Abstract
During mitosis in the yeast Saccharomyces cerevisiae, Kar9p directs one spindle pole body (SPB) toward the incipient daughter cell by linking the associated set of cytoplasmic microtubules (cMTs) to the polarized actin network on the bud cortex. The asymmetric localization of Kar9p to one SPB and attached cMTs is dependent on its interactions with microtubule-associated proteins and is regulated by the yeast Cdk1 Cdc28p. Two phosphorylation sites in Kar9p were previously identified. Here, we propose that the two sites are likely to govern Kar9p function through separate mechanisms, each involving a distinct cyclin. In the first mechanism, phosphorylation at serine 496 recruits Kar9p to one SPB. A phosphomimetic mutation at serine 496 bypasses the requirement of BIK1 and CLB5 in generating Kar9p asymmetry. In the second mechanism, Clb4p may target serine 197 of Kar9p for phosphorylation. This modification is required for Kar9p to direct cMTs to the bud. Two-hybrid analysis suggests that this phosphorylation may attenuate the interaction between Kar9p and the XMAP215-homologue Stu2p. We propose that phosphorylation at serine 197 regulates the release of Kar9p from Stu2p at the SPB, either to clear it from the mother-SPB or to allow it to travel to the plus end.
INTRODUCTION
Positioning of the mitotic spindle is important for asymmetric cell divisions in numerous developmental paradigms. In the budding yeast Saccharomyces cerevisiae, spindle positioning is accomplished through interactions between cytoplasmic microtubules (cMTs) and the cell cortex (Sullivan and Huffaker, 1992; Carminati and Stearns, 1997; Adames and Cooper, 2000; Yeh et al., 2000). cMTs are anchored to the spindle through the SPB, which serves as the centrosome equivalent in fungal cells (Byers and Goetsch, 1975; Byers, 1981).
Alignment of the spindle along the long axis of division and its placement at the mother-bud neck requires the linker protein Kar9p. Kar9p initially localizes to the SPB, and it is transported from the pole toward the plus ends of cMTs by the kinesin Kip2p (Liakopoulos et al., 2003; Maekawa et al., 2003; Moore et al., 2006). The association of Kar9p with cMT plus ends enables those microtubules to interact with the cortical actin network through its interaction with the myosin Myo2p (Beach et al., 2000; Miller et al., 2000; Yin et al., 2000). Myo2p then delivers Kar9p and the attached cMT end to the bud via transport along polarized actin cables (Hwang et al., 2003). Because Kar9p is necessary for linking microtubule ends to cortical myosin, the association of Kar9p with cMTs emanating from one of the two SPBs allows only that spindle pole to be oriented toward the bud (Liakopoulos et al., 2003; Maekawa et al., 2003; Moore et al., 2006). The asymmetric positioning of the poles is therefore dependent on the restriction of Kar9p to one SPB and set of cMTs (Liakopoulos et al., 2003; Moore et al., 2006).
Loading Kar9p onto the SPB is an important early step in the Kar9p mechanism that precedes the localization of Kar9p to cMT plus ends (Liakopoulos et al., 2003). This process is influenced by two microtubule-associated proteins (MAPs): Bim1p/Yeb1p, the yeast homologue of EB1; and Bik1p, the yeast homologue of CLIP-170 (Lee et al., 2000; Miller et al., 2000, 2006; Moore et al., 2006). Kar9p expressed at endogenous levels is not detected at the SPB in the absence of Bim1p (Liakopoulos et al., 2003), suggesting that Bim1p plays a central role in loading Kar9p onto SPBs. However, the observation that overexpressed Kar9p localizes to SPBs in the absence of Bim1p indicates that Kar9p may associate with additional factors at the SPB (Miller et al., 2000). The second MAP, Bik1p, also localizes to the SPBs and microtubule plus ends (Carvalho et al., 2004; Moore et al., 2006). Bik1p is not required for the association of Kar9p with SPBs, but it contributes to Kar9p asymmetry by restricting it to one SPB (Moore et al., 2006).
Kar9p also interacts with a third MAP, Stu2p, the yeast homologue of XMAP215/TOGp (Miller et al., 2000). Stu2p is localized primarily at the SPBs, but it is also found at cMT plus ends and on spindle microtubules (Kosco et al., 2001, Wolyniak et al., 2006). Stu2p plays an important role in anchoring the minus-ends of cMTs to the SPB (Wang and Huffaker, 1997; Kosco et al., 2001; Usui et al., 2003), binding tubulin dimers (Al-Bassam et al., 2006) and regulating microtubule dynamics (Kosco et al., 2001; van Breugel et al., 2003). However, the contribution of Stu2p to the function of Kar9p has remained unclear (Miller et al., 2000).
