Abstract
Major histocompatibility complex (MHC) class I and class II molecules bind to and display peptidic antigens acquired from pathogens that are recognized by lymphocytes coordinating and executing adaptive immune responses. The two classes of MHC proteins have nearly identical tertiary structures and were derived from a common ancestor that probably existed not long before the emergence of the cartilaginous fish. Class I and class II genes are genetically linked in tetrapods but are not syntenic in teleost fish, a phylogenetic taxon derived from the oldest vertebrate ancestor examined to date. Cartilaginous fish (sharks, skates, and rays) are in the oldest taxon of extant jawed vertebrates; we have carried out segregation analyses in two families of nurse sharks and one family of the banded houndshark that revealed a close linkage of class IIα and β genes both with each other and with the classical class I (class Ia) gene. These results strongly suggest that the primordial duplication giving rise to classical class I and class II occurred in cis, and the close linkage between these two classes of genes has been maintained for at least 460 million years in representatives of most vertebrate taxa.
Major histocompatibility complex (MHC) class I and class II molecules function to display foreign peptides to T cells and are thus fundamental components of the adaptive immune system (reviewed in ref. 1). Class I and class II proteins are composed of four extracellular domains, with two membrane-proximal domains belonging to the Ig superfamily, and two membrane-distal domains that form the peptide-binding region (PBR) (2, 3). The PBR/peptide complex is recognized by T cells via their T cell receptors, leading to induction of acquired immunity. Class II molecules are composed of MHC-encoded α and β chains, each having two extracellular domains (half of the PBR and one Ig domain) whereas MHC-encoded class I heavy chains are constituted of three domains encompassing the entire PBR and one Ig domain [the noncovalently associated β2-microglobulin contributes the other Ig domain of class I molecules; its gene was probably translocated out of the MHC early in vertebrate evolution (4)]. Class I and class II genes are very polymorphic in most species, and PBR amino acid residues that interact with peptides are under Darwinian positive selection (5, 6). Class I molecules are expressed ubiquitously and associate with peptides generated in the cytosol by the multicatalytic proteasome whereas class II proteins bind to lysosomally generated peptides and are expressed only by B cells, antigen presenting cells, and the thymus (reviewed in ref. 1). In addition to the classical class I, or class Ia, genes described above, there are also nonclassical, or class Ib, genes whose products are structurally similar to class Ia but are generally nonpolymorphic, may or may not be MHC-linked, and can bind to molecules besides peptides (7). There is universal agreement that class I and class II genes are derived from a common ancestor, believed by most investigators, but not all (8), to be class II-like (9–11).
Class I and class II genes have been isolated from representatives of almost all major jawed vertebrate taxa, including cartilaginous fish, bony fish, amphibians, birds, and mammals (reviewed in refs. 12 and 13). Interestingly, there has been no hint of their presence (or the existence of any other defining component of the adaptive immune system) in jawless fish or invertebrates, suggesting that the MHC arose rather abruptly in a jawed vertebrate ancestor, probably a placoderm (14). One hypothesis suggests that genome-wide duplications played a role in the emergence of the MHC and the entire adaptive immune system (15), as genes linked to class I and class II are found in four paralogous clusters in mammalian genomes (16, 17). In all tetrapod species examined to date, including several mammals, the bird Gallus (18), and the amphibian Xenopus (13, 19), class I and class II genes are closely linked. However, among older taxa, all investigated bony fish species, including the zebrafish (20), carp, salmon (21), and trout (22), the presumed classical class I and class II genes are not linked and even are found on different chromosomes. It was proposed that one of two scenarios occurred in vertebrate evolution (20, 23): (i) class I and class II genes arose on different paralogous chromosomes (likely the ones mentioned above) in a jawed vertebrate ancestor and “clustered” together in a tetrapod ancestor, or (ii) the genes were originally in the same linkage group but were rent apart in a recent teleost ancestor and now lie on different chromosomes in this single vertebrate lineage. The two scenarios can be distinguished with studies of cartilaginous fish, the oldest class of extant jawed vertebrates (24). Thus, to examine the primordial condition of the MHC regarding class Ia/class II linkage, we carried out family studies in members of two divergent orders of sharks separated by over 100 million years (24).
Materials and Methods
Animals.
