Abstract
DNA nanostructures based on programmable DNA molecular recognition have been developed, but the nanoelectronics of using DNA is still challenging. A more rapid charge-transfer (CT) process through the DNA nanoassembly is required for further development of programmable DNA nanoelectronics. In this article, we present direct absorption measurements of the long-range CT over a 140-Å DNA assembly based on a GC repetitive sequence constructed by simply mixing DNA building blocks. We show that a CT through DNA nanoscale assembly is possible and programmable with the designed DNA sequence.
Keywords: transient absorption measurement, nanostructure hole transfer, DNA oxidation, nanotechnology
DNA has been used extensively to form nanoscale structures that may be used as nanotechnology devices in the future (1–3). Although many DNA nanostructures have been constructed (4, 5), the realization of DNA-based molecular nanoelectronics is still challenging because its physical and chemical properties continue to remain unclear (6).
There are many reports on the construction of desired structures using DNA. Structurally controlled DNA motifs, called DNA tiles, have been used as building blocks for creating DNA nanostructures. Such DNA tiles are linked together with a branched junction, called sticky-end DNAs, which are used for self-assembling nanostructures, such as two- or three-dimensional DNA nanostructures (1, 4, 7). These self-assembling nanostructures are produced simply by mixing the short single strands of the DNA. In addition, a DNA automated synthetic method has made it possible to synthesize site-specific functionalized DNAs, such as a photosensitizer-modified DNA (7). By using these functionalized DNAs as building blocks, the creation of functionalized DNA nanostructures could be accomplished, leading to DNA frontier nanotechnologies and nanoelectronics (8).
Charge transfer (CT) in a duplex B-DNA has been studied intensively (9–14). Particularly, the CT mechanism has been investigated by various experimental methods and theoretical calculations (15). Giese and coworkers (16, 17) demonstrated that the CT between guanine (G) sites occurs via a multihopping mechanism. Furthermore, they revealed that the CT between Gs separated by (A:T)n (n ≥ 4) sequences can take place because the adenines (As) also act as the charge carriers (18). An alternative mechanism in which the delocalized charge is transported by polarons has been proposed by Schuster and coworkers (19). More recently, Barton and O'Neill (20, 21) proved that CT through a DNA is described as a conformational gated hopping among stacked domains. Although somewhat controversial, DNA sequences as well as conformational dynamics and local flexibility seem to contribute to the CT through DNA. Previously, we reported the direct observation of CT between Gs through a DNA in which the long-range CT between Gs across A/T base pairs occurs in a slow time scale, microsecond-to-millisecond range (22). A more rapid CT is required for further development of programmable DNA nanoelectronics.
Despite these advances in understanding the CT through a DNA, the CT through DNA nanoscaled assembly has not been clarified. A further understanding not only of the CT through DNA nanoscaled assembly but also of the kinetic mechanisms, which provide information about the rapid CT sequences, are of fundamental importance to create functionalized nanometer-scaled DNA wires and arrays. Recently, Xu et al. reported the conductance for GC repetitive [(GC)n] sequences by using direct electrical measurements, and they indicated that the conduction mechanism of the (GC)n sequences is mostly through the highest occupied molecular orbital (HOMO) (23, 24). Inspired by these findings, we assumed that the photoinduced CT through (GC)n sequences would proceed rapidly because every G that acts as a charge carrier has neighboring Gs. Reporting on the CT through (GC)n sequences is challenging because of the difficulty of CT detection with a biological assay (25). We now report the photoinduced long-range CT of over 140 Å through a programmable DNA nanoscaled assembly based on rapid CT through the (GC)n sequence by using time-resolved transient-absorption measurements.
Results and Discussion
The Mechanisms of the CT Through the (GC)n.
