown for 16 h at 37°C. (A) No induction; (B) peptide induction alone; (C) protein induction alone; (D) peptide and protein induction. Each quadrant is labeled with the protein and peptide, respectively, whose expression is induced. Peptide 920 was selected on LpxA; peptide 1337 was selected on DnaN.
DISCUSSION
We have demonstrated a protein knockout method by isolating peptides from phage display libraries that are specifically directed to essential bacterial proteins and by showing that intracellular expression of these peptides inhibits bacterial growth. In contrast, intracellular expression of peptides with the same amino acid composition but a scrambled sequence did not cause inhibition of growth. There was no correlation between growth inhibition and level of expression for all tested peptide-GST fusions. Therefore, the growth-inhibitory effects seen are due neither to mere protein overexpression nor to the amino acid composition of the fusions but, instead, require the specific amino acid sequence of the peptides identified via phage display. Furthermore, overexpression of target protein relieved inhibition caused by the corresponding peptide but not a different peptide for three of four proteins tested (DnaN, LpxA, and SecA). These findings support the hypothesis that intracellular expression of peptides directed to essential bacterial targets can interfere with cell growth. That this approach is successful with proteins known to be essential would predict that it will successfully identify essential proteins of unknown function encoded in microbial genomes.
It is not surprising that peptides selected to bind a specific protein would inhibit the function of that protein intracellularly. The phage display process selects peptides that bind active or functional sites rather than random sites on protein surfaces (21, 26). Although this phenomenon is not completely understood, it is known that active sites possess physicochemical properties such as deep, flexible clefts that have evolved to bind ligands (28, 48). In addition, peptide binding to these clefts is energetically favorable (11). Accordingly, the activity of many enzymes can be inhibited by peptides in vitro, and this inhibition is quite specific. Enzymes inhibited in this way include prolyl- and tyrosyl-tRNA synthetases (24), β-glucosidase (24), hexokinase (24), glycogen phosphorylase a (24), alcohol dehydrogenase (24), enzyme I of the phosphotransferase system (31), rRNA methyltransferase (19), human immunodeficiency virus HIV integrase (37), membrane dipeptidase (38), NADPH-dependent oxidase (12), protein phosphatase I (49), Src kinase (34), urease (23), and diphosphocytidine methylerythritol synthase (A. Jacobi, A., H. Baum, T. Nakano, R. R. Annand, S. Anderson, J. Breunig, S. Bowes, P. Goldenblatt, S. S. Ashraf, H. Grøn, P. Hamilton, D. J. Christensen, and H. Loferer, Program Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2124, 2001). Biologically relevant associations between proteins and peptides have also been seen in crystal structures. Peptides selected by phage display on the DM2 oncoprotein resembled the portion of the p53 substrate seen in the crystal structure to bind in the DM2 binding pocket (3, 4, 27). A peptide selected by phage display against another target not only inhibited that target's biochemical activity but also was found in the target's active site in a cocrystal (P. Bernasconi, Abstr. HTS Appl. Agribusiness, SBS North Carolina Regional Meeting, 2002). These examples make clear that peptides can bind to active sites and inhibit function in vitro and therefore could inhibit function intracellularly.
However, it is difficult to draw conclusions if intracellular peptide expression does not lead to growth inhibition. We obtained this result with peptides isolated against the proteins dihydrofolate reductase (DHFR) and deoxyxylulose reductoisomerase (Dxr). Mutants defective in folA (encoding DHFR) are slow growing on rich media (22), and mutants defective in yaeM (encoding Dxr) are not viable on rich media unless specifically supplemented with methylerythritol (45). In both cases, the peptides we isolated by phage display were competed from the protein by antimicrobial drugs specific for that protein; additionally, one of the Dxr-specific peptides inhibited the biochemical activity of Dxr (reference 20 and data not shown). When growth inhibition by intracellular expression of such peptides is not seen, there are a number of possible explanations. These include nonessentiality of the protein, binding of the peptide to a site not relevant to the essential function of the protein, protection of the protein from peptide inhibition by substrate or by assembly into multienzyme complexes, and insufficient expression of the peptide. Furthermore, even if intracellular peptide-protein complexes are efficiently formed, a low level of residual activity from unbound protein may suffice for growth. Any of these factors could also cause incomplete growth inhibition, as seen in the present study, for example, with the SecA peptide. If sufficient numbers of peptides were tested, this ambiguity might be resolved for a given protein. This technique is thus most useful when a clear growth inhibition phenotype is observed with a minimal number of peptides and when protein-specific rescue is demonstrated.