Nurse sharks (Ginglymostoma cirratum, order Orectolobiformes) were captured off of Little Torch Key, Florida in October, 1996 and October, 1998. Two broods of 17 (family 1) and 39 (family 2) pups were delivered by Caesarian section and were maintained in aerated seawater. Banded houndsharks Triakis scyllium (order Carcharhiniformes) were captured off the coast of Japan, and one family typed for the class Ia gene was found to be the offspring of one father and mother (25). One to three milliliters of blood was obtained from each pup. Genomic DNA was prepared from nucleated erythrocytes as described (26).
cDNA Library Screen.
The exon encoding the α3 domain of the horned shark Heterodontus francisci class Ia cDNA Hefr-20 (GenBank accession no. AF028559) was used to select class I clones from a nurse shark spleen cDNA library under low stringency conditions. The probe was PCR amplified from cloned DNA by using the primer set 5′-TGA TAG TAA CCG ACT GTC CTG-3′ and 5′-ACC ACC ATC TTC TCC TTC AG-3′.
Southern Blots.
Five micrograms of genomic DNA was digested with various enzymes to obtain useful restriction fragment length polymorphism (RFLP). Probes encompassed either entire cDNA clones, entire coding regions, or exons encoding various domains or 3′ untranslated regions (UT) of nurse shark class IIα (27), class IIβ (28), and class Ia genes (this paper); the specific regions used for probes are noted in figure legends. Probes were generated either by liberating inserts from plasmid DNA or by PCR amplification of specific regions from plasmid clones. The procedures for Southern blotting under low and high stringency conditions with 32P-dCTP random-primed probes have been described (29).
Nurse Shark Genomic PCR.
In the first nurse shark family of 17 pups, the exons encoding the PBR of the class IIα and β genes were PCR amplified and sequenced to confirm the Southern blot analyses. Genomic DNA (200–500 ng) was amplified with the primers 5′-ACC TTT CGG TTC CGG GGT CCC-3′ and 5′-TCT CTC TG/CT CT/CT TG/CT CTC A/TT/CA/T-3′ for the α genes (accession no. M89950), and 5′-CGG GAT GTT GTG GTG TTC ACA-3′ and 5′-C TTG CAT AGA TGT GTG TTT AA-3′ for the β genes (accession no. L20274). The PBR exons (and intervening intron) of the class I genes were amplified with primers 5′-TCT CAC AGT CTC CGG TAT TT-3′ (α1 domain exon) and 5′-TC ACA GCC GCA CAT CAG C-3′ (α2 domain exon). Conditions were 94°C for 4 min, followed by 35 cycles of 94°C for 1 min, 52°C for 1 min, 72°C for 4 min, and a final extension at 72°C for 15 min. DNA fragments were cloned into the pCR2.1 TA-cloning vector (Invitrogen) and were sequenced by the Biopolymer Laboratory of the University of Maryland by using an automated DNA sequencing system (Applied Biosystems).
Results
Cloning and Genomic Analysis of the Nurse Shark Class Ia Gene.
Our previous studies reported the isolation of class Ia (Hefr-20) and class Ib (Hefr-19) genes from the horned shark (29), class Ia genes (Trsc-UAA) from the banded houndshark (25), and a predicted class Ib gene from the nurse shark (Gici-11) (29). Because we had not unearthed the expected nurse shark class Ia gene in the first set of experiments, we used the exon encoding the α3 domain of horned shark class Ia as probe to isolate additional nurse shark cDNA clones. A clone differing in sequence from Gici-11 was isolated and named Gici-UAA (Fig. 1); it was the only class Ia-like clone encountered from this library screen, and its gene was shown to have class Ia-like high expression in intestine, spleen, and kidney in Northern blotting experiments (not shown). The deduced protein sequence of Gici-UAA displays class Ia features, notably canonical evolutionarily conserved peptide-binding residues that anchor the termini of associated peptides at both ends of the class I PBR (Fig. 1; ref. 30). The class Ia genes from the three divergent shark species (Hefr-20, Trsc-UAA, Gici-UAA) are more similar to each other than they are to class Ib genes (Hefr-19 and Gici-11), especially in the C-terminal halves of the PBR α1 and α2 domains and the cytoplasmic region (Fig. 1), suggesting with these limited data that, in contrast to mammalian class I genes, class Ia and class Ib lineages are exceptionally old in the cartilaginous fish (31).
