Abstract
Lactococcus lactis LMA12-4 is a pTR2030 transconjugant that has been used as an industrial starter culture because of its resistance to phages predominant in cheese plants. Plasmid pTR2030 interferes with susceptible phages in this host strain via two mechanisms, restriction and modification (R/M) and abortive infection (Hsp). After prolonged use of LMA12-4 transconjugants in the industry, two different bacteriophages, designated nck202.φ48 (φ48) and nck202.φ50 (φ50), were isolated which could produce plaques on LMA12-4 containing pTR2030. In this study, these two phages were characterized and compared with a third phage, nck202.φ31 (φ31), which is susceptible to both the R/M and Hsp activities encoded by pTR2030. Phage φ48 was not susceptible to inhibition by Hsp, whereas φ50 was unaffected by either the R/M or Hsp mechanisms. All three were small isometric-headed phages, but small differences were noted between the phages in the structural details of the tail base plate, susceptibility to chloroform treatment, and requirements for calcium infectivity. The phage genomes were all between 29.9 and 31.9 kb in length. Phages φ31 and φ48 harbored cohesive ends, whereas the phage φ50 genome was circularly permuted, terminally redundant, and carried a putative packaging initiation site. DNA-DNA hybridization experiments conducted between the phages revealed a common region in φ48 and φ50 that may correlate with the resistance of the two phages to the Hsp-abortive infection induced by pTR2030. Phage φ50 also harbored DNA sequences that shared homology to pTR2030 in the region where R/M activities have been localized on the plasmid. Molecular characterization of the three phages localized regions within the genomes of the pTR2030-resistant phages that may be responsible for circumventing plasmid-encoded Hsp and R/M defense mechanisms in lactococci.