The protein knockout method does not depend on prior knowledge of function and thus is well suited for use with genomics targets. Knowledge of function is often lacking for open reading frames (ORFs) discovered in genome sequencing, and even 20% of ORFs in the genome of the well-studied bacterium E. coli have no known function (43). Another advantage of this method is the direct demonstration of essentiality on the protein level—the level at which an antimicrobial drug would act. Once essentiality of a protein is demonstrated, the method could also be used as a genetic tool to acquire functional information. For example, extragenic suppressors of peptide-induced death could be isolated. Function could also be addressed by the use of transcription profiling or proteomics approaches once it is known that the protein in question is essential.
Peptides shown to knock out function of a specific protein intracellularly can also be used as tools for drug screening (6). For example, such a peptide could be competed from a target protein by small molecules suitable for drug development. In one instance, a peptide that specifically binds E. coli Dxr was used in a high-throughput “surrogate ligand” displacement assay: 32,000 compounds from a diverse historical library were screened in this way, and 30 compounds were identified that inhibited Dxr activity with 50% inhibitory concentrations under 20 μM (20). This kind of peptide competition assay has been used to format other high-throughput screens for difficult-to-assay targets, such as DnaN, the sliding-clamp subunit of DNA polymerase III (D. J. Christensen, E. B. Gottlin, R. E. Benson, and P. T. Hamilton, Program Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. 2131, 2001.) The same format could also be used to discover drugs directed to targets of unknown function. For this purpose, the peptide and the target would both be validated for drug discovery by the results of peptide overexpression on cell growth.
Acknowledgments
We thank Erin Anderson, Kellie Duke, and E. Sturgis Payne for excellent technical assistance.
This work was supported in part by National Institutes of Health grant 5 R43 AI 46036-02.
REFERENCES
- 1.Blum, J. H., S. L. Dove, A. Hochschild, and J. J. Mekalanos. 2000. Isolation of peptide aptamers that inhibit intracellular processes. Proc. Natl. Acad. Sci. USA 97:2241-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohman, K., and L. A. Isaksson. 1980. A temperature-sensitive mutant in prolinyl-tRNA ligase of Escherichia coli K-12. Mol. Gen. Genet. 177:603-605. [DOI] [PubMed] [Google Scholar]
- 3.Bottger, A., V. Bottger, C. Garcia-Echeverria, P. Chene, H. K. Hochkeppel, W. Sampson, K. Ang, S. F. Howard, S. M. Picksley, and D. P. Lane. 1997. Molecular characterization of the hdm2-p53 interaction. J. Mol. Biol. 269:744-756. [DOI] [PubMed] [Google Scholar]
- 4.Bottger, V., A. Bottger, S. F. Howard, S. M. Picksley, P. Chene, C. Garcia-Echeverria, H. K. Hochkeppel, and D. P. Lane. 1996. Identification of novel mdm2 binding peptides by phage display. Oncogene 13:2141-2147. [PubMed] [Google Scholar]
- 5.Caponigro, G., M. R. Abedi, A. P. Hurlburt, A. Maxfield, W. Judd, and A. Kamb. 1998. Transdominant genetic analysis of a growth control pathway. Proc. Natl. Acad. Sci. USA 95:7508-7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, D. J., E. B. Gottlin, R. E. Benson, and P. T. Hamilton. 2001. Phage display for target-based antibacterial drug discovery. Drug Discovery Today 6:721-727. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, B. A., P. Colas, and R. Brent. 1998. An artificial cell-cycle inhibitor isolated from a combinatorial library. Proc. Natl. Acad. Sci. USA 95:14272-14277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen, M. L. 2000. Changing patterns of infectious disease. Nature 406:762-767. [DOI] [PubMed] [Google Scholar]
- 9.Colas, P., B. Cohen, T. Jessen, I. Grishina, J. McCoy, and R. Brent. 1996. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature 380:548-550. [DOI] [PubMed] [Google Scholar]