The asymmetric localization of Kar9p to one SPB and attached microtubules is regulated by the yeast Cdk1 Cdc28p (Liakopoulos et al., 2003; Maekawa and Schiebel, 2004; Moore et al., 2006). The cyclins Clb5p and Clb4p have also been implicated in generating SPB and Kar9p asymmetry, because deletion of either gene results in an increased localization of Kar9p to both SPBs (Segal et al., 1998, 2000; Liakopoulos et al., 2003; Maekawa and Schiebel, 2004; Moore et al., 2006). Two Cdc28p-dependent phosphorylation sites have been identified in Kar9p, at serines 197 and 496 (Liakopoulos et al., 2003; Maekawa and Schiebel, 2004; Moore et al., 2006). Serine 496 lies within a region of homology to the adenomatous polyposis coli (APC) tumor suppressor protein, and phosphorylation of this residue is thought to represent a conserved means of regulating the association of both proteins with microtubules and microtubule organizing centers (Trzepacz et al., 1997; Liakopoulos et al., 2003; Honnappa et al., 2005). Kar9p phosphorylation is likely to be translated into its selective association with one SPB by exerting local effects on interactions between Kar9p and SPB-bound factors. Two models could explain this regulation. In the first scenario, kinase activity at one SPB would alter the interaction of Kar9p with an SPB-associated protein at that pole. This is similar to the model of Liakopoulos et al. (2003), in which they propose that the phosphorylation of Kar9p at the mother-bound SPB disrupts the interaction between Kar9p and the EB1 homologue Bim1p, thereby preventing Kar9p from loading onto that pole. In an alternative model, Kar9p could be phosphorylated irrespective of its proximity to either SPB, but a selective factor that is sensitive to this phosphorylation would only be present at one pole. In this model, phosphorylation would enable Kar9p to act on this preexisting asymmetry at the SPBs. Kar9p would either be recruited to the pole to which this factor was bound or, alternatively, be repelled from that pole.
In this work, we test these models by examining the effects of phosphorylation at serines 197 and 496 on Kar9p localization and its interactions with the MAPs Bim1p and Stu2p. Mimicking phosphorylation at serine 496 restores the asymmetric localization of Kar9p in cells lacking Clb5p or Bik1p. These data support the model that a phosphorylation event on Kar9p enables it to recognize an intrinsic asymmetry between the SPBs. The mislocalization of Kar9p in the absence of Clb4p is not suppressed by mimicking phosphorylation at serine 496, but it is instead partially suppressed by mimicking phosphorylation at serine 197. Western blot analysis is consistent with the idea that Clb4p is required for the phosphorylation of Kar9p at serine 197 but not serine 496. Mutations preventing the phosphorylation of serine 197 are synthetically lethal with mutants of the dynein pathway, suggesting that this event is necessary for Kar9p function. Finally, we show that the interaction of Kar9p with Stu2p is attenuated by phosphorylation at serine 197. These results illustrate a novel function for Stu2p and imply that Stu2p may play a central role in regulating Kar9p function at the SPB.
MATERIALS AND METHODS
Yeast Strains and Growth Conditions
S. cerevisiae strains and plasmids used in this study are listed in Table 1. Primers and oligonucleotides used for strain constructions can be found in Supplemental Table S1. Cells were grown in yeast peptone dextrose (YPD) or synthetic complete (SC) media as described previously (Miller et al., 2000).
Table 1.