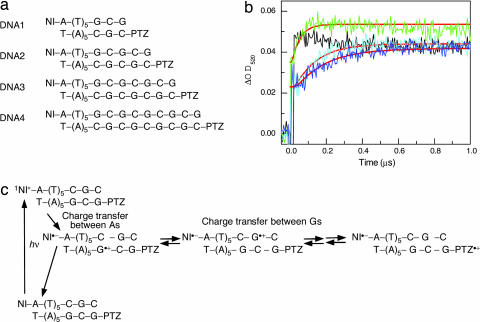
To clarify the long-range CT process through a programmable DNA nanoscaled assembly based on the (GC)n sequence, naphthalimide (NI)- and phenothiazine (PTZ)-modified DNAs were synthesized. Irradiation of NI with a 355-nm laser causes charge separation between the NI and an adjacent A to give an NI radical anion and an A radical cation (A•+) (26). When NI is attached to the consecutive A sequences, after the fast CT between As, the charge is trapped at the nearest G, to produce a charge-separated state within the laser flash duration of 4 ns. Subsequently, the charge can migrate to more distal Gs (27). After charge hopping between Gs, the charge on G can be accessible to the PTZ and be trapped at the PTZ [Eox = 0.76 V versus normal hydrogen electrode (NHE)], which has an oxidation potential lower than G. Thus, the formation of PTZ radical cation (PTZ•+) can be implicated as the CT process between Gs.
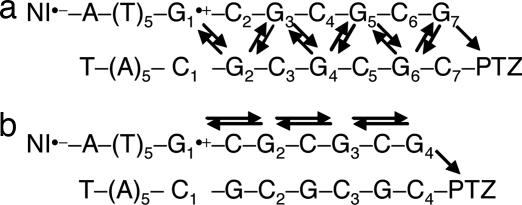
First, several DNAs with (GC)n sequences were synthesized, and the CT through the (GC)n sequence was examined by monitoring the formation of PTZ•+ at 520 nm as shown in Fig. 1. Decreasing the number of repetitive GCs produced an accelerated attainment of the charge on the PTZ through the CT between Gs because of the short distance for migration, and the CT through the shortest sequence of DNA1 (Fig. 1a) was so rapid that we could not detect it by using the nanosecond transient-absorption measurement. For further quantification of the actual CT rate, we examined in detail the CT process by using Matlab software based on kinetic modeling. The two following models were assumed: CT between adjacent interstrand Gs (Fig. 2a) and CT between intrastrand Gs (Fig. 2b). The rate constants of the CT between Gs (kht) were obtained from a fit to the measured transient absorption (Table 1). A kinetic analysis provided values of kht = (3.6 − 4.0) × 108 s−1 (CT between adjacent interstrand Gs; Fig. 2a) and kht = (1.0 − 1.2) × 108 s−1 (CT between intrastrand Gs; Fig. 2b) at 35°C. Using these rate constants, we obtained the mobility of the positive charge (μ) undergoing a hopping motion between G bases as already described (28). We previously have reported the rate constants of CT between Gs across A/T base pairs in which the most rapid CT between Gs was observed for the GAG sequence, kht = 5.1 × 107 s−1 at 38°C; μ = 8.7 × 10−6 cm2 V−1 s−1. Compared with a previous report, the μ value obtained in the present study was ≈1.8- to ≈2.3-fold greater than that for the GAG sequence. Thus, the CT rate through the (GC)n sequence certainly is rapid under both kinetic models. Previously, Lewis et al. (29) reported that the CT between Gs depends on the intervening bridge base (A, C, and thymine) between Gs. They described that the lower oxidation potential [Eox(A) = 1.94 > Eox(C) = 2.1 ∼ Eox(thymine) = 2.1 V versus the Na+/H+ exchanger (NHE) in CH3CN] (30) of the bridge base produces a small tunneling energy gap. Actually, they showed that the intrastrand CT between Gs occurs ≈40-fold more rapidly for the GAGG sequence than for GTGG sequence (12). They also displayed the dynamics of intrastrand versus interstrand CT between Gs across the A/T base pairs, in which the intrastrand CT for the GAGG sequence proceeds only ≈7-fold more rapidly than the interstrand CT for the GACC sequence (12). The finding that the intrastrand CT between Gs (Fig. 2b), the CT for the GCG sequence (bridge base = C) proceeded more rapidly than for the GAG sequence (bridge base = A) with a smaller tunneling energy gap conflicted with Lewis's report (29). Thus, it is suggested strongly that the CT through the (GC)n sequence mainly proceeds through the CT between the interstrand Gs (Fig. 2a). Because the consecutive G sequences might be a candidate for a more rapid CT between Gs, observations will be possible with the pico- or femtosecond transient-absorption measurement.