Figure 1.
Gici-UAA is a class Ia gene. Alignment of the five shark class I genes cloned to date [and HLA-A2 (46)], with Gici-UAA, Hefr-20 (GenBank accession no. AF028559), and Trsc-UAA (accession no. AF034316) postulated to be class Ia genes and Hefr-19 (accession no. AF028558) and Gici-11 (accession no. AF028557) class Ib genes. The eight PBR amino acid residues found in most vertebrate class Ia sequences that bind to peptide termini (e.g., see refs. 25, 29, and 30) are noted with an asterisk. The percent identity to the Gici-UAA sequence for the leader, α1, α2, α3, and cytoplasmic domains (following the hydrophobic transmembrane region) is noted at the right; a bracket indicates identity between the two class Ib sequences. The relatively low level of identity between Gici-UAA and Hefr-20 in the α1 domains may be attributable to an insertion in Hefr-20 near the beginning of the α-helical region of this domain that threw things a bit out of kilter.
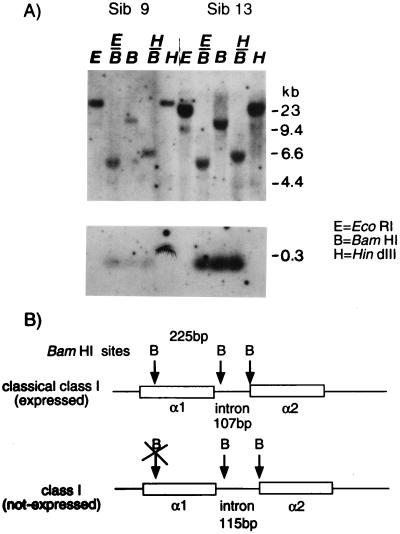
Southern blots of genomic DNA with probes encoding the Gici-UAA α3 domain (see Fig. 3) and α2 domain (not shown) suggested the presence of at least two class Ia-like genes in the nurse shark genome; indeed, in addition to Gici-UAA, we isolated another, related class I gene (78% nucleotide identity; GenBank accession no. AF220360) by PCR from genomic DNA with α1- and α2-specific oligonucleotide primers based on the Gici-UAA sequence. We could not, however, detect expression of this gene or any other gene closely related to Gici-UAA in the spleen. A probe encoding the Gici-UAA PBR α1 domain, in contrast to the α2 and α3 domain probes, yielded only one or two bands under high stringency conditions, strongly suggesting that only one gene, Gici-UAA, was detected (see Figs. 3 and 4; two bands indicate heterozygocity in α1 domain hybridizations, revealed in the segregation analyses). We further confirmed that no other class Ia-like gene except Gici-UAA was detected in our experiments: the sequence of Gici-UAA, but not the other, related class I gene, has a BamHI site in the exon encoding the α1 domain, generating a unique 225 base pair (bp) fragment (Fig. 2B). Indeed, this restriction fragment was obtained in Southern blotting from all sharks tested with the α1 domain probe (two animals, sibs 9 and 13 shown in Fig. 2A), leaving little doubt that the expressed class Ia gene Gici-UAA is the only one detected in our experiments (hybridizations with the nonclassical gene described above did not detect any of the major hybridizing bands in Fig. 2). Thus, we initiated family studies to examine whether the single expressed nurse shark class Ia gene was linked to nurse shark class II α and β genes.
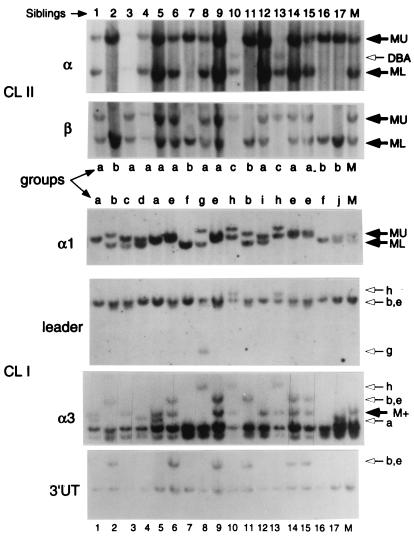
Figure 3.