S. cerevisiae strains and plasmids used in this study
| Genotype/comments | Source | |
|---|---|---|
| Yeast strains | ||
| yRM1757/PJ69-4 | MATatrp1901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ | James et al., 1996 |
| LYS2:GAL1-HIS3 GAL2-ADE2 met3::GAL7-lacZ | ||
| yRM396/MS4589 | MATakar9Δ-1::LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | Miller and Rose, 1998 |
| yRM469/MS4321 | MATajnm1Δ::LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | Miller and Rose, 1998 |
| yRM565 | MATabik1Δ::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | Moore et al., 2006 |
| yRM1094/MS4903 | MATadyn1Δ::LEU2 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | Miller and Rose, 1998 |
| yRM2060/MS7310 | MATabim1Δ::KanR leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | Miller et al., 2000 |
| yRM2147/MS1556 | MATaleu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | M. D. Rose, Princeton University |
| yRM2146/MS52 | MATα leu2-3 leu2-112 ura3-52 trp1Δ1 | M. D. Rose, Princeton University |
| yRM2057 | MATabim1Δ::KanR trp1-901 leu2-3 leu2-112 ura3-52 his3Δ200 gal4Δ gal80Δ LYS2:GAL1-HIS3 GAL2-ADE2 met3::GAL7-lacZ | This study |
| yRM3886 | MATα KAR9-3GFP::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | Moore et al., 2006 |
| yRM4357 | MATα KAR9-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | Moore et al., 2006 |
| yRM4366 | MATaKAR9-tap::URA3 bar1Δ::LEU2 leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | Moore et al., 2006 |
| yRM4369 | MATα KAR9-3GFP::TRP1 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM4598 | MATα KAR9-3GFP::TRP1 bik1Δ::TRP1 kip2Δ::URA3 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | Moore et al., 2006 |
| yRM5122 | MATaKAR9-3GFP::TRP1 clb5Δ::URA3 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | Moore et al., 2006 |
| yRM5509 | MATaclb5Δ::URA3 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM5664 | MATakar9-S496E-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 | This study |
| yRM5833 | MATakar9-S496E-3GFP::TRP1 clb5Δ::URA3 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 | This study |
| yRM5862 | MATakar9-S496E-3GFP::TRP1 bik1Δ::TRP1 kip2Δ::URA3 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| yRM6007 | MATakar9-S496A-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | Moore et al., 2006 |
| yRM6008 | MATakar9-S197E-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6015 | MATα kar9-S496A-tap::URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | Moore et al., 2006 |
| yRM6026 | MATα clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6094 | MATaKAR9-tap::URA3 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6099 | MATaKAR9-3GFP::TRP1 clb4Δ::KanR SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6110 | MATα kar9-S197A-3GFP::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6122 | MATakar9-S496E-3GFP::TRP1 clb4Δ::KanR SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| yRM6142 | MATα kar9-S496A-tap::URA3 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 | This study |
| yRM6144 | MATakar9-S197A-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6148 | MATakar9-S197A,S496A-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6152 | MATaclb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| yRM6168 | MATα kar9-S197A-tap::URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | Moore et al., 2006 |
| yRM6170 | MATα kar9-S197A,S496A-tap::URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | Moore et al., 2006 |
| yRM6202 | MATα kar9-S197E-3GFP::TRP1 clb4Δ::KanR SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6211 | MATakar9-S197A-tap::URA3 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 | This study |
| yRM6235 | MATakar9-S197A,S496A-tap::URA3 clb4Δ::KanR leu2-3 leu2-112 ura3-52 ade2-101 | This study |
| yRM6246 | MATaSPC110-DsRed::KanR clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM6275 | MATα kar9-A196E,S197E-3GFP::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6276 | MATα kar9-A196E,S197E-tap::URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6278 | MATα kar9-A196E,S197E-3GFP::TRP1 clb4Δ::KanR SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6281 | MATα kar9-A196E,S197E-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6285 | MATα kar9-A196E,S197E-3GFP::TRP1 clb5Δ::URA3 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 ade2-101 | This study |
| yRM6399 | MATα kar9-S496E-tap::URA3 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6518 | MATaKAR9-3GFP::TRP1 stu2-13::URA3 leu2-3 leu2-112 ura3-52 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6601 | MATaKAR9-3GFP::TRP1 stu2-13::URA3 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 his3Δ200 ade2-101 | This study |
| yRM6602 | MATα kar9-S496A-3GFP::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6900 | MATakar9-Δ2::HIS3 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 | This study |
| yRM6901 | MATakar9-S496E-3GFP::TRP1 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM6902 | MATaKAR9-3GFP::TRP1 SPC110-CFP::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 ade2-101 | This study |
| yRM6903 | MATα kar9-A196E,S197E-3GFP::TRP1 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM6904 | MATα kar9-S197A,S496A-3GFP::TRP1 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM6905 | MATα kar9-S197A-3GFP::TRP1 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM6906 | MATakar9-S496A-3GFP::TRP1 SPC110-DsRed::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 his3Δ200 [pAFS92 GFP-TUB1::URA3] | This study |