- 10.Coleman, K., M. Athalye, A. Clancey, M. Davison, D. J. Payne, C. R. Perry, and I. Chopra. 1994. Bacterial resistance mechanisms as therapeutic targets. J. Antimicrob. Chemother. 33:1091-1116. [DOI] [PubMed] [Google Scholar]
- 11.Conte, L. L., C. Chothia, and J. Janin. 1999. The atomic structure of protein- protein recognition sites. J. Mol. Biol. 285:2177-2198. [DOI] [PubMed] [Google Scholar]
- 12.DeLeo, F. R., L. Yu, J. B. Burritt, L. R. Loetterle, C. W. Bond, A. J. Jesaitis, and M. T. Quinn. 1995. Mapping sites of interaction of p47-phox and flavocytochrome b with random-sequence peptide phage display libraries. Proc. Natl. Acad. Sci. USA 192:7110-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbrizio, E., L. Le Cam, J. Polanowska, M. Kaczorek, N. Lamb, R. Brent, and C. Sardet. 1999. Inhibition of mammalian cell proliferation by genetically selected peptide aptamers that functionally antagonize E2F activity. Oncogene 18:4357-4363. [DOI] [PubMed] [Google Scholar]
- 14.Fauci, A. 2001. Infectious diseases: considerations for the 21st century. Clin. Infect. Dis. 32:675-685. [DOI] [PubMed] [Google Scholar]
- 15.Fraser, C. M., J. A. Eisen, and S. L. Salzberg. 2000. Microbial genome sequencing. Nature 406:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galloway, S. M., and C. R. Raetz. 1990. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J. Biol. Chem. 265:6394-6402. [PubMed] [Google Scholar]
- 17.Geyer, C. R., and R. Brent. 2000. Selection of genetic agents from random peptide aptamer expression libraries. Methods Enzymol. 328:171-208. [DOI] [PubMed] [Google Scholar]
- 18.Geyer, C. R., A. Colman-Lerner, and R. Brent. 1999. “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc. Natl. Acad. Sci. USA 96:8567-8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannattasio, R. B., and B. Weisblum. 2000. Modulation of erm methyltransferase activity by peptides derived from phage display. Antimicrob. Agents Chemother. 44:1961-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottlin, E. B., R. E. Benson, S. Conary, B. Antonio, K. Duke, E. S. Payne, S. A. Ashraf, and D. J. Christensen. 2003. High throughput screen for inhibitors of 1-deoxy-d-xylulose 5-phosphate reductoisomerase by surrogate ligand competition. J. Biomol. Screen. 8:332-339. [DOI] [PubMed]
- 21.Grøn, H., and R. Hyde-DeRuyscher. 2000. Peptides as tools in drug discovery. Curr. Opin. Drug Discovery Dev. 3:636-645. [PubMed] [Google Scholar]
- 22.Herrington M. B., and N. T. Chirwa. 1999. Growth properties of a folA null mutant of Escherichia coli K12. Can. J. Microbiol. 45:191-200. [PubMed] [Google Scholar]
- 23.Houimel, M., J. P. Mach, I. Corthesy-Theulaz, B. Corthesy, and I. Fisch. 1999. New inhibitors of Helicobacter pylori urease holoenzyme selected from phage-displayed peptide libraries. Eur. J. Biochem. 262:774-780. [DOI] [PubMed] [Google Scholar]
- 24.Hyde-DeRuyscher, R., L. Paige, D. Christensen, N. Hyde-DeRuyscher, A. Lim, Z. Fredericks, J. Kranz, P. Gallant, J. S. Zhang, S. Rocklage, D. Fowlkes, P. Wendler, and P. Hamilton. 2000. Detection of small-molecule enzyme inhibitors with peptides isolated from phage-displayed combinatorial peptide libraries. Chem. Biol. 7:17-25. [DOI] [PubMed] [Google Scholar]
- 25.Itaya, M. 1995. An estimation of minimal genome size required for life. FEBS Lett. 362:257-260. [DOI] [PubMed] [Google Scholar]
- 26.Kay, B. K., and P. T. Hamilton. 2001. Identification of enzyme inhibitors from phage-displayed combinatorial peptide libraries. Comb. Chem. High Throughput Screen. 4:535-543. [DOI] [PubMed] [Google Scholar]
- 27.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 28.Laskowski, R. A., N. M. Luscombe, M. B. Swindells, and J. M. Thornton. 1996. Protein clefts in molecular recognition and function. Protein Sci. 5:2438-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.March, P. E., C. G. Lerner, J. Ahnn, X. Cui, and M. Inouye. 1988. The Escherichia coli Ras-like protein (Era) has GTPase activity and is essential for cell growth. Oncogene 2:539-544. [PubMed] [Google Scholar]
- 30.Moellering, R. C., Jr. 1998. Antibiotic resistance: lessons for the future. Clin. Infect. Dis. 27(Suppl. 1):S135-S140. [DOI] [PubMed] [Google Scholar]
- 31.Mukhija, S., and B. Erni. 1997. Phage display selection of peptides against enzyme I of the phosphoenolpyruvate-sugar phosphotransferase system (PTS). Mol. Microbiol. 25:1159-1166. [DOI] [PubMed] [Google Scholar]
- 32.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakamura, Y. 1978. RNA polymerase mutant with altered sigma factor in Escherichia coli. Mol. Gen. Genet. 165:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Nishi, T., R. J. Budde, J. S. McMurray, N. U. Obeyesekere, N. Safdar, V. A. Levin., and H. Saya. 1996. Tight-binding inhibitory sequences against pp60(c-src) identified using a random 15-amino-acid peptide library. FEBS Lett. 399:237-240. [DOI] [PubMed] [Google Scholar]
- 35.Oliver, D. B., and J. Beckwith. 1982. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell 30:311-319. [DOI] [PubMed] [Google Scholar]
- 36.Oram, M., R. Kuroda, and L. M. Fisher. 1992. Escherichia coli DNA gyrase: genetic analysis of gyrA and gyrB mutations responsible for thermosensitive enzyme activity. FEBS Lett. 312:61-65. [DOI] [PubMed] [Google Scholar]
- 37.Puras Lutzke, R. A., N. A. Eppens, P. A. Weber, R. A. Houghten, and R. H. Plasterk. 1995. Identification of a hexapeptide inhibitor of the human immunodeficiency virus integrase protein by using a combinatorial chemical library. Proc. Natl. Acad. Sci. USA 92:11456-11460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajotte, D., and E. Ruoslahti. 1999. Membrane dipeptidase is the receptor for a lung-targeting peptide identified by in vivo phage display. J. Biol. Chem. 274:11593-11598. [DOI] [PubMed] [Google Scholar]
- 39.Rosamond, J., and A. Allsop. 2000. Harnessing the power of the genome in the search for new antibiotics. Science 287:1973-1976. [DOI] [PubMed] [Google Scholar]
- 40.Sakakibara, Y., and T. Mizukami. 1980. A temperature-sensitive Escherichia coli mutant defective in DNA replication: dnaN, a new gene adjacent to the dnaA gene. Mol. Gen. Genet. 178:541-553. [DOI] [PubMed] [Google Scholar]
- 41.Sandrock, T., M. Poritz, M. Kim, M. J. Feldhaus, B. Roth, G. Caponigro, and A. Kamb. 2002. Expression levels of transdominant peptides and proteins in Saccharomyces cerevisiae. Yeast 19:1-7. [DOI] [PubMed] [Google Scholar]
- 42.Schmid, M. B. 1998. Novel approaches to the discovery of antimicrobial agents. Curr. Opin. Chem. Biol. 2:529-534. [DOI] [PubMed] [Google Scholar]
- 43.Serres, M. H., S. Gopal, L. A. Nahum, P. Liang, T. Gaasterland, and M. Riley. 2001. A functional update of the Escherichia coli K-12 genome. Genome Biol. 2 [Online.] http://genomebiology.com/2001/2/9/research/0035. [DOI] [PMC free article] [PubMed]
- 44.Sparks, A. B., N. B. Adey, S. Cwirla, and B. K. Kay. 1996. Screening phage-displayed random peptide libraries, p. 227-253. In B. K. Kay, J. Winter, and J. McCafferty (ed.), Phage display of peptides and proteins, a laboratory manual. Academic Press, Inc., San Diego, Calif.
- 45.Takahashi, S., T. Kuzuyama, H. Watanabe, and H. Seto. 1998. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-d-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc. Natl. Acad. Sci. USA 95:9879-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao, J., P. Wendler, G. Connelly, A. Lim, J. Zhang, M. King, T. Li, J. A. Silverman, P. R. Schimmel, and F. P. Tally. 2000. Drug target validation: lethal infection blocked by inducible peptide. Proc. Natl. Acad. Sci. USA 97:783-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker, J. R., J. R. Roth, and E. Altman. 2001. An in vivo study of novel bioactive peptides that inhibit the growth of Escherichia coli. J. Pept. Res. 58:380-388. [DOI] [PubMed] [Google Scholar]
- 48.Zavodszky, P., J. Kardos, A. Svingor, and G. A. Petsko. 1998. Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins. Proc. Natl. Acad. Sci. USA 95:7406-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, S., and E. Y. Lee. 1997. A protein phosphatase-1-binding motif identified by the panning of a random peptide display library. J. Biol. Chem. 272:28368-28372. [DOI] [PubMed] [Google Scholar]