Linkage of class Ia and class IIα and β genes in the first nurse shark family of 17 offspring. The sibling number is noted above and below the blots. α1 domain, α3 domain, leader, and 3′ UT class Ia probes (CL I) revealed 10 segregating groups (lowercase letters a–j) whereas probes encompassing entire cDNA clones for the class IIα and β genes (CL II) revealed three groups (a–c). MU and ML refer to maternal upper and lower bands in the class IIα, class IIβ, and class Ia α1 blots; M+ indicates the presence (or absence) of a band in the class Ia α3 hybridizations. Lowercase letters to the right of the class Ia blots indicate the positions of paternal alleles in the different groups. DBA found in siblings 10 and 13 refers to a second class IIα locus that segregates as an allele of DAA (see text and ref. 32). Restriction enzymes were HindIII (class IIα and -β and class Ia α1 domain and leader) and BamHI (class Ia α3 domain and 3′UT). Approximate molecular sizes of discriminatory bands are as follows. Class IIα: MU, 10 kb, ML, 7kb; class IIβ: MU, 6 kb, ML, 4 kb; class I α1: MU, 21 kb, ML, 17 kb; class I leader: common band, 4 kb, g, 3 kb; class I α3: M+, 12 kb; class I 3′UT b: e group, 15 kb, nonpolymorphic band, 6.5 kb.
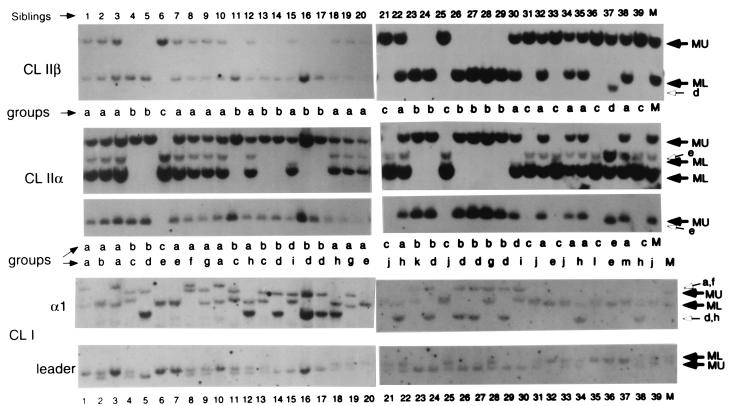
Figure 4.
Linkage of class Ia and class IIα and -β genes in the second nurse shark family of 39 offspring. The designations of diploid groupings and alleles are the same as in Fig. 3. Note that the class Ia leader hybridizations are much more discriminatory on this blot and are designated as U or L according to which band cosegregates with the U/L bands revealed with the α1 domain probe. The restriction enzymes used and the approximate sizes of the bands are the same as in Fig. 3.
Figure 2.
The class Ia gene appears to be the only nurse shark gene that hybridizes to the Gici-UAA α1 probe on Southern blots. Genomic DNA digested with BamHI results in a band of 225 bp (A) predicted to be in the expressed class Ia sequence, but not in a Gici-UAA-related class I gene isolated by PCR (B).
Family Studies.
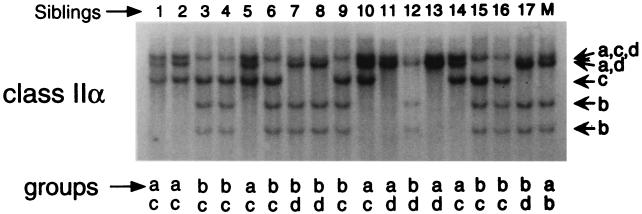
Two nurse shark families (17 and 39 sharks) were surveyed for RFLP with class IIα, IIβ, and class Ia probes (Figs. 3 and 4). It was fortunate that the mothers of both families were MHC heterozygotes with readily detectable class Ia and class II RFLP. Class Ia probes specific for the α3 domain, leader, 3′UT, or especially the α1 domain were more revealing than class II probes in delineating the MHC type or segregating “group” of each offspring (lowercase letters above or below each lane in Figs. 3 and 4). Ten class I diploid groups were detected in the first family of sharks (Fig. 3, groups a–j), showing multiple paternity of the family (at least three fathers). Several of the offspring formed identical diploid groups that matched perfectly with the class II groupings (Table 1). Furthermore, maternal haplotypes could also be tracked, matching for the three genes tested (Table 2). Because of the relatively low level of class II polymorphism (RFLP)—only three or four groups were detected in all of our experiments—we sequenced the class II α1 and β1 exons after PCR amplification of the alleles from genomic DNA. The predicted maternal class II allele, either DAA*01 or DAA*03 for α (32) or DAB*02 or DAB*01 for β, was found to match perfectly to the “upper (U)” or “lower (L)” RFLP positions for each offspring (see supplemental Figs. 6 and 7 and Table 3, published on the PNAS web site, www.pnas.org). There was only one discordant case (offspring 3), likely a recombinant between the two class II loci.