| yRM6907 | MATakar9-Δ2::HIS3 leu2-3 leu2-112 ura3-52 ade2-101 his3Δ200 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6908 | MATα kar9-S197A,S496A-3GFP::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6909 | MATα kar9-S496E-3GFP::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6910 | MATα clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6911 | MATα KAR9-3GFP::TRP1 clb5Δ::hphMX4 leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6912 | MATα KAR9-3GFP::TRP1 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6913 | MATaKAR9-3GFP::TRP1 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6914 | MATα kar9-Δ2::HIS3 clb4Δ::KanR leu2-3 leu2-112 ura3-52 ade2-101 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6915 | MATα kar9-S197A-3GFP::TRP1 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6916 | MATakar9-A196E,S197E-3GFP::TRP1 clb4Δ::KanR leu2-3 leu2-112 ura3-52 trp1Δ1 ade2-101 [pAFS125C CFP-TUB1::URA3] | This study |
| yRM6917 | MATα STU2-13myc::KanMX6 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6918 | MATα kar9-A196E,S197E-tap::URA3 STU2-13myc::KanMX6 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6919 | MATα kar9-S197A-tap::URA3 STU2-13myc::KanMX6 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6920 | MATaKAR9-tap::URA3 bar1Δ::LEU2 leu2-3 leu2-112 ura3-52 trp1Δ1 his3Δ200 | This study |
| yRM6921 | MATα kar9-S496A-tap::URA3 STU2-13myc::KanMX6 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6922 | MATα kar9-S197A,S496A-tap::URA3 STU2-13myc::KanMX6 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6923 | MATα kar9-S496E-tap::URA3 STU2-13myc::KanMX6 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| yRM6959 | MATakar9-A196E S197E-3GFP::TRP1 SPC110-CFP::KanR bik1Δ::TRP1 leu2-3 leu2-112 ura3-52 trp1Δ1 | This study |
| Plasmids | ||
| pRM1493/pMR4150 | GBDU-KAR9 URA3 2μ | Miller et al., 2000 |
| pRM1916/pMR4769 | GAD-STU2649-888aa LEU2 2μ AmpR | Miller et al., 2000 |
| pRM2023/pMR4653 | GAD-BIM1187-276aa LEU2 2μ AmpR | Miller et al., 2000 |
| pRM2345 | GBDU-C3 URA3 2μ with the BglII restriction site in the polylinker replaced with SacI | Moore et al., 2006 |
| pRM2432 | GBDU-KAR91-316aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2480 | GBDU-KAR91-279aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2632 | GBDU-KAR9391-644aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2633 | GBDU-KAR9200-644aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2634 | GBDU-KAR9200-399aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2719 | GBDU-KAR9391-470aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2720 | GBDU-KAR9200-470aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2730 | GBDU-KAR9117-644aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2862 | GBDU-KAR9391-540aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2863 | GBDU-KAR9471-644aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2888 | GBDU-KAR9471-580aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2889 | GBDU-KAR9471-613aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2909 | GBDU-KAR9534-580aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM2951 | GBDU-KAR9117-297aa URA3 2μ AmpR | Moore et al., 2006 |
| pRM3175/AVA0258 | A template for TAP tagging with his6x-HA-3C-ProteinA K.l. URA3 | E. Phizicky, University of Rochester Medical School |
| pRM3662 | KAR9117-644aa-3GFP TRP1 AmpR integration plasmid | Moore et al., 2006 |
| pRM4335/pTY24 | pFA6a-DsRed.T1.N1-Kan-Mx6 AmpR | Yeast Resource Center |
| pRM4340/pDH3 | pFA6a-CFP-Kan-Mx6 AmpR | Yeast Resource Center |
| pRM5595 | kar9-S496E117-644aa-3GFP TRP1 AmpR integration plasmid | This study |
| pRM5617 | GBDU-kar9-S197E URA3 2μ | This study |
| pRM5619 | GBDU-kar9-S496E URA3 2μ | This study |
| pRM5777 | GBDU-kar9-S496A URA3 2μ | This study |
| pRM5778 | kar9-S496A117-644aa-3GFP TRP1 AmpR integration plasmid | Moore et al., 2006 |
| pRM5779 | kar9-S197E117-644aa-3GFP TRP1 AmpR integration plasmid | This study |
| pRM6048 | kar9-S197A117-644aa-3GFP TRP1 AmpR integration plasmid | Moore et al., 2006 |
| pRM6049 | kar9-S197A,S496A117-644aa-3GFP TRP1 AmpR integration plasmid | Moore et al., 2006 |
| pRM6050 | GBDU-kar9-S197A URA3 2μ | This study |
| pRM6113 | GBDU-kar9-S197A,S496A URA3 2μ | This study |
| pRM6251 | kar9-A196E,S197E117-644aa-3GFP TRP1 AmpR integration plasmid | This study |
| pRM6255 | GBDU-kar9-A196E,S197E URA3 2μ | This study |
| pRM6294 | GBDU-kar9-S197A,S496E URA3 2μ | This study |
| pRM6295 | GBDU-kar9-A196E,S197E,S496A URA3 2μ | This study |
| pAFS125C | pCFP-TUB1::URA3 AmpR | A. Straight, Harvard Medical School |
kar9 Mutants
Point mutations were introduced into the KAR9–3GFP plasmid (pRM3662) by site-directed mutagenesis by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotides used in each mutagenesis are listed in Table S1. The resulting plasmids were verified by sequencing. These were then integrated at the endogenous KAR9 locus of the wild-type strain MS52/yRM2146 after digestion with the ClaI endonuclease. These green fluorescent protein (GFP)-tagged integrants were used to obtain the TAP-tagged versions of these mutants. The TAP tag cassette consists of 6x-histidine, hemagglutinin (HA), and protein A epitopes followed by the URA3 marker from Kluyveromyces lactis, and it was a gift from Eric Phizicky (University of Rochester, Rochester, NY). For each mutant, this cassette was amplified by polymerase chain reaction (PCR) and transformed into the corresponding GFP-tagged strains, replacing the 3GFP tag and TRP+ marker.