Fig. 1.
CT through (GC)n sequences. (a) Sequences of NI- and PTZ-modified DNAs. (b) Time profiles of the transient absorption of PTZ•+ monitored at 520 nm during the 355-nm laser flash photolysis of an Ar-saturated solution of NI- and PTZ-modified DNAs at 10°C for DNA1 (black), DNA2 (green), DNA3 (cyan), and DNA4 (blue), respectively. The smooth red curves superimposed on the rise curves are the fit to the kinetic model depicted in Fig. 2a. (c) Kinetic scheme for photo-induced one-electron oxidation of adenine and subsequent CT between guanines.
Fig. 2.
Kinetic schemes for the CT through (GC)n sequences. Two models were assumed: CT between adjacent interstrand Gs (a) and CT between intrastrand Gs (b).
Table 1.
Rate constants (kht), mobility of positive charge (μ), activation energy (Ea), and reorganization energy (λ) for charge transfer between Gs
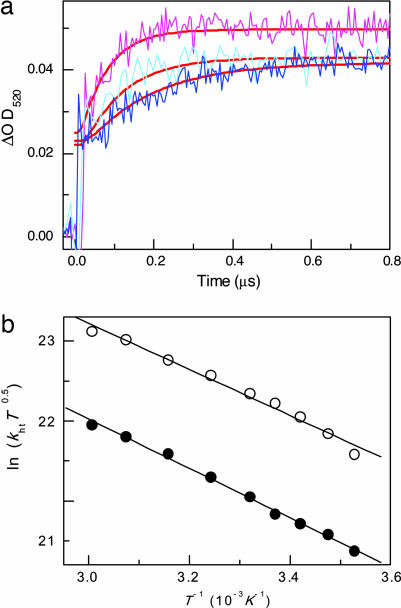
For the purpose of further elucidating the mechanisms of the CT through the (GC)n sequence, we investigated the temperature dependence of the CT (31). The transient absorption of PTZ•+ was monitored at 520 nm between 10°C and 60°C for DNA3 and DNA4. The increased temperature produced an accelerated attainment of the charge on the PTZ, as expected according to the electron-transfer theory as shown in Fig. 3a. A similar CT rate acceleration was observed for DNA3. The activation energy, Ea, and the reorganization energy, λ, obtained for the CT between Gs are defined in terms of Eqs. 1 and 2.
where T, kb, and ΔG denote the absolute temperature, Boltzmann constant, and thermodynamic driving force, respectively (31). Based on the assumption that the CT between Gs occurs at ΔG = 0, λ can be estimated. A plot of ln(khtT0.5) versus T−1 gave the Ea value for the CT between Gs (Table 1). Interestingly, the Ea and λ values were higher for the (GC)n sequence than for the GAG sequence for either kinetic modeling. These results might correspond to the relatively rigid DNA structures of the (GC)n sequence. Thus, as described by Barton and O'Neill (20) and Fiebig and colleagues (32), our results further support the fact that the CT through DNA might be related to the dynamic structures of DNA and affected by the conformational changes.
Fig. 3.
Temperature dependence on CT through (GC)n sequences. (a) Temperature dependence of the time profiles of transient absorption of PTZ•+ for DNA4, monitored at 520 nm during the 355-nm laser flash photolysis of an Ar-saturated solution of NI- and PTZ-modified DNAs at 10 (blue), 28(cyan), and 44°C (pink), respectively. The smooth red curves superimposed on the rise curves are the fit to the kinetic model depicted in Fig. 2a. (b) Effects of temperature on the rate constants for the CT. Plots of ln(khtT0.5) versus T−1 for DNA 4. Rate constants were obtained from a fit with model a (○) and model b (●), respectively.