Table 1.
Class Ia diploid groups match those of class IIα and β
| Family 1 groups
|
Family 2 groups
|
||||||
|---|---|---|---|---|---|---|---|
| Frequency | I | IIα | IIβ | Frequency | I | IIα | IIβ |
| 2 | a | a | a | 3 | a | a | a |
| 2* | b | b | b | 3 | c | b | b |
| 4* | e | a | a | 8 | d | b | b |
| 2 | f | b | b | 2 | e† | c | c |
| 2 | h | c | c | 3 | e† | a | a |
| 5 | h | a | a | ||||
| 2 | i | d | a | ||||
| 5 | j | c | c | ||||
These two groups are likely to have the same paternal haplotype based on the sequences of the paternal class IIα allele and unique class Ia α3 domain exon and 3′ UT bands.
†Group e may have two fathers.
Table 2.
Segregation of maternal haplotypes reveals class Ia/II linkage
| Family 1†
|
Family 2‡
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Animal | IIα | IIβ | I-α1/α3 | Animal | IIα | IIβ | I-α1 | Animal | IIα | IIβ | I-α1 |
| Mother | L(01) | U(02) | U/+ | Mother | U | L | U | ||||
| U(03) | L(01) | L/− | L | U | L | 24 | U | L | U | ||
| 2 | U(03) | L(01) | L/− | 4 | U | L | U | 25 | L | U | L |
| 7 | U(03) | L(01) | L/− | 5 | U | L | U | 26 | U | L | U |
| 10 | L(01) | U(02) | U/+ | 6 | L | U | L | 27 | U | L | U |
| 11 | U(03) | L(01) | L/− | 11 | U | L | U | 29 | U | L | U |
| 13 | L(01) | U(02) | U/+ | 14 | U | L | U | 31 | L | U | L |
| 16 | U(03) | L(01) | L/− | 16 | U | L | U | 33 | L | U | L |
| 17 | U(03) | L(01) | L/− | 17 | U | L | U | 36 | L | U | L |
| 21 | L | U | L | 37 | L | U | L | ||||
| 39 | L | U | L | ||||||||
The offspring shown here had only a single band for class IIα and β and clearly identifiable maternal class Ia bands. L, lower band; U, upper band. Numbers in the first family refer to class IIα (DAA*01 or 03) and class IIβ (DAB*01 or 02) alleles. +, presence of band in Fig. 3; −, absence of band in Fig. 3.
† Because there are at least two genes that hybridize to the Gici-UAA α3 domain probe, the polymorphic band (+/−) detected in Fig. 3 may be class II-linked but not necessarily Gici-UAA itself.
‡ Although we are certain that the α1 domain probe hybridizes to the expressed class I gene, the leader-hybridizing fragment in Fig. 4 is only deduced to be part of Gici-UAA.
The paternal class IIα genes of offspring 10 and 13 were encoded by two DBA genes (Table 3) that segregate like alleles of DAA. In the previous report of nurse shark class IIα polymorphism, it was believed that DBA and DAA were different class IIα loci (32); rather, our studies here suggest that these are ancient genes that now segregate as alleles, but they should properly be regarded as pseudoalleles. Further studies are needed to clarify this point.
The second family of 39 sharks segregated into 13 detectable groups (at least four fathers) by the class Ia typing (Fig. 4). Three class Ia groups, d, h, and j, were frequent and segregated perfectly with three different class II α and β groupings (Table 1). Several potential recombinants were found as well, but these might also be attributable to a larger number of paternal alleles than are accounted for by the RFLP. The maternal haplotypes were again discriminating, and RFLP of class Ia, class IIα, and class IIβ were absolutely concordant (Table 2).
Finally, the nurse shark class IIα gene was used as a probe on a genomic Southern blot on the houndshark family of 17 pups (Fig. 5), shown beforehand to segregate into four groups with the Trsc-UAA class Ia probe (25). In this family, we were able to designate bands with the maternal and paternal alleles (mother, a/b; father, c/d). The four segregating groups matched the class Ia typings, demonstrating close linkage.
Figure 5.
Linkage of class IIα to class Ia genes in the banded houndshark. Sibling number is noted at the top of the blot, and the maternal and paternal alleles are designated on the right side. The segregating diploid groups are noted at the bottom of the figure and match the previous class Ia typings (25). Note that the haplotype of the top band (a, c, d) was deduced from the relative intensities of the signals in the segregants and is somewhat ambiguous; the a and d haplotypes have the same RFLP with this enzyme but not with others (not shown). The entire coding region of the nurse shark class IIα gene was used in this experiment, and EcoRV was used for digestions. Approximate molecular sizes of bands: a, c, and d, 23 kb; c, 16 kb; top b, 9 kb; bottom b, 7 kb.
Discussion
Previous studies of the cartilaginous fish MHC showed that the nurse shark class IIα (32) and houndshark class Ia (25) genes are highly polymorphic and under Darwinian positive selection similar to their mammalian homologues. Here we show that the MHC consisted of class Ia and class II genes ever since the common ancestor of cartilaginous fish and all other vertebrates existed between 460 and 540 million years ago (24). These data strongly suggest that one gene duplicated from the other in cis and the two loci subsequently remained closely linked in most vertebrate taxa.
In contrast to all other vertebrates, every teleost species examined so far carries unlinked class Ia and class II genes (23). It was suggested that this unusual feature might have been the primordial condition, with class I and class II arising on two paralogous chromosomes and being “brought together” in a tetrapod ancestor. Our work makes this proposal untenable and suggests the more likely possibility (also recognized by the teleost workers), that the situation in bony fish is a derived characteristic. Even if recent mitochondrial DNA studies suggesting that cartilaginous fish have a terminal position in the piscine phylogenetic tree are true (33), then the class II and class I genes would have had to come together rapidly in a “functional cluster” (34) twice in vertebrate evolution. Other investigators, using nuclear gene sequences, have instead confirmed the paradigmatic phylogenetic tree based on the fossil record (35); furthermore, phylogenetic analyses based on mitochondrial sequences frequently have been called into question (36), and such genes seem to mutate very slowly in the cartilaginous fish (37). Thus, it is much more likely, and simpler to imagine, that class I and class II genes were linked in the primordial MHC typified by the shark. The “nonlinkage” in teleosts may have occurred by differential silencing of MHC genes after genome-wide chromosomal duplication events proposed for a teleost-specific ancestor (38).
Another comparison to be made with teleost and all other vertebrates is the relative stability of the class I and class II genes. In mammals, class I genes are plastic, fluctuating in numbers and function in different species, whereas class II genes are more stable (reviewed in ref. 1). In teleosts, the situation is reversed, with class II being plastic and found on multiple chromosomes whereas class I appears to be more conserved (20, 39). In addition, class II gene number in the teleostean cichlid fishes differs greatly from haplotype to haplotype (40). The close association of class Ia to the transporter (TAP) and proteasome genes in teleosts [and birds (41) and likely amphibians (42)] indicates coevolution of such genes and may account for the relative stability of class I in these species whereas teleost class II, probably being linked to no other genes involved in MHC peptide presentation, is capable of rapid diversification (20, 42). In the nurse shark, both class II and class I genes seem rather stable, although studies in other species must be done; tight linkage to the other MHC genes probably maintains this stability in both classes. In addition, with the little data we have so far obtained, cartilaginous fish class Ia and class Ib lineages seem old, suggesting little expansion and contraction of class Ia-like genes as is found in mammals (31).
Is there a functional advantage to maintaining class I and class II genes together? This has been the important question since class II genes (Ir genes) were shown to be linked to class I genes (the major transplantation loci) over 30 years ago (43). In mammals, linkage disequilibrium studies would suggest this is indeed the case; perhaps, for coordination of effective immune responses, certain combinations of class Ia and class II alleles are advantageous (44). Will the sharks also have a defined “class I region,” as is certainly true in zebrafish (20) and chicken (41) and most likely in Xenopus (42)? Will there be a cluster of shark class Ib genes that is not closely linked to the functional MHC, as is found in chickens (45) and frogs (46)? Detailed studies of shark MHC, in comparison to all other vertebrates, should help define the primordial condition of this complex.
Supplementary Material
Acknowledgments
We thank Penny Conner for technical support, Mike Criscitiello for help in the marathon fishing expedition, and Pierre Pontarotti for critical reading of the manuscript. M.F.F. and Y.O. are supported by National Institutes of Health grant AI27877. K.H. and K.O. are supported by PROBRAIN and Fujita Health University; K.H. is also supported by the Japan Society for the Promotion of Science and Ministry of Education, Science, Sports, and Culture.
Abbreviations
- PBR
peptide binding region
- UT
untranslated region
- RFLP
restriction fragment length polymorphism
Footnotes
References
- 1.Margulies D H. In: Fundamental Immunology. Paul W E, editor. Philadelphia: Lippincott; 1999. pp. 263–285. [Google Scholar]
- 2.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Nature (London) 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 3.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Nature (London) 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 4.Klein J, Figueroa F. Crit Rev Immunol. 1986;6:295–386. [PubMed] [Google Scholar]
- 5.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 6.Hughes A L, Nei M. Proc Natl Acad Sci USA. 1989;86:958–962. doi: 10.1073/pnas.86.3.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hughes A L, Yeager M, Ten Elshof A E, Chorney M J. Immunol Today. 1999;20:22–26. doi: 10.1016/s0167-5699(98)01377-2. [DOI] [PubMed] [Google Scholar]
- 8.Flajnik M F, Canel C, Kramer J, Kasahara M. Immunogenetics. 1991;33:295–300. doi: 10.1007/BF00216688. [DOI] [PubMed] [Google Scholar]
- 9.Kaufman J F, Auffray C, Korman A J, Shackelford D A, Strominger J L. Cell. 1984;13:1–13. doi: 10.1016/0092-8674(84)90068-0. [DOI] [PubMed] [Google Scholar]
- 10.Klein J, O'hUigin C. Curr Opin Genet Dev. 1993;3:923–930. doi: 10.1016/0959-437x(93)90015-h. [DOI] [PubMed] [Google Scholar]
- 11.Hughes A L, Yeager M. BioEssays. 1997;19:777–786. doi: 10.1002/bies.950190907. [DOI] [PubMed] [Google Scholar]
- 12.Du Pasquier L, Flajnik M F. In: Fundamental Immunology. Paul W E, editor. Philadelphia: Lippincott; 1999. pp. 605–650. [Google Scholar]
- 13.Flajnik M F, Ohta Y, Namikawa-Yamada C, Nonaka M. Immunol Rev. 1999;167:59–67. doi: 10.1111/j.1600-065x.1999.tb01382.x. [DOI] [PubMed] [Google Scholar]
- 14.Marchalonis J J, Schluter S F. Scand J Immunol. 1990;32:13–20. doi: 10.1111/j.1365-3083.1990.tb02886.x. [DOI] [PubMed] [Google Scholar]
- 15.Kasahara M. Immunol Rev. 1998;166:159–175. doi: 10.1111/j.1600-065x.1998.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 16.Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T. Proc Natl Acad Sci USA. 1996;93:9096–9101. doi: 10.1073/pnas.93.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rached L A, McDermott M F, Pontarotti P. Immunol Rev. 1999;167:33–45. doi: 10.1111/j.1600-065x.1999.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 18.Pink J R L, Droege W, Hala K, Miggiano V C, Ziegler A. Immunogenetics. 1977;5:203–216. [Google Scholar]
- 19.Du Pasquier L, Chardonnens X, Miggiano V C. Immunogenetics. 1975;1:482–494. [Google Scholar]
- 20.Bingulac-Popovich J, Figueroa F, Sato A, Talbot W S, Johnson S L, Gates M, Postelthwaite J H, Klein J. Immunogenetics. 1997;46:129–134. doi: 10.1007/s002510050251. [DOI] [PubMed] [Google Scholar]
- 21.Stet R J M, Johnstone R, Parham P. Hereditas. 1997;127:169–170. [Google Scholar]
- 22.Hansen J D, Strassburger P, Thorgaard G H, Young W P, Du Pasquier L. J Immunol. 1999;163:774–786. [PubMed] [Google Scholar]
- 23.Klein J, Sato A. Scand J Immunol. 1998;47:417–425. doi: 10.1046/j.1365-3083.1998.00292.x. [DOI] [PubMed] [Google Scholar]
- 24.Carroll R L. Vertebrate Paleontology and Evolution. New York: Freeman; 1988. pp. 62–83. [Google Scholar]
- 25.Okamura K, Ototake M, Nakanishi T, Kurosawa Y, Hashimoto K. Immunity. 1997;7:777–790. doi: 10.1016/s1074-7613(00)80396-9. [DOI] [PubMed] [Google Scholar]
- 26.Wong W M, Au D M Y, Lam V H S, Tam J W O, Cheng L Y L. Nucleic Acids Res. 1990;18:5573–5577. doi: 10.1093/nar/18.18.5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasahara M, Vasquez M, Sato K, McKinney E C, Flajnik M F. Proc Natl Acad Sci USA. 1992;89:6688–6692. doi: 10.1073/pnas.89.15.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartl S, Weissman I L. Proc Natl Acad Sci USA. 1994;91:262–266. doi: 10.1073/pnas.91.1.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartl S, Baish M A, Flajnik M F, Ohta Y. J Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- 30.Madden D R, Gorga J C, Strominger J L, Wiley D C. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 31.Bartl S. Immunol Rev. 1998;166:317–331. doi: 10.1111/j.1600-065x.1998.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 32.Kasahara M, McKinney E C, Flajnik M F, Ishibashi T. Eur J Immunol. 1993;23:2160–2165. doi: 10.1002/eji.1830230917. [DOI] [PubMed] [Google Scholar]
- 33.Rasmussen A-S, Arnason U. Proc Natl Acad Sci USA. 1999;96:2177–2182. doi: 10.1073/pnas.96.5.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeager M, Hughes A L. Immunol Rev. 1999;167:45–58. doi: 10.1111/j.1600-065x.1999.tb01381.x. [DOI] [PubMed] [Google Scholar]
- 35.Hedges S B. In: Major Events in Early Vertebrate Evolution: Paleontology, Phylogeny, and Development. Ahlberg P E, editor. London: Taylor and Francis; 2000. , in press. [Google Scholar]
- 36.Strauss E. Science. 1999;283:1436–1438. [Google Scholar]
- 37.Martin A P, Naylor G J P, Palumbi S R. Nature (London) 1992;357:153–155. doi: 10.1038/357153a0. [DOI] [PubMed] [Google Scholar]
- 38.Amores A, Force A, Yan Y-L, Joly L, Amemiya C, Fritz A, Ho R K, Langeland J, Prince V, Wang Y-L, et al. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 39.McConnell T J, Godwin U B, Norton S F, Nairn R S, Kazianis S, Morizot D C. Genetics. 1998;149:1921–1934. doi: 10.1093/genetics/149.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malaga-Trillo E, Zaleska-Rutczynska Z, McAndrew B, Vincek V, Klein J. Genetics. 1998;149:1527–1537. doi: 10.1093/genetics/149.3.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufman J, Jacob J, Shaw I, Walker B, Milne S, Beck S, Salomonsen J. Immunol Rev. 1999;167:101–117. doi: 10.1111/j.1600-065x.1999.tb01385.x. [DOI] [PubMed] [Google Scholar]
- 42.Nonaka M, Namikawa C, Kato Y, Sasaki M, Salter-Cid L, Flajnik M F. Proc Natl Acad Sci USA. 1997;94:5789–5791. doi: 10.1073/pnas.94.11.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDevitt H O, Tyan M L. J Exp Med. 1968;128:1–11. [PMC free article] [PubMed] [Google Scholar]
- 44.Klein J. Natural History of the Major Histocompatibility Complex. New York: Wiley; 1986. pp. 619–621. [Google Scholar]
- 45.Briles W E, Goto R, Auffray C, Miller M A. Immunogenetics. 1993;37:408–414. doi: 10.1007/BF00222464. [DOI] [PubMed] [Google Scholar]
- 46.Flajnik M F, Kasahara M, Shum B P, Salter-Cid L, Taylor E, Du Pasquier L. EMBO J. 1993;12:4385–4396. doi: 10.1002/j.1460-2075.1993.tb06123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koller B, Orr H. J Immunol. 1985;134:2727–2733. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.