Deletion of CLB4
To delete CLB4 from our strain background, the CLB4 locus was amplified from the clb4Δ strain YLR210W of the American Type Culture Collection (Manassas, VA) deletion collection. The amplified region contained the kanMX4-selectable marker and ∼200 base pairs of homologous sequence flanking both ends of the CLB4 open reading frame. This was transformed into the wild-type strain MS52/yRM2146, and transformants were selected on rich media containing Geneticin (G-418) (Invitrogen, Carlsbad, CA). Integration of the kanMX4 cassette at the CLB4 locus was confirmed by PCR.
Fluorescence Microscopy
SPC110 was tagged with cyan fluorescent protein (CFP) or red fluorescent protein (DsRed) as described previously (Moore et al., 2006). The CFP tag was constructed using the pDH3/pRM4340 template, and the DsRed tag was constructed using pTY24/pRM4335 (both gifts of the Yeast Resource Center, University of Washington, Seattle, WA). Microtubules were labeled with CFP-Tub1p by the integration of pAFS125C at the URA3 locus.
Microscopy was carried out on a motorized Zeiss Axioplan 2 microscope equipped with a 100× Plan-Neofluor lens (1.3 numerical aperture [NA]) (Carl Zeiss, Thornwood, NY), a cooled charged-coupled device camera (ORCA-ER; Hamamatsu, Hamamatsu City, Japan), and Chroma (Chroma Technology, Brattleboro, VT) and/or Zeiss (Carl Zeiss) filter sets. Images were acquired and processed using Openlab 3.5.2 software (Improvision, Lexington, MA).
For fluorescence intensity quantification, images were captured on an Olympus IX70 scope with a 100× Plan-Apo lens (1.4NA) (Olympus. Melville, NY), and CoolSNAP HQ camera (Roper Scientific, Duluth, GA) using QED software (QED Imaging, Pittsburgh, PA). Intensity measurements were determined using Image J (Wayne Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/), and analyzed using Microsoft Excel (Microsoft, Redmond, WA). These values were corrected for background fluorescence by subtracting the minimum pixel intensity of the region of the cytosol containing either the plus or minus end of the microtubule.
Preparation of Bim1p Antibodies
A his6x-BIM1 plasmid (pRM 3014) was used to produce his6x-Bim1p in bacteria. This was used for the production of antisera essentially as described for Bik1p (Moore et al., 2006).
Western Blotting and Affinity Purification
Protein extracts were prepared from cultures grown to mid-exponential phase. Cells were washed once with water, resuspended in ice-cold B150 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 0.2% Triton X-100) with protease inhibitors (protease inhibitor cocktail [Sigma-Aldrich, St. Louis, MO] and 1 mM phenylmethylsulfonyl fluoride), and lysed by vortexing with glass beads. Crude extracts were then clarified by centrifugation at 16,000 × g for 20 min. The supernatant was transferred to a fresh tube and centrifuged at 16,000 × g for an additional 20 min.
Thirty micrograms of each protein extract sample was run on 8% SDS-PAGE, which provided optimal resolution for the 96-kDa Kar9p-tap protein. Gels ran under a reduced current at 15 mA per gel. Kar9p-tap was detected with α-HA (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:200. Chicken α-actin was used at 1:20,000.
For affinity purification of Kar9p-tap, 20 μl of IgG-Sepharose (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) was added to 1 ml of 3 mg/ml protein extract and incubated on a rotisserie at 4°C for 12 h. The precipitate was then washed 16 times with cold B150 buffer. Bound proteins were eluted by the addition of 50 μl of 3× Laemmli buffer and boiled for 3 min. Then, 15 μl of each sample was analyzed after 12% SDS-PAGE and Western blotting.
Drug Treatments
For hydroxyurea (HU; Sigma-Aldrich) treatment, cultures were grown to early exponential phase in YPD media. HU was added to a final concentration of 100 mM at 30°C for 2 h. Bud morphology was used to demonstrate that >98% of cells were arrested in late S phase.