The CT Through the DNA Nanoscaled Assembly Based on the (GC)n Sequence.
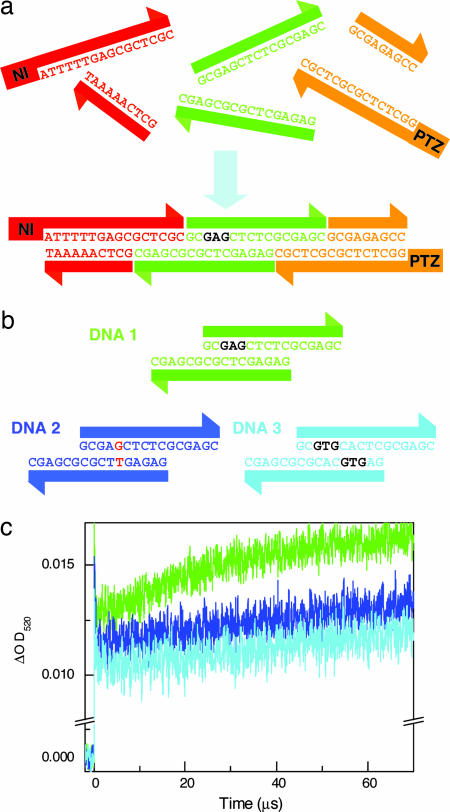
To create DNA nanometer-scaled wires and arrays, an understanding not only of the kinetic mechanisms of the rapid CT but also of the CT through the DNA nanoscaled assembly is required. As already mentioned, we have demonstrated that the CT rate through the (GC)n sequence certainly is rapid. Next, we examined the long-range CT through DNA nanoscaled assembly mainly containing the (GC)n sequence. We constructed a DNA nanoscaled assembly by using three DNA blocks: a charge-injection block (NI-labeled block), charge-conductive block, and charge-detection block (PTZ-labeled block) as shown in Fig. 4a. These blocks are designed for the effective charge injection with a consecutive A sequence and a rapid charge migration with the (GC)n sequence. Three blocks are connected by two sticky ends. After hybridization, the CT through the DNA constructed by the three DNA building blocks was observed by monitoring the formation of the PTZ•+. As is evident in Fig. 4c, the formation of PTZ•+ was observed for the DNA nanoscaled assembly constructed by simply mixing the three DNA blocks. The DNA block system makes it possible to achieve a CT over 140 Å through the DNA nanoscaled assembly based on the (GC)n sequences. The CT through the DNA nanoscaled assembly based on the (GC)n sequence certainly occurred, but the CT rate through the sticky-end DNA seems to be somewhat slower. This might reflect the disruption of the base stacking because of the local conformational fluctuation of the sticky-end DNA. Manipulating with the modifying enzymes, such as DNA ligase, should make the CT through the DNA building blocks much faster (33, 34). Considering that the CT process depends on the DNA sequence, the conductivity through the DNA nanoscaled assembly could be modulated by changing only a couple of DNA bases in the charge-conductive block. Thus, we replaced one or two DNA base of the charge-conductive block as shown in Fig. 4b. Fig. 4c displays the transient-absorption spectrum for such sequences. Our DNA nanoscaled assembly constructed by three blocks shows that the DNA conductivity is modulated by replacing only one or two DNA bases of the conductive block. Consequently, the transient-absorption measurement revealed that the DNA conductivity through the DNA nanoscaled assembly based on the (GC)n sequence could be designed by using programmable DNA blocks.
Fig. 4.
CT through nanoscaled DNA assembly. (a) Schematic representation of CT through nanoscaled DNA assembly based on sticky-end DNAs. Constructed DNA contain three blocks: charge-injection block (red), charge-conductive block (green), and charge-detection block (orange). These blocks are connected by sticky-end DNAs. (b) Sequence of conductive building blocks. (c) Time profiles of the transient absorption of PTZ•+ monitored at 520 nm during the 355-nm laser flash photolysis of an Ar-saturated solution of NI- and PTZ-modified DNAs at room temperature for DNA1 (green), DNA2 (blue), and DNA3 (cyan), respectively.
Conclusion
The time-resolved transient-absorption measurement demonstrated that the CT rate through the DNA with the (GC)n sequence actually is rapid. The DNA block system makes it possible to achieve the CT over 140 Å through the DNA nanoscaled assembly based on the rapid CT through the (GC)n sequences. Moreover, the CT through the nanoscaled DNA assembly sequence is programmable by using DNA blocks. The activation energy for the (GC)n sequence is relatively higher, which might correspond to the rigid structure. As previously described by Barton and O'Neill (20) and Fiebig and colleagues (32), the conformational flexibility and structures of the DNA promises to be important for the CT.
DNA nanostructures are constructed by cross-over DNAs as well as junction sticky-end DNAs. Previously, Barton and colleagues (35) and Sen and colleagues (36) investigated the effects of cross-over DNA four-way junctions. In addition, a recent study reported G-based nanowires constructed by using a poly(G) DNA strand (37). Further study of the focuses on CT through the cross-over DNA and G consecutive sequences will open the door to future nanoelectronics.
Materials and Methods
DNA Synthesis.
All reagents for DNA synthesis were purchased from Glen Research (Sterling, VA). Cyanoethyl phosphoramidites of N-(3-hydroxy-propyl)-1,8-naphthalimide and 10-(2-hydroxyethyl)phenothiazine were synthesized as previously reported (38–40). DNA used in this study was synthesized on an Applied Biosystems (Foster City, CA) DNA synthesizer with standard solid-phase techniques and purified on a Jasco (Tokyo, Japan) HPLC with a reverse-phase C-18 column with an acetonitrile/50 mM ammonium formate gradient. Duplex solutions (100 μM in 20 mM sodium phosphate buffer, pH 7.0, and 100 mM NaCl) were prepared by mixing equimolar amounts of the desired DNA complements and gradually annealing with cooling from 80°C to room temperature. Cooling was performed over 16 h in the case of DNA nanoscaled assembly. DNAs conjugated with NI and PTZ at the 5′ end were synthesized according to a previous procedure (27, 38, 41).
Laser Flash Photolysis Experiments.
Nanosecond transient-absorption measurements were performed as described in ref. 27. The third-harmonic oscillation (355 nm, FWHM of 4 ns, 20 mJ per pulse) from a Q-switched Nd:YAG laser (Surelight; Continuum, Santa Clara, CA) was used for the excitation light. A xenon flash lamp (XBO-450; Osram, Munich, Germany) was focused into the sample solution as the probe light for the transient-absorption measurement. Time profiles of the transient absorption in the UV-visible region were measured with a Nikon G250 monochromator equipped with a photomultiplier (R928; Hamamatsu Photonics, Hamamatsu City, Japan) and digital oscilloscope (TDS-580D; Tektronix, Inc., Tokyo, Japan).
Kinetic Modeling Procedures.
Analysis of the experimental data for all sequences was performed by using Matlab software according to procedures reported in ref. 42. In brief, a kinetic model of multistep CT in DNA is shown in Fig. 2. The charge-recombination process can be ignored because the charge-separated state persisted over several hundred microseconds when NI and the nearest G are separated by six A-T base pairs. A simultaneous differential equation can be obtained as shown in Eq. 3,
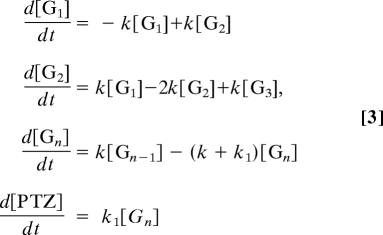 |
where [Gi] (i = 1 − n) and [PTZ] correspond to the charge population at G and PTZ sites, respectively, k is the CT rate constant between Gs, and k1 is the rate constant for the CT form G•+ to PTZ.
Acknowledgments
This work was supported partly by a grant-in-aid for Scientific Research (Project 17105005, Priority Area 417, 21st Century Center of Excellence Research, and others) from the Japanese Ministry of Education, Culture, Sports, Science, and Technology.
Abbreviations
- CT
charge transfer
- (GC)n
GC repetitive
- NI
naphthalimide
- PTZ
phenothiazine
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Seeman NC. Biochemistry. 2003;42:7259–7269. doi: 10.1021/bi030079v. [DOI] [PubMed] [Google Scholar]
- 2.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 3.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 4.Paukstelis PJ, Nowakowski J, Birktoft JJ, Seeman NC. Chem Biol. 2004;11:1119–1126. doi: 10.1016/j.chembiol.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 5.Mathieu F, Liao SP, Kopatscht J, Wang T, Mao CD, Seeman NC. Nano Lett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez-Navarro C, Moreno-Herrero F, de Pablo PJ, Colchero J, Gomez-Herrero J, Baro AM. Proc Natl Acad Sci USA. 2002;99:8484–8487. doi: 10.1073/pnas.122610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endo M, Seeman NC, Majima T. Angew Chem Int Ed. 2005;44:6074–6077. doi: 10.1002/anie.200501034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamoto A, Tanaka K, Saito I. J Am Chem Soc. 2003;125:5066–5071. doi: 10.1021/ja0294008. [DOI] [PubMed] [Google Scholar]
- 9.Giese B. Acc Chem Res. 2000;33:631–636. doi: 10.1021/ar990040b. [DOI] [PubMed] [Google Scholar]
- 10.Schuster GB. Acc Chem Res. 2000;33:253–260. doi: 10.1021/ar980059z. [DOI] [PubMed] [Google Scholar]
- 11.Delaney S, Barton JK. J Org Chem. 2003;68:6475–6483. doi: 10.1021/jo030095y. [DOI] [PubMed] [Google Scholar]
- 12.Lewis FD, Liu JQ, Zuo XB, Hayes RT, Wasielewski MR. J Am Chem Soc. 2003;125:4850–4861. doi: 10.1021/ja029390a. [DOI] [PubMed] [Google Scholar]
- 13.Shafirovich V, Cadet J, Gasparutto D, Dourandin A, Huang WD, Geacintov NE. J Phys Chem B. 2001;105:586–592. [Google Scholar]
- 14.Kelley SO, Barton JK. Science. 1999;283:375–381. doi: 10.1126/science.283.5400.375. [DOI] [PubMed] [Google Scholar]
- 15.Jortner J, Bixon M, Voityuk AA, Rosch N. J Phys Chem A. 2002;106:7599–7606. [Google Scholar]
- 16.Bixon M, Giese B, Wessely S, Langenbacher T, Michel-Beyerle ME, Jortner J. Proc Natl Acad Sci USA. 1999;96:11713–11716. doi: 10.1073/pnas.96.21.11713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakatani K, Dohno C, Saito I. J Am Chem Soc. 1999;121:10854–10855. [Google Scholar]
- 18.Giese B, Amaudrut J, Kohler AK, Spormann M, Wessely S. Nature. 2001;412:318–320. doi: 10.1038/35085542. [DOI] [PubMed] [Google Scholar]
- 19.Liu CS, Hernandez R, Schuster GB. J Am Chem Soc. 2004;126:2877–2884. doi: 10.1021/ja0378254. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill MA, Barton JK. J Am Chem Soc. 2004;126:13234–13235. doi: 10.1021/ja0455897. [DOI] [PubMed] [Google Scholar]
- 21.O'Neill MA, Barton JK. J Am Chem Soc. 2004;126:11471–11483. doi: 10.1021/ja048956n. [DOI] [PubMed] [Google Scholar]
- 22.Takada T, Kawai K, Fujitsuka M, Majima T. Proc Natl Acad Sci USA. 2004;101:14002–14006. doi: 10.1073/pnas.0402756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu B, Zhang P, Li X, Tao N. Nano Lett. 2004;4:1105–1108. [Google Scholar]
- 24.Xu MS, Tsukamoto S, Ishida S, Kitamura M, Arakawa Y, Endres RG, Shimoda M. Appl Phys Lett. 2005;87:083902. [Google Scholar]
- 25.Abdou IM, Sartor V, Cao HC, Schuster GB. J Am Chem Soc. 2001;123:6696–6697. doi: 10.1021/ja004274y. [DOI] [PubMed] [Google Scholar]
- 26.Takada T, Kawai K, Cai XC, Sugimoto A, Fujitsuka M, Majima T. J Am Chem Soc. 2004;126:1125–1129. doi: 10.1021/ja035730w. [DOI] [PubMed] [Google Scholar]
- 27.Kawai K, Osakada Y, Takada T, Fujitsuka M, Majima T. J Am Chem Soc. 2004;126:12843–12846. doi: 10.1021/ja0475813. [DOI] [PubMed] [Google Scholar]
- 28.Senthilkumar K, Grozema FC, Guerra CF, Bickelhaupt FM, Lewis FD, Berlin YA, Ratner MA, Siebbeles LDA. J Am Chem Soc. 2005;127:14894–14903. doi: 10.1021/ja054257e. [DOI] [PubMed] [Google Scholar]
- 29.Lewis FD, Liu JQ, Weigel W, Rettig W, Kurnikov IV, Beratan DN. Proc Natl Acad Sci USA. 2002;99:12536–12541. doi: 10.1073/pnas.192432899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seidel CAM, Schulz A, Sauer MHM. J Phys Chem. 1996;100:5541–5553. [Google Scholar]
- 31.Davis WB, Hess S, Naydenova I, Haselsberger R, Ogrodnik A, Newton MD, Michel-Beyerle ME. J Am Chem Soc. 2002;124:2422–2423. doi: 10.1021/ja017304l. [DOI] [PubMed] [Google Scholar]
- 32.Valis L, Wang Q, Raytchev M, Buchvarov I, Wagenknecht H-A, Fiebig T. Proc Natl Acad Sci USA. 2006;103:10192–10195. doi: 10.1073/pnas.0600957103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang XP, Wenzler LA, Qi J, Li XJ, Seeman NC. J Am Chem Soc. 1998;120:9779–9786. [Google Scholar]
- 34.LaBean TH, Yan H, Kopatsch J, Liu FR, Winfree E, Reif JH, Seeman NC. J Am Chem Soc. 2000;122:1848–1860. [Google Scholar]
- 35.Odom DT, Dill EA, Barton JK. Nucleic Acids Res. 2001;29:2026–2033. doi: 10.1093/nar/29.10.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fahlman RP, Sharma RD, Sen D. J Am Chem Soc. 2002;124:12477–12485. doi: 10.1021/ja020495n. [DOI] [PubMed] [Google Scholar]
- 37.Kotlyar AB, Borovok N, Molotsky T, Cohen H, Shapir E, Porath D. Adv Mater. 2005;17:1901–1905. [Google Scholar]
- 38.Kawai K, Kawabata K, Tojo S, Majima T. Bioorg Med Chem Lett. 2002;12:2363–2366. doi: 10.1016/s0960-894x(02)00404-3. [DOI] [PubMed] [Google Scholar]
- 39.Kawai K, Takada T, Tojo S, Majima T. Tetrahedron Lett. 2002;43:89–91. [Google Scholar]
- 40.Tierney MT, Grinstaff MW. J Org Chem. 2000;65:5355–5359. doi: 10.1021/jo0055133. [DOI] [PubMed] [Google Scholar]
- 41.Takada T, Kawai K, Fujitsuka M, Majima T. Chem Eur J. 2005;11:3835–3842. doi: 10.1002/chem.200500052. [DOI] [PubMed] [Google Scholar]