Full text
PDF

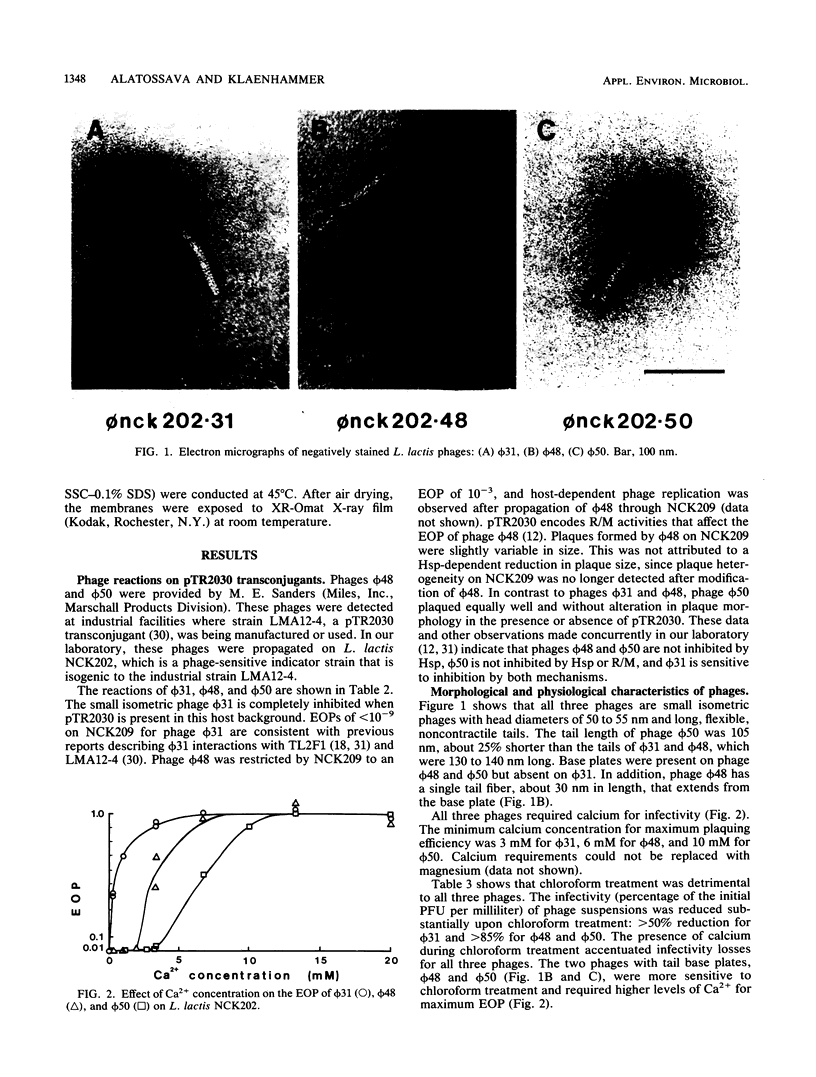
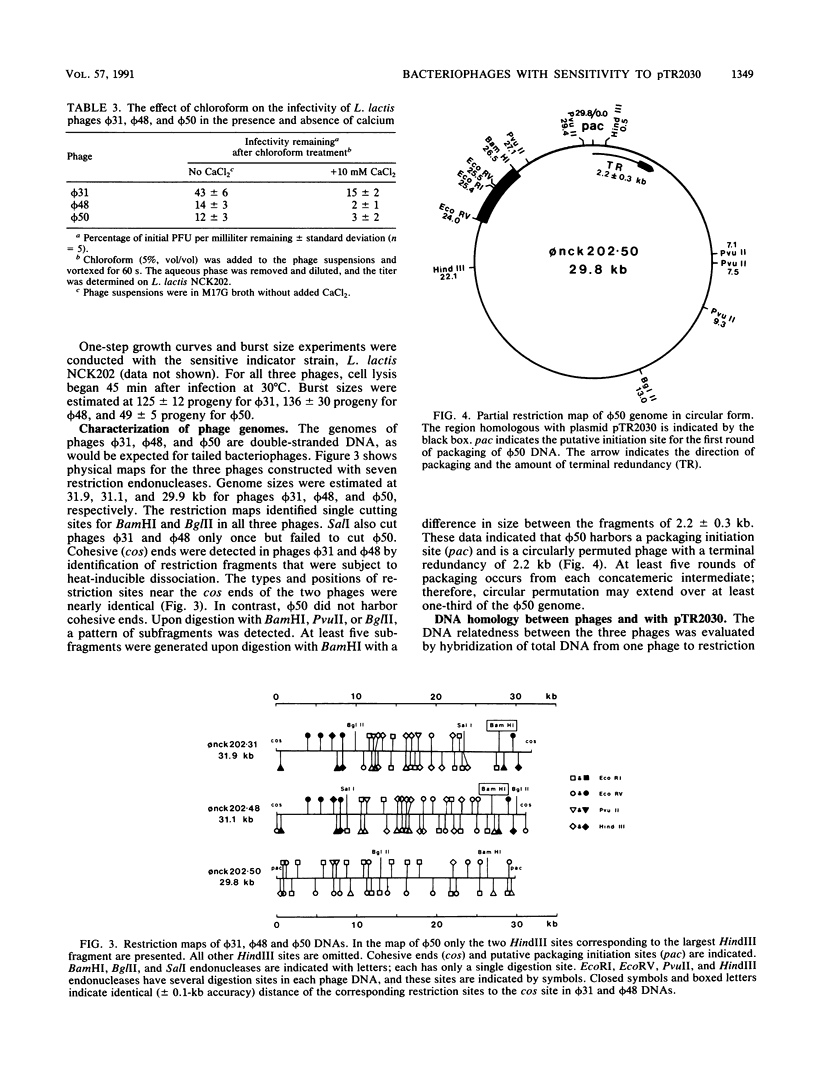
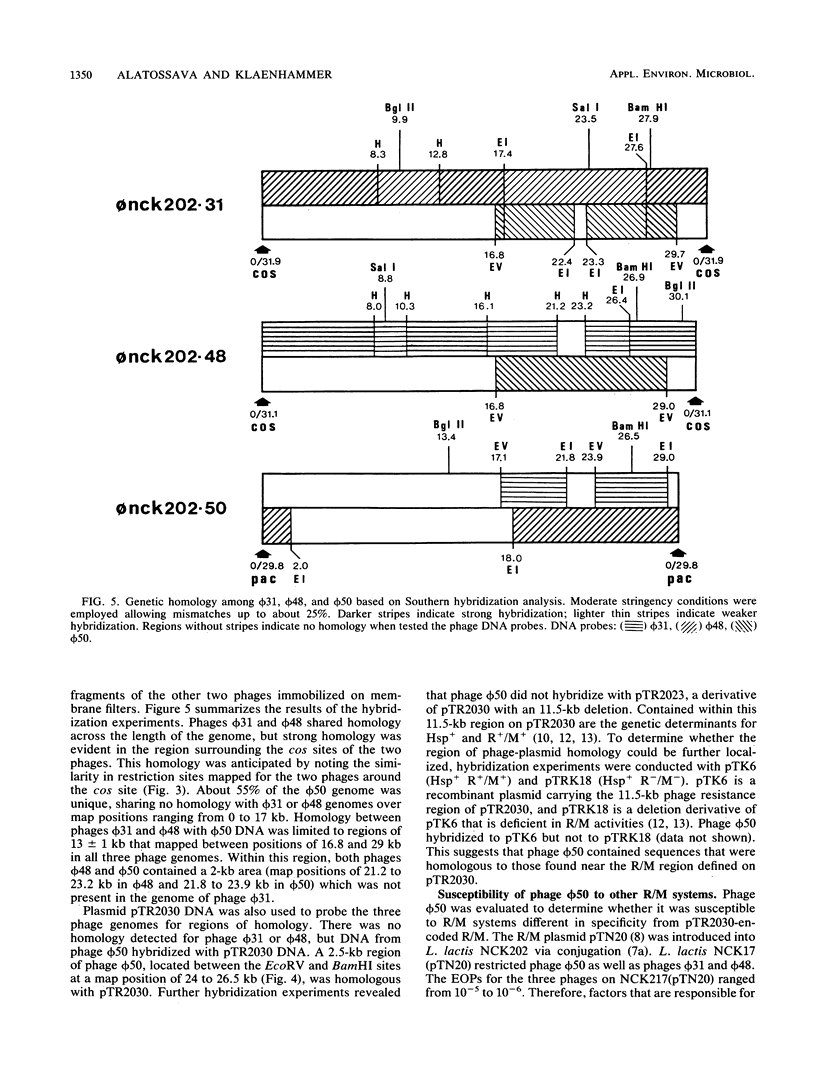



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alatossava T. Factors affecting in vitro DNA ejection of the Lactobacillus lactis bacteriophage LL-H. J Gen Virol. 1982 Mar;59(Pt 1):173–175. doi: 10.1099/0022-1317-59-1-173. [DOI] [PubMed] [Google Scholar]
- Alatossava T., Jütte H., Seiler H. Transmembrane cation movements during infection of Lactobacillus lactis by bacteriophage LL-H. J Gen Virol. 1987 Jun;68(Pt 6):1525–1532. doi: 10.1099/0022-1317-68-6-1525. [DOI] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froseth B. R., Harlander S. K., McKay L. L. Plasmid-mediated reduced phage sensitivity in Streptococcus lactis KR5. J Dairy Sci. 1988 Feb;71(2):275–284. doi: 10.3168/jds.S0022-0302(88)79555-7. [DOI] [PubMed] [Google Scholar]
- Higgins D. L., Sanozky-Dawes R. B., Klaenhammer T. R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988 Aug;170(8):3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Massey I. J., Klaenhammer T. R. Rapid method to characterize lactococcal bacteriophage genomes. Appl Environ Microbiol. 1991 Jan;57(1):283–288. doi: 10.1128/aem.57.1.283-288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J Bacteriol. 1990 Nov;172(11):6419–6426. doi: 10.1128/jb.172.11.6419-6426.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Miller L. A., Klaenhammer T. R. Nucleotide sequence and distribution of the pTR2030 resistance determinant (hsp) which aborts bacteriophage infection in lactococci. Appl Environ Microbiol. 1990 Jul;56(7):2255–2258. doi: 10.1128/aem.56.7.2255-2258.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Heap H. A., Limsowtin G. K. Resistance against Industrial Bacteriophages Conferred on Lactococci by Plasmid pAJ1106 and Related Plasmids. Appl Environ Microbiol. 1989 Jun;55(6):1537–1543. doi: 10.1128/aem.55.6.1537-1543.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Plasmid pTR2030 Inhibits Lytic Infection of r(1)t Temperate Bacteriophage but Not Induction of r(1)t Prophage in Streptococcus cremoris R1. Appl Environ Microbiol. 1987 Feb;53(2):385–389. doi: 10.1128/aem.53.2.385-389.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L. Isolation and examination of transducing bacteriophage particles from Streptococcus lactis C2. J Dairy Sci. 1976 Mar;59(3):396–404. doi: 10.3168/jds.s0022-0302(76)84219-1. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevots F., Mata M., Ritzenthaler P. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl Environ Microbiol. 1990 Jul;56(7):2180–2185. doi: 10.1128/aem.56.7.2180-2185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Restriction and modification in group N streptococci: effect of heat on development of modified lytic bacteriophage. Appl Environ Microbiol. 1980 Sep;40(3):500–506. doi: 10.1128/aem.40.3.500-506.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E. Phage resistance in lactic acid bacteria. Biochimie. 1988 Mar;70(3):411–422. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Streptococcus cremoris M12R transconjugants carrying the conjugal plasmid pTR2030 are insensitive to attack by lytic bacteriophages. Appl Environ Microbiol. 1985 Oct;50(4):851–858. doi: 10.1128/aem.50.4.851-858.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Takesue S. The requirement for calcium in infection with Lactobacillus phage. J Gen Virol. 1972 Oct;17(1):19–30. doi: 10.1099/0022-1317-17-1-19. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]