Cell Synchronization
To generate populations of cells that progressed synchronously through the cell cycle, we isolated quiescent stationary phase cells by using the methods of Allen et al. (2006). Cultures were grown in YPD for 7 d at 30°C, pelleted, and resuspended in 10 mM Tris, pH 7.5. Approximately 2 × 109 cells from each culture were spun through a Percoll gradient (GE Healthcare) at 400 × g for 60 min at 20°C. Low-density fractions were collected, pelleted, and washed with Tris buffer. To release from stationary phase, these cells were then resuspended in 5 ml of YPD and returned to 30°C. At 150 min after the introduction of fresh media, the majority of cells in each strain contained short bipolar spindles. Aliquots were collected at 20-min intervals and fixed for 10 min in 3.7% formaldehyde followed by three washes with phosphate-buffered saline.
Two-Hybrid Assay
The two-hybrid system of (James et al., 1996) was used. To generate KAR9 phosphomutants for two-hybrid analysis, point mutations were introduced into the DBD-KAR9 plasmid (pRM1493) by using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The oligonucleotide pairs used in each mutagenesis are listed in Supplemental Table S1. Each resulting DBD-KAR9 mutant was sequenced to verify the absence of additional errors.
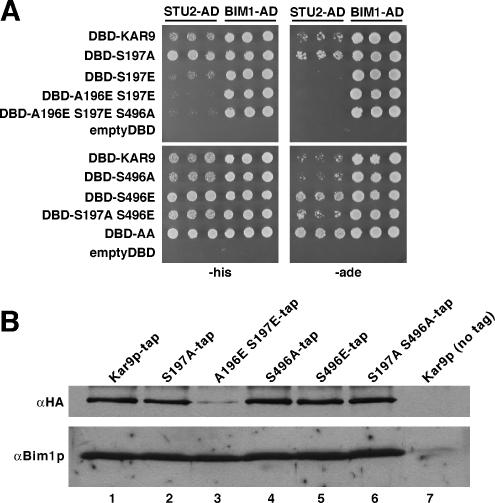
The bim1Δ two-hybrid reporter strain was generated using methods similar to those reported in Miller and Rose (1998). The kanMX2 cassette was amplified by PCR to include 75 base pairs of homology to the sequences flanking the chromosomal BIM1 open reading frame. This disruption fragment was transformed into the wild-type two-hybrid reporter strain PJ69-4A (yRM1757), and integrants were selected on rich media containing the drug G-418 (Invitrogen). The disruption of BIM1 was confirmed by PCR. DBD and AD plasmids were transformed into the bim1Δ reporter strain and selected for growth on plates lacking uracil and leucine. Interactions were assayed by transferring cells with a multiprong transfer device to SC plates lacking uracil and leucine, histidine (−His), and adenine (−Ade). Growth on histidine-deficient media was scored after incubation at 30°C 3 d, whereas growth on adenine-deficient media was scored after 7 d.
RESULTS
Mimicking Phosphorylation at Serine 496 Suppresses the Kar9p Localization Defect Present in clb5Δ and bik1Δ kip2Δ Strains
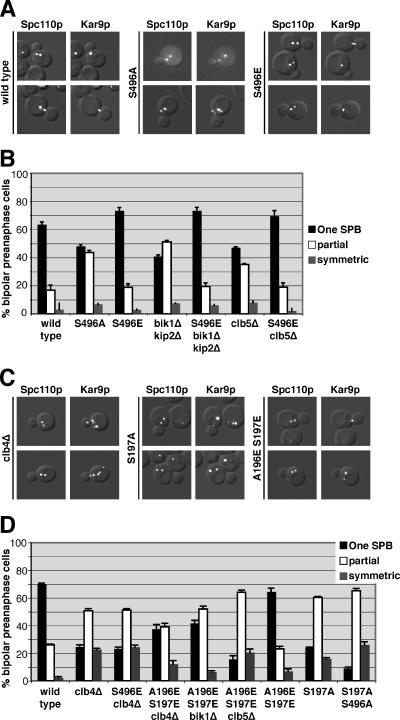
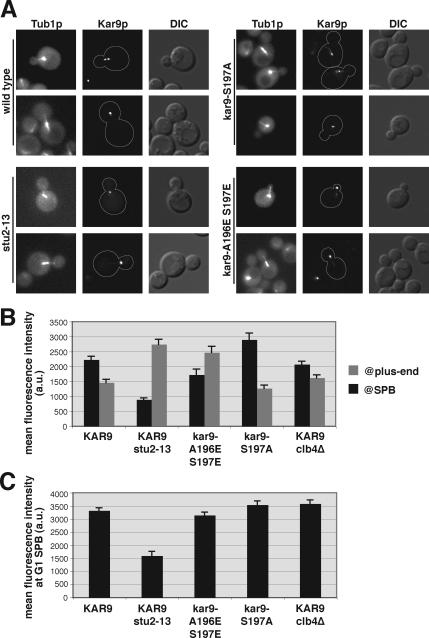
To investigate the role that phosphorylation of Kar9p at serine 496 plays in regulating its association with SPBs, we replaced serine 496 with a glutamic acid residue to mimic the negative charge of phosphorylation. kar9-S496E fused to three copies of GFP was then integrated at the chromosomal KAR9 locus, leaving the only source of Kar9p in a constitutively pseudophosphorylated state. The SPB localization of wild-type or Kar9p-S496E-3GFP was scored in cells containing Spc110p-CFP–labeled SPBs. In 63% of cells expressing wild-type Kar9p-3GFP, GFP localization was detected on only one pole of preanaphase cells with short bipolar spindles (Figure 1A, quantified in B). Similar to wild type, Kar9p-S496E-3GFP localized to one pole in 73% of cells (Figure 1B). Consistent with a previous report (Liakopoulos et al., 2003), replacement of serine 496 with an alanine residue to prevent phosphorylation at this site resulted in an increase of Kar9p localization to both SPBs (Figure 1B). This confirms that this phosphorylation event is important for restricting Kar9p to one pole.
Figure 1.
Phosphomimetic mutants suppress the mislocalization of Kar9p in bik1Δkip2Δ and clb5Δ and clb4Δ strains. (A) Examples of wild-type Kar9p, Kar9p-S496A, and Kar9p-S496E fused to 3GFP in cells containing SPBs labeled with Spc110p-CFP. Composites are shown of differential interference contrast microscopy with either CFP or GFP single images. (B) Quantification of Kar9p localization to the SPBs for wild-type (yRM4357), Kar9p-S496A (yRM6007), Kar9p-S496E (yRM5664), Kar9p in bik1Δkip2Δ (yRM4598), Kar9p-S496E in bik1Δkip2Δ (yRM5862), Kar9p in clb5Δ (yRM5119), and Kar9p-S496E in clb5Δ (yRM5833) cells with short bipolar spindles. “One SPB” represents cells in which Kar9p-3GFP was observed at only one of the two SPBs. “Partial” denotes cells in which Kar9p-3GFP was detected at both SPBs, but a more intense GFP signal was seen at one pole. “Symmetric” represents cells in which equally intense GFP signal was observed at both poles. In addition to its SPB localization, Kar9p-3GFP signal was also apparent at regions adjacent to the SPB and as distinct punctae away from the SPB. Based on our previous work, this presumably represents Kar9p associated with the lateral sides and plus ends of cMTs, respectively (Moore et al., 2006). For our experiments, we scored only GFP signal that coincided with the Spc110p-CFP or was immediately adjacent to it as SPB-localized Kar9p. At least 250 cells were scored for each strain. Cells were grown in SC media −Trp at 30°C. Error bars denote the SEM of the percentages calculated in at least five separate counts. (C) Examples of wild-type Kar9p-3GFP localization in clb4Δ cells, and Kar9p-S197A-3GFP and Kar9p-A196E S197E-3GFP in wild-type cells containing SPBs labeled with Spc110p-CFP. (D) Quantification of Kar9p localization to the SPBs for wild-type (yRM4357), Kar9p in clb4Δ (yRM6099), Kar9p-S496E in clb4Δ (yRM6122), Kar9p-A196E S197E in clb4Δ (yRM6278), Kar9p-A196E S197E in bik1Δ (yRM6959), Kar9p-A196E S197E in clb5Δ (yRM6285), Kar9p-A196E S197E (yRM6281), Kar9p-S197A (yRM6144), and Kar9p-S197A S496A (yRM6148) cells with short bipolar spindles. Cells were scored as described for B. A Fisher's exact test was used to compare the frequency at which Kar9p was localized to one or both poles in clb4Δ and S197A strains, generating a p value of 0.704. This suggests that there is not a significant difference between the data sets for clb4Δ and S197A. Comparing the data for Kar9p and Kar9p-A196E S197E in the clb4Δ background generated a p value of <0.0001, indicating that the localization of Kar9p-A196E S197E is significantly different from that of wild-type Kar9p.
We previously showed that Kar9p localizes to both SPBs in cells deleted for BIK1 (Moore et al., 2006). We attributed this to the role Bik1p plays in promoting the phosphorylation of Kar9p by facilitating an interaction between Kar9p and the cyclin Clb5p (Moore et al., 2006). If Bik1p functions upstream of the phosphorylation of Kar9p at serine 496 by Clb5p-Cdc28p, then mimicking this phosphorylation should restore the restriction of Kar9p to one SPB in the absence of Bik1p. To test this, we assayed the localization of Kar9p-S496E-3GFP in a bik1Δkip2Δ background. This double mutant exhibits the same Kar9p localization defect seen in the bik1Δ single mutant except that Kar9p is retained at the SPBs rather than being transported onto the microtubule by the Kip2p motor (Maekawa et al., 2003; Moore et al., 2006). This provides a more rigorous test of SPB localization by increasing the SPB-bound pool of Kar9p. Wild-type Kar9p localized to one SPB in 41% of bik1Δkip2Δ cells (Figure 1, quantified in B). In contrast, Kar9p-S496E-3GFP was detected at only one pole in 73% of these cells. Thus, the S496E mutation suppressed the defect in the bik1Δkip2Δ mutant (Figure 1B). Similarly, we observed that S496E restored the asymmetric localization of Kar9p to one SPB in clb5Δ cells (Figure 1B). This supports the model that Bik1p and Clb5p act upstream in the mechanism to phosphorylate Kar9p at serine 496.
Mimicking Phosphorylation at Serine 197 Partially Suppresses the Mislocalization of Kar9p in clb4Δ Mutants
The cyclin Clb4p has also been implicated in Kar9p phosphorylation and the restriction of Kar9p to one SPB (Liakopoulos et al., 2003; Maekawa et al., 2004). Consistent with these reports, we observed that Kar9p asymmetry was lost in clb4Δ cells. Kar9p-3GFP was present at only one SPB in 27% of clb4Δ cells with short bipolar spindles (Figure 1C, quantified in D). To test whether pseudophosphorylation at serine 496 might suppress this defect, we scored Kar9p-S496E-3GFP localization in clb4Δ. Kar9p-S496E-3GFP was observed at one SPB in 23% of clb4Δ cells. Thus, unlike clb5Δ, mimicking phosphorylation at serine 496 did not restore Kar9p localization to wild-type frequencies in clb4Δ. This raised the possibility that Clb4p may affect Kar9p localization through a separate mechanism. Liakopoulos et al. (2003) identified serine 197 as an additional Cdc28p phosphorylation site in Kar9p. To investigate the role of phosphorylation at this site, we mutated serine 197 to alanine. As expected, this resulted in the mislocalization of Kar9p to both poles (Figure 1C, quantified in D). Kar9p-S197A-3GFP was detected at only one pole in 24% of wild-type cells with short bipolar spindles, whereas 76% of cells exhibited GFP signal at both poles. The severity of this defect was similar to that of wild-type Kar9p-3GFP in cells lacking Clb4p. This is consistent with the possibility that Clb4p promotes the phosphorylation of this residue. We therefore tested whether a phosphomimetic mutation at serine 197 (S197E) could suppress the mislocalization seen in the clb4Δ mutant. In clb4Δ cells, Kar9p-S197E-3GFP localized to both poles at a frequency similar to that observed for wild-type Kar9p in the clb4Δ background (Figure 1D). Thus, the pseudophosphorylation at serine 197 did not suppress the Kar9p localization defect seen in clb4Δ. One explanation for this result is based on the fact that a glutamic acid residue introduces only one negative charge, whereas a phosphate moiety carries two negative charges. To determine whether an additional negative charge in this area would suppress the mislocalization defect in clb4Δ, we introduced a second glutamic acid residue at alanine 196 immediately adjacent to the serine 197, creating A196E S197E. Forty-three percent of clb4Δ cells expressing this allele displayed Kar9p localization at one pole. Thus, Kar9p-A196E S197E partially suppressed the SPB mislocalization defect of clb4Δ.
In contrast, the Kar9p-A196E S197E mutant did not suppress either bik1Δ or clb5Δ (Figure 1D). However, it did exacerbate the localization defect seen in clb5Δ. This additive defect is consistent with the idea that Clb5p acts separately from serine 197.
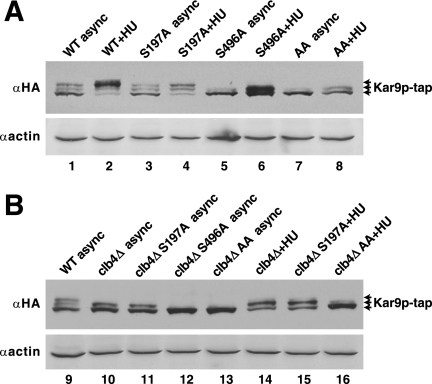
Clb4p and Serine 197
To determine whether Clb4p phosphorylates serine 197, we examined the effect of clb4Δ on the phosphorylation pattern of Kar9p-tap by immunoblotting. As reported previously, three bands of Kar9p-tap were detected in extracts from asynchronous culture of wild-type cells (Figure 2A, lane 1), and the two slower migrating bands were identified as phosphorylated isoforms by phosphatase treatment (Moore et al., 2006). Treatment with hydroxyurea to arrest cells in S phase enriched for the slowest migrating band (Figure 2A, lane 2) (Moore et al., 2006). To investigate whether Clb4p contributes to the phosphorylation of Kar9p at either serine 197 or 496, we first sought to identify which bands were dependent upon phosphorylation at these sