Abstract
The crystal structure of (4S)-limonene synthase from Mentha spic ata, a metal ion-dependent monoterpene cyclase that catalyzes the coupled isomerization and cyclization of geranyl diphosphate, is reported at 2.7-Å; resolution in two forms liganded to the substrate and intermediate analogs, 2-fluorogeranyl diphosphate and 2-fluorolinalyl diphosphate, respectively. The implications of these findings are described for domain interactions in the homodimer and for changes in diphosphate–metal ion coordination and substrate binding conformation in the course of the multistep reaction.
Keywords: crystal structure, geranyl diphosphate, linalyl diphosphate, monoterpene cyclase, monoterpene synthase
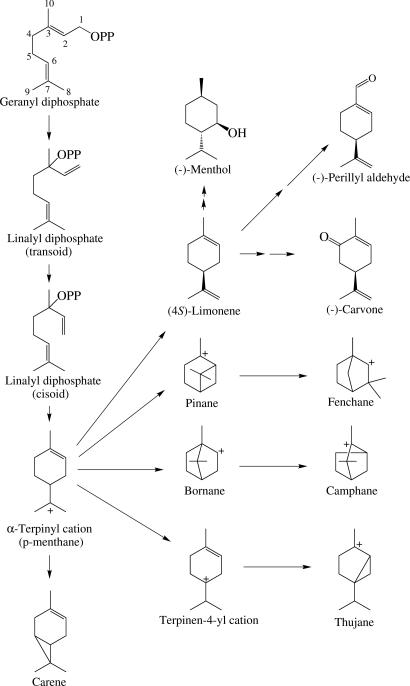
Monoterpenoid (C10), sesquiterpenoid (C15), and diterpenoid (C20) synthases catalyze the electrophilic cyclizations of their respective acyclic precursors, geranyl diphosphate (GPP), farnesyl diphosphate, and geranylgeranyl diphosphate, to the various terpene skeletons. Cyclization proceeds by ionization of the substrate followed by internal addition, often involving secondary cyclizations and rearrangements, before termination of the reaction by deprotonation or nucleophile capture (1–3). In the case of the monoterpene synthases, a preliminary ionization and isomerization of the geranyl precursor to (3R)- or (3S)-linalyl diphosphate (LPP) (depending on the initial folding of the substrate) is required to overcome the geometric impediment to cyclization imposed by the 2,3-trans-double bond of GPP (Fig. 1). After the isomerization to enzyme-bound LPP, a second ionization initiates C6–C1 cyclization to the α-terpinyl cation (a universal intermediate in monoterpene cyclizations). Bicyclic monoterpene skeletons are generated from this intermediate by internal addition to the remaining double bond (Fig. 1).
Fig. 1.
Monoterpene cyclization reactions and the formation of (−)-(4S)-limonene and derived products.
The mechanistically simplest of all monoterpene cyclizations is that leading to limonene, in that the reaction cycle is completed by methyl group deprotonation in the α-terpinyl cation leading directly to the olefin product (Fig. 1). (4R)- and (4S)-limonene are among the most common monoterpenes, occurring in Citrus oils, conifer turpentines, and the essential oils of a broad range of other plant genera (4). Notably, limonene also serves as the precursor of many oxygenated derivatives found in the essential oils of a number of agronomic species, including (−)-menthol in peppermint, (−)-carvone in spearmint, (+)-carvone in caraway and dill, and (−)-perillaldehyde in perilla (Fig. 1) (5, 6).
(4S)-LS from mint (Mentha) species was first described in 1983 (7). The enzyme from the oil glands of spearmint (Mentha spicata) (8, 9) was purified, characterized, and microsequenced (10), ultimately allowing isolation and functional expression of the corresponding cDNA from several members of the mint (Lamiaceae) family, including spearmint (11), peppermint (12), and perilla (13). LS is typical of angiosperm monoterpene cyclases in being localized to plastids (14); in requiring a divalent metal ion (Mg2+ or Mn2+) for substrate binding and catalysis; in having a size of ≈65 kDa, a pH optimum near neutrality, and a pI of ≈5; in having a Km value for the geranyl substrate in the low micromolar range with kcat < 1 sec−1; and in exhibiting sensitivity to cysteine-, methionine- and histidine-directed reagents (10, 15–17). The recombinant enzyme from spearmint expressed in Escherichia coli (and from which the plastidial targeting sequence has been deleted) is kinetically comparable with the native enzyme, and both are of high fidelity in producing 94% (−)-(4S)-limonene with ≈2% each of myrcene, (−)-α-pinene and (−)-β-pinene (17).
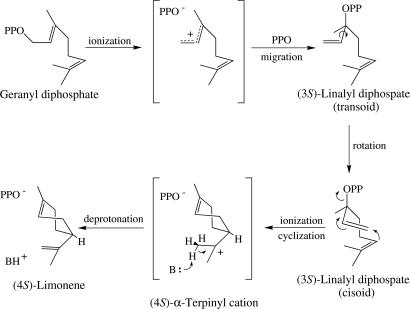
The stereochemistry and mechanism of the cyclization to (4S)-limonene have been studied in some detail and are thought to involve binding of the right-handed (in the screw sense) conformer of GPP and ionization, with syn-migration of the diphosphate (PPi) moiety to C3 to yield (3S)-LPP (Fig. 2); this isomerization appears to be the rate-limiting step of the reaction (15). After rotation about C2–C3, enzyme-bound (3S)-LPP ionizes in anti-endo conformation to promote the C6–C1, anti-SN′ cyclization to the (4S)-α-terpinyl cation, with net retention of configuration at C1 of the original geranyl substrate (due to the rotation of the vinyl group to present the methylene face from which the diphosphate moiety departed in the prior isomerization step) (18). Deprotonation from the adjacent methyl of the α-terpinyl cation via cisoid anti-elimination affords the corresponding olefin (4S)-limonene (Fig. 2) (19, 20) that debinds from the enzyme (in an as-yet-undefined fashion) to complete the reaction cycle. This transformation constitutes one of the least complicated of all terpenoid cyclization reactions (21), and it is one of very few that has ample precedent in biomimetic solvolysis studies (22–24). The widespread occurrence of limonene also implies an equally broad distribution of LSs in the plant kingdom. For these reasons, LS has become a model for this class of enzymes (25). In this paper, we describe the crystal structures of (4S)-LS from mint cocrystallized with 2-fluoroGPP (FGPP) and 2-fluoroLPP (FLPP), and we report on the implications of these structures for the coupled isomerization–cyclization reactions catalyzed by monoterpene synthases.
Fig. 2.
Traditional proposed stereochemical mechanism for the isomerization and cyclization of GPP to (4S)-limonene.
Results and Discussion
Structures of Enzyme–Ligand Complexes.
Numerous studies of monoterpene synthase reactions have established that the stereo chemical course of the reaction (leading to a particular enantiomer) is determined by the initial binding conformation of the geranyl substrate (1). To better understand how LS enforces binding of GPP in the appropriate conformation leading to (−)-(4S)-limonene, we sought an inert substrate analog that would bind to LS and mimic the initial enzyme–substrate complex with GPP bound in the presumed right-handed helical folding (Fig. 2). Several fluorinated prenyl diphosphates, in which an allylic hydrogen has been replaced with fluorine, are potent inhibitors of terpenoid synthases and the mechanistically related prenyltransferases (26–29). One of these compounds, FGPP, was chosen as a cocrystallization ligand because it is structurally very similar to GPP, and it was presumed to be essentially nonreactive because of the electron-withdrawing influence of the α-fluorine substituent in suppressing ionization of the C1-diphosphate ester. The tertiary allylic isomer, FLPP, was chosen as an additional ligand because of its close resemblance to the enzyme-bound linalyl intermediate of the reaction (see Fig. 1; note that the numbering for the LPP skeleton is the same as for GPP). Extended incubation of LS with each analog under typical assay conditions (15–17) yielded detectable levels of the same fluorinated olefin, which had a gas chromatographic retention time and mass spectrum consistent with fluorolimonene. Thus, both fluoro-analogs were slowly turned over to product, with the rate of FLPP ≈10 times that of FGPP.
Cocrystallization of LS with either FGPP or FLPP and Mn2+ ion yielded crystals of the I4 space group, with two molecules of LS in the asymmetric unit and diffracting to 2.7-Å; resolution. Crystals were obtained under identical conditions by using both FGPP and FLPP, and both crystals were of apparently identical quality. Crystals also were obtained with Mg2+ in the place of Mn2+, but these were of lesser quality and of a different crystal form. LS can use either Mg2+ or Mn2+ in the cyclization reaction with similar kinetics and identical product outcomes (15), including the ratios of minor products (D.C.H. and R.B.C., unpublished data). Therefore, Mn2+ was used throughout this study.
The structure of LS was determined by molecular replacement using a search model derived by fitting the LS amino acid sequence to the published coordinates of (+)-bornyl diphosphate synthase (BPPS) from Salvia (30). BPPS has been determined to be a homodimer, both in solution (31) and in the crystal state (30), whereas LS has been observed as a monomer in solution (10). In the present structures, LS crystallized with two molecules in the asymmetric unit, in the same relative orientation as observed for the BPPS dimer (30), thus raising the possibility that LS may also form functional dimers in solution, at least transiently, which may account for the atypical reaction kinetics observed for this enzyme at higher substrate (>30 μM) concentrations (17).
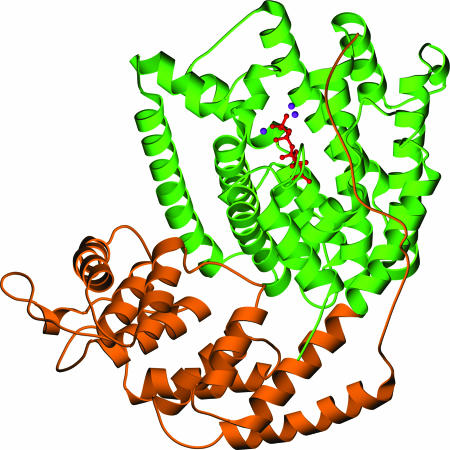
The fold of LS is similar to that of BPPS (30) and 5-epi-aristolochene synthase from tobacco (32) in having two helical domains: a C-terminal domain containing the active site and an N-terminal domain of uncertain function (Fig. 3). Also consistent with the previous structures is the N-terminal strand, which precedes the N-terminal domain but which folds back across the C-terminal domain to form part of the active site “cap” that presumably shields reactive carbocation intermediates from solvent. The presence of the apparently nonfunctional N-terminal domain in LS and similar terpenoid synthases (33) may be a consequence of their evolutionary history. A study of intron conservation patterns in terpenoid synthase genes has indicated that modern terpenoid synthases evolved from an ancestral enzyme in which both domains were catalytically functional (34). A modern example of such a dual-function enzyme is abietadiene synthase, in which both the N-terminal and C-terminal domains catalyze distinct steps in the cyclization to the diterpene olefin (35, 36). In enzymes such as LS, which appear to use only the C-terminal domain, the N-terminal domain may thus be a relic; however, there is a significant level of sequence conservation in this region of the known terpenoid synthases (34), beyond what might be expected for an evolutionary relic, suggesting some yet undiscovered role.
Fig. 3.
Ribbon diagram of LS cocrystallized with FLPP. The N-terminal domain is orange, the C-terminal domain is green, FLPP is red, and Mn2+ ions are purple.
The four LS chains (two from each structure) are similar in overall conformation but show considerable localized structural differences, suggesting general flexibility that is reflected in an average pairwise rms deviation of 0.68 Å; for all Cα atoms. Both the BPPS (30) and 5-epi-aristolochene synthase (32) structures have shown that these enzymes change from an open to a closed conformation of the active site upon ligand binding. The structures presented here, although both in the closed, ligand-bound conformation, still display conformational flexibility. A flexible structure, particularly in the active site, would be expected for an enzyme that binds two structurally distinct prenyl diphosphates (GPP and the isomerized intermediate LPP) during the course of the reaction. This apparent flexibility may relate to the role of the tandem arginine residues (R58 and R59) located in the N-terminal strand. This motif is found in many terpenoid synthases of plant origin and is thought to mark the approximate cleavage site of the plastid targeting sequence (17, 37). In the truncated form of LS used here, the tandem arginine residues are located immediately after the added N-terminal methionine. A previous truncation study with LS (17) has implicated the tandem arginine residues in the initial isomerization of GPP to LPP, owing to the inability of LS truncated beyond R58 to isomerize and cyclize GPP while still functioning with LPP as substrate for the cyclization step. In the present structures, neither arginine residue makes contact with reactants, suggesting that their role in the conversion of GPP to LPP is indirect. Nevertheless, it is suggested from the data on the present structures that the tandem arginine residues may play a structural role; R58 participates in an electrostatic interaction with the side chain of conserved E363, and R59 appears to form a bifurcated hydrogen bond to the backbone carbonyl of residue 357 and to the Y435 side chain. The combination of these weak interactions seems to anchor the N-terminal strand to the outside of the active site and thus stabilize the required closed conformation. These interactions remain in place in both structures despite the apparent flexibility of the surrounding regions. Anchoring of the termini, either through disulfide bonds (38) or electrostatic interaction (39), has been suggested as an element of protein thermostability, and the role of the tandem arginine residues may thus be to help maintain a stable, closed active site while still allowing the flexibility necessary for binding two significantly different prenyl diphosphates and variably shaped intermediates during the course of the reaction. A possible explanation for the results of earlier truncation studies is that GPP binding is more dependent on the stabilizing interactions of the N-terminal arginine pair than is LPP binding, and the former interaction is thus precluded in the absence of these arginine residues. Further support for an indirect role of the tandem arginine residues is derived from the observation that terpenoid synthases lacking this motif can still catalyze the analogous initial reaction steps (40). Therefore, these enzymes must use other means to achieve the functionality of the tandem arginine residues of LS and similar enzymes.
Observable electron density is present in the active site regions for both the FGPP- and FLPP-cocrystallized structures. The positions of the fluorinated analogs and Mn2+ ions can be clearly established, as can the positions of the amino acid side chains involved in ligand binding. Interpretation of the data for LS cocrystallized with FLPP was straightforward, and the density is consistent with bound (3S)-FLPP in the right-handed helical (anti-endo) conformation predicted for the LPP intermediate by the stereochemical model of the reaction (Fig. 2) (1, 25). The FLPP used here was racemic, and the electron density also can accommodate the (3R)-enantiomer in the left-handed helical conformation. Indeed, the density was better accounted for when both enantiomers of FLPP were simultaneously modeled into the active site than when either enantiomer was modeled alone, suggesting that the observed density represents a mixture of both enantiomers bound in anti-endo form. LS has been shown to use preferentially the (3S)-enantiomer of LPP, but this enzyme also can use (3R)-LPP at a much reduced rate to produce (+)-(4R)-limonene (15). Similar preferential, but not exclusive, utilization of one LPP enantiomer has been observed for other monoterpene synthases (41–43), suggesting that the lack of strong enantioselectivity for this normally enzyme-bound intermediate is a general property of the enzyme type.
Interpreting the structural data for LS cocrystallized with FGPP was far more difficult because, surprisingly, a model of FGPP could not be made to fit the active site electron density without significant bond distortion or without crowding the diphosphate into the metal ions in an unnatural coordination arrangement. Only when the FGPP model was replaced with a model of FLPP, which is shorter in overall length, could the ligand be made to fit the density while maintaining realistic bonding and coordination parameters. This unexpected result suggested that the FGPP used for crystallization was isomerized by the enzyme to bound FLPP, despite the rate suppression of this step imparted by the 2-fluoro substituent. The conformation of this enzymatically generated FLPP, however, is different from that of FLPP bound from solution in that the C4–C5 bond is rotated ≈150° from the position anticipated for helical (endo) folding, thus resulting in an extended conformation in which C6 is directed away from C1 (Fig. 4a). A model of (S)-FLPP in this extended conformation is in good agreement with the observed density except for the C1–C2 vinyl group with fluorine at C2, which is not represented by the density. The C2–C3 single bond of the linalyl isomer allows free rotation of the vinyl group, and there do not appear to be any steric restrictions to this rotation in the crystal structure. The absence of density for these atoms may thus be due to thermal rotation around C2–C3.
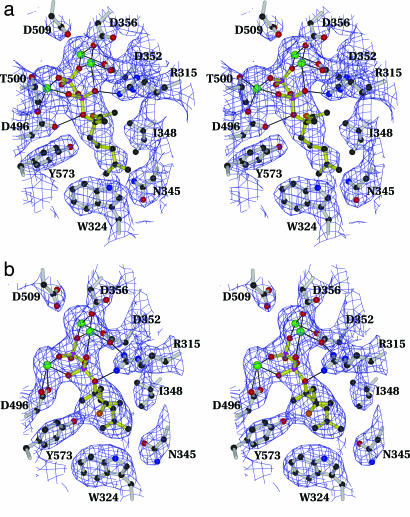
Fig. 4.
Stereodiagrams depicting bound ligand in chain A of LS cocrystallized with FGPP that has been enzymatically converted to bound FLPP (a) and chain A of LS cocrystallized with FLPP (b). Both models are superimposed on the respective 2Fo − Fc maps contoured at 1σ. Fluorine atoms are orange, Mn2+ ions are green, and all other atoms are standard colors. Inferred hydrogen bonds are indicated by black lines. Note that FLPP bound from solution is in the helical anti-endo conformation necessary for cyclization. Slightly altered ligand conformations were observed in the corresponding B chains.
Slight differences in the conformation of the bound ligand in each of the four active sites (two from each structure) became apparent when all of the chains were superimposed. In both active sites of LS cocrystallized with FGPP, the enzymatically generated FLPP is in the extended conformation, but the conformation in chain B is shifted slightly toward that of the helical (anti-endo) folding observed for LS cocrystallized with FLPP. There also are differences in ligand conformation between the two active sites of LS cocrystallized with FLPP; the ligand in chain A exhibits a nearly ideal anti-endo helical folding (Fig. 4b), but in chain B the helical conformation is somewhat distorted and slightly rotated, although it still appears to be in a conformation that is competent for cyclization. These differences in ligand position also are reflected in the hydrogen bonding between LS and the bound ligand in both structures. In both chains of LS cocrystallized with FLPP, the phosphoester oxygen of the ligand forms a hydrogen bond to invariant R315, as was observed for BPPS (Fig. 4b) (30). A similar hydrogen bonding pattern is observed for chain B of LS cocrystallized with FGPP; however, in chain A of this structure, the diphosphate has shifted to a position in which the phosphoester oxygen forms a hydrogen bond with D496 on the opposite side of the active site (Fig. 4a). Differences in the usually invariant position and orientation of the diphosphate moiety have also been observed in the D100E mutant of tricodiene synthase, a sesquiterpene synthase of microbial origin (44), indicating at least some variability in diphosphate binding. The present results suggest that changes in diphosphate positioning may occur at different stages of the multistep reaction and thus represent an additional example of the active-site plasticity required for binding the various reaction intermediates.
Mechanistic Implications.
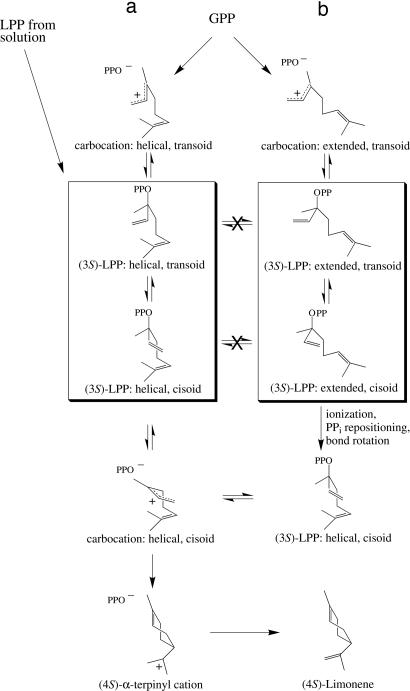
The observation of the two different, origin-dependent conformations of FLPP suggests that the conformation of LPP generated enzymatically from GPP in the normal reaction also may differ from that of LPP bound from solution in reactions in which LPP is used as an alternate substrate. This difference may explain why pinene synthase from Salvia, for example, produces a somewhat altered product mixture with LPP as substrate compared with that generated from GPP as substrate (21). The possibility remains that the presence of the fluorine atom at C2 may cause FGPP to bind in an unnatural conformation; however, a survey of related fluorinated compounds (45) suggests that the single fluorine substitution at the C2 site not involved in hydrogen bonding would cause only minor steric and geometric perturbations in the geranyl analog. Unfortunately, because FGPP was converted to FLPP during the crystallization process, the initial binding conformation of GPP remains unknown, but it almost certainly resembles the extended conformation of the enzymatically generated FLPP more than the conformation of FLPP bound from solution. However, the enzymatically generated (3S)-FLPP (extended form) does not appear to represent an intermediate that is competent to cyclize because ionization in this form would yield a carbocation in which electrophilic C1 is too distant from the C6–C7 double bond (because of the C4–C5 rotation angle) to permit ring closure. Two bond rotations must occur before such an intermediate can cyclize (C6 to C1) to the (4S)-α-terpinyl cation. One bond rotation is about C2–C3 to the cisoid conformer, which has long been recognized as a requirement for cyclization (18), and the other rotation is about C4–C5 to bring C6 close enough to the relocated C1 for bond closure to the cyclohexanoid system. Because these two distinct conformations of FLPP have been captured in the crystal structures, it does not appear that they are rotationally interchangeable without an intervening ionization and diphosphate repositioning. This process is seemingly hindered in the present case because of steric and/or electronic constraints imposed by the C2 fluorine substituent of the substrate analog. If the extended form is an accurate portrayal of initial substrate binding, then the second ionization and repositioning to achieve the cyclization-competent conformation would appear to be the slow step of the isomerization component of the coupled reaction sequence, rather than the initial ionization of GPP (in helical form) with syn-migration of the PPi to LPP (in helical form) as previously thought (1, 18, 25).
Fig. 5 depicts the traditional proposal for the reaction cycle involving helical conformations throughout (Fig. 5a) and that involving initial binding of the extended conformation of GPP with PPi migration in this form (Fig. 5b). Achievement of the cyclization-competent form from the intermediate in Fig. 5b requires either a protein conformational change not possible in the crystal state or an additional ionization and repositioning of the PPi involving movement of the C3–O phosphoester bond and rotation about C2–C3 and C4–C5 to the anti-endo conformation as a prelude to the final ionization-cyclization step. This proposal implies that the helical, cyclization-competent conformation of LPP, predicted from biochemical studies (1, 18, 25), is achieved directly only when LPP is bound from solution when it can adopt this thermodynamically stable form. This conformational difference in binding, coupled with the inherently faster solvolytic rate of the tertiary allylic system (22–24), readily explains the faster rate of enzymatic cyclization of LPP than GPP by LS (15) and other monoterpene synthases (1, 18). It has been suggested (44) that terpenoid synthase reactions are under kinetic rather than thermodynamic control, which is consistent with the present implication that achieving the cyclization-competent intermediate from GPP is a significant component of overall reaction rate. Further structural studies with additional substrate and intermediate analogs and with site specific mutants of LS may clarify the role of the extended intermediate in monoterpene cyclase catalysis.
Fig. 5.
LS reaction. (a) The traditional rendition of the coupled isomerization–cyclization reaction from the helical folding of GPP. (b) The (4S)-LS reaction sequence with additional steps involving conformational changes from the extended form of GPP. Ligand conformations represented by the two crystal structures are boxed.
Materials and Methods
Materials and General Procedures.
2-FGPP (26) was prepared from 6-methyl-5-hepten-2-one by using procedures (46) similar to those for the synthesis of related fluorinated prenyl diphosphates (47). (±)-2-Fluorolinalool obtained by solvolysis of 2-fluorogeranyl methanesulfonate (48) was converted to 2-FLPP (27) by pyrophosphorylation according to modified methods (49) based on the Danilov–Cramer procedure (50). 2-FLPP was separated from mono- and polyphosphates on a Dowex column (51). The two fluoroanalogs were characterized by using their 3H, 19F, and 31P NMR spectra.
The cDNA for spearmint LS was cloned into the pSBET (52) vector, expressed in E. coli, and purified as previously described (17). This version of the enzyme was truncated at R58, the approximate cleavage site for the plastid-targeting sequence (17, 37). Crystallization trials were performed by using vapor diffusion from hanging drops at 293 K. Substrate analogs were mixed with the enzyme before the crystallization setup. A typical crystallization involved 5 μl of protein solution (12 mg/ml protein/4 mM substrate analog) mixed with 5 μl of precipitant solution [300 mM BisTris (pH 6.8)/7.0% polyethylene glycol 8000/25 mM MnCl2] and suspended over a reservoir containing 1 ml of precipitant solution. Crystals with dimensions of 0.5 × 0.7 × 0.8 mm appeared in 4–10 days from a heavy background precipitation. These crystals belong to the tetragonal space group, I4, with unit cell dimensions of a = 200.48, b = 200.48, and c = 123.41 Å; (for the FGPP complex) and a = 198.66, b = 198.66, c = 122.67 Å; (for the FLPP complex), with two molecules in the asymmetric unit (Table 1).
Table 1.
Crystallographic data for LS structures
| FGPP complex | FLPP complex | |
|---|---|---|
| Data | ||
| Beamline | ALS 8.2.1 | ALS 8.2.1 |
| Wavelength, Å | 1.07812 | 1.07812 |
| Resolution, Å | 50–2.7 | 50–2.7 |
| Space group | I4 | I4 |
| Cell dimenstions, Å | ||
| a | 200.48 | 198.66 |
| b | 200.48 | 198.66 |
| c | 123.41 | 122.67 |
| Asymmetric unit | 2 molecules | 2 molecules |
| Total observations | 292,212 | 289,179 |
| Unique reflections | 28,338 | 26,748 |
| Completeness, % | 99.5 | 99.7 |
| Rsym* | 0.062 (0.164) | 0.059 (0.159) |
| Refinement | ||
| Resolution, Å | 10–2.7 | 10–2.7 |
| No. of reflections | 59,212 | 53,233 |
| Rcryst† | 0.208 | 0.206 |
| Rfree‡ | 0.241 | 0.234 |
| rmsd bonds, Å | 0.018 | 0.019 |
| rmsd angles, ° | 3.2 | 3.4 |
| No. of atoms | ||
| Protein, ligand, ion§, and buffer¶ | 9,089 | 9,054 |
| Water | 92 | 111 |
*Values in parentheses refer to the highest shell. Rsym = Σ∣Ih − 〈Ih〉∣/ΣIh, where 〈Ih〉 is the average intensity over symmetry equivalent reflections.
†Rcryst = Σ∣Fobs − Fcalc∣/ΣFobs, where summation is over the data used for refinement.
‡Rfree was calculated as for Rcryst by using 5% of the data that was excluded from refinement.
§Manganese ions.
¶Bis-Tris buffer.
Data Collection and Structure Determination.
X-ray diffraction data were collected under a liquid nitrogen temperature stream at 100 K on beamline 8.2.1 at the Berkeley Advanced Light Source. Before data collection, crystals were flash-frozen in liquid nitrogen after soaking in cryoprotectant (25% glycerol in reservoir solution) for 1 min. Data were processed with the HKL software suite (53). Initial phases were determined by molecular replacement with the software package AMoRe (54), and a search model was made by fitting the R58-truncated LS amino acid sequence to the coordinates of (+)-BPPS complexed with product (Protein Data Bank ID code 1N24) (30) by using the Swiss-model server (www.expasy.ch). The rigid body refinement of the initial position was performed by using 15- to 3.5-Å; data and produced an R value of 0.37. After making several cycles of positional and temperature factor refinements with the program X-PLOR (55) and creating a series of simulated annealing omit maps, most residues were fitted against the electron density. To improve the model, iterative rounds of adjustments were made using the program O (56), and refinements were performed using the program X-PLOR (55). Especially, a series of omit maps were made to validate the position and conformation of substrate analogs and residues near the active site. Data and refinement statistics are summarized in Table 1. Fitting of structures for comparison was performed with the McLachlan algorithm (57) as implemented in the program ProFit (www.bioinf.org.uk/software/profit). The final structure coordinates were deposited in the Protein Data Bank. Structure figures were prepared by using the programs Molscript (58), Bobscript (59, 60), and Raster3d (61).
Acknowledgments
We thank C. Ralston (Berkeley Advanced Light Source, beamline 8.2.1). This work was supported by National Institutes of Health Grants GM31354 (to R.C.) and GM13956 (to R.M.C.) and by U.S. Department of Agriculture Grant 2006-03339 (to C.K.)
Abbreviations
- LS
limonene synthase
- BPPS
bornyl diphosphate synthase
- GPP
geranyl diphosphate
- FGPP
2-fluoroGPP
- LPP
linalyl diphosphate
- FLPP
2-fluoroLPP.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors reported have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2ONG and 2ONH).
References
- 1.Wise ML, Croteau R. In: Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Steroids. Cane DE, editor. Vol 2. Oxford, UK: Elsevier Science; 1999. pp. 97–153. [Google Scholar]
- 2.Cane DE. In: Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Steroids. Cane DE, editor. Vol 2. Oxford, UK: Elsevier Science; 1999. pp. 155–200. [Google Scholar]
- 3.MacMillan J, Beale MH. In: Comprehensive Natural Products Chemistry: Isoprenoids Including Carotenoids and Steroids. Cane DE, editor. Vol 2. Oxford, UK: Elsevier Science; 1999. pp. 217–243. [Google Scholar]
- 4.Guenther B. The Essential Oils. Vol 2. Huntington, NY: RE Kreiger; 1975. pp. 22–27. [Google Scholar]
- 5.Karp F, Mihaliak CA, Harris JL, Croteau R. Arch Biochem Biophys. 1990;276:219–226. doi: 10.1016/0003-9861(90)90029-x. [DOI] [PubMed] [Google Scholar]
- 6.Bouwmeester HJ, Gershenzon J, Konings MCJM, Croteau R. Plant Physiol. 1998;117:901–912. doi: 10.1104/pp.117.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kjonaas R, Croteau R. Arch Biochem Biophys. 1983;220:79–89. doi: 10.1016/0003-9861(83)90389-2. [DOI] [PubMed] [Google Scholar]
- 8.Gershenzon J, Maffei M, Croteau R. Plant Physiol. 1989;89:1351–1357. doi: 10.1104/pp.89.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCaskill D, Gershenzon J, Croteau R. Planta. 1992;187:445–454. doi: 10.1007/BF00199962. [DOI] [PubMed] [Google Scholar]
- 10.Alonso WR, Rajaonarivony JIM, Gershenzon J, Croteau R. J Biol Chem. 1992;267:7582–7587. [PubMed] [Google Scholar]
- 11.Colby SM, Alonso WR, Katahira EJ, McGarvey DJ, Croteau R. J Biol Chem. 1993;268:23016–23024. [PubMed] [Google Scholar]
- 12.Lange BM, Wildung MR, Stauber EJ, Sanchez C, Pouchnik D, Croteau R. Proc Natl Acad Sci USA. 2000;97:2934–2939. doi: 10.1073/pnas.97.6.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuba A, Yazaki K, Tabata M, Honda G, Croteau R. Arch Biochem Biophys. 1996;332:280–287. doi: 10.1006/abbi.1996.0343. [DOI] [PubMed] [Google Scholar]
- 14.Turner G, Gershenzon J, Nielson EE, Froehlich JE, Croteau R. Plant Physiol. 1999;120:879–886. doi: 10.1104/pp.120.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajaonarivony JIM, Gershenzon J, Croteau R. Arch Biochem Biophys. 1992;296:49–57. doi: 10.1016/0003-9861(92)90543-6. [DOI] [PubMed] [Google Scholar]
- 16.Rajaonarivony JIM, Gershenzon J, Miyazaki J, Croteau R. Arch Biochem Biophys. 1992;299:77–82. doi: 10.1016/0003-9861(92)90246-s. [DOI] [PubMed] [Google Scholar]
- 17.Williams DC, McGarvey DJ, Katahira EJ, Croteau R. Biochemistry. 1998;37:12213–12220. doi: 10.1021/bi980854k. [DOI] [PubMed] [Google Scholar]
- 18.Croteau R. Chem Rev. 1987;87:929–954. [Google Scholar]
- 19.Pyun H-J, Coates RM, Wagschal KC, McGeady P, Croteau R. J Org Chem. 1993;58:3998–4009. [Google Scholar]
- 20.Coates RM, Elmore CS, Croteau RB, Williams DC, Morimoto H, Williams PG. Chem Commun. 1997:2079–2080. [Google Scholar]
- 21.Croteau R, Satterwhite DM. J Biol Chem. 1989;264:15309–15315. [PubMed] [Google Scholar]
- 22.Cori O, Chayet L, Perez LM, Bunton CA, Cori M. J Org Chem. 1986;51:1310–1317. [Google Scholar]
- 23.Cramer F, Rittersdorf W. Tetrahedron. 1967;23:3015–3022. [Google Scholar]
- 24.Haley RC, Miller JA, Wood HCS. J Chem Soc C. 1969:264–268. [Google Scholar]
- 25.Davis EM, Croteau R. Top Curr Chem. 2000;209:53–95. [Google Scholar]
- 26.Poulter CD, Argyle JC, Mash EA. J Biol Chem. 1978;253:7227–7233. [PubMed] [Google Scholar]
- 27.Croteau R. Arch Biochem Biophys. 1986;251:777–782. doi: 10.1016/0003-9861(86)90390-5. [DOI] [PubMed] [Google Scholar]
- 28.Poulter CD, Satterwhite DM. Biochemistry. 1977;16:5470–5478. doi: 10.1021/bi00644a012. [DOI] [PubMed] [Google Scholar]
- 29.Poulter CD, Wiggins PL, Le AT. J Am Chem Soc. 1981;103:3926–3927. [Google Scholar]
- 30.Whittington DA, Wise ML, Urbansky M, Coates RM, Croteau RB, Christianson DW. Proc Natl Acad Sci USA. 2002;99:15375–15380. doi: 10.1073/pnas.232591099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise ML, Savage TJ, Katahira E, Croteau R. J Biol Chem. 1998;273:14891–14899. doi: 10.1074/jbc.273.24.14891. [DOI] [PubMed] [Google Scholar]
- 32.Starks CM, Back K, Chappell J, Noel JP. Science. 1997;277:1815–1820. doi: 10.1126/science.277.5333.1815. [DOI] [PubMed] [Google Scholar]
- 33.Christianson DW. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 34.Trapp SC, Croteau RB. Genetics. 2001;158:811–832. doi: 10.1093/genetics/158.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters RJ, Flory JE, Jetter R, Ravn MM, Lee H-J, Coates RM, Croteau RB. Biochemistry. 2000;39:15592–15602. doi: 10.1021/bi001997l. [DOI] [PubMed] [Google Scholar]
- 36.Peters RJ, Ravn MM, Coates RM, Croteau RB. J Am Chem Soc. 2001;123:8974–8978. doi: 10.1021/ja010670k. [DOI] [PubMed] [Google Scholar]
- 37.Bohlmann J, Meyer-Gauen G, Croteau R. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karlström M, Stokke R, Steen IH, Birkeland N-K, Ladenstein R. J Mol Biol. 2005;345:559–577. doi: 10.1016/j.jmb.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 39.Karlström M, Steen IH, Madern D, Fedöy A-E, Birkeland N-K, Ladenstein R. FEBS J. 2006;273:2851–2868. doi: 10.1111/j.1742-4658.2006.05298.x. [DOI] [PubMed] [Google Scholar]
- 40.Aubourg S, Lecharny A, Bohlmann J. Mol Genet Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 41.Satterwhite DM, Wheeler CJ, Croteau R. J Biol Chem. 1985;260:13901–13908. [PubMed] [Google Scholar]
- 42.Croteau R, Satterwhite DM, Cane DE, Chang CC. J Biol Chem. 1986;261:13438–13445. [PubMed] [Google Scholar]
- 43.Croteau R, Satterwhite DM, Cane DE, Chang CC. J Biol Chem. 1988;263:10063–10071. [PubMed] [Google Scholar]
- 44.Vedula LS, Rynkiewicz MJ, Pyun H-J, Coates RM, Cane DE, Christianson DW. Biochemistry. 2005;44:6153–6163. doi: 10.1021/bi050059o. [DOI] [PubMed] [Google Scholar]
- 45.O'Hagan D, Rzepa HS. Chem Commun. 1997:645–652. [Google Scholar]
- 46.Zhao Y. Urbana: University of Illinois at Urbana-Champaign; 2005. PhD thesis. [Google Scholar]
- 47.Jin Y, Williams DC, Croteau R, Coates RM. J Am Chem Soc. 2005;127:7834–7842. doi: 10.1021/ja050592r. [DOI] [PubMed] [Google Scholar]
- 48.Poulter CD, King C-H. J Am Chem Soc. 1982;104:1422–1424. [Google Scholar]
- 49.Assink BK. Urbana: University of Illinois at Urbana-Champaign; 1999. MS thesis. [Google Scholar]
- 50.Danilov LL, Mal'tsev SD, Sibarv VN. Sov J Bioorg Chem. 1988;14:712–714. [Google Scholar]
- 51.Coates RM, Kang HY. J Org Chem. 1987;52:2065–2074. [Google Scholar]
- 52.Shenk PM, Baumann S, Mattes R, Steinbiss H-H. BioTechniques. 1995;19:196–200. [PubMed] [Google Scholar]
- 53.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic; 1996. [DOI] [PubMed] [Google Scholar]
- 54.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 55.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunsteve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 56.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 57.McLachlan AD. Acta Crystallogr A. 1982;38:871–873. [Google Scholar]
- 58.Kraulis PJ. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 59.Esnouf RM. Acta Crystallogr D. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- 60.Esnouf RM. J Mol Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 61.Merritt EA, Bacon DJ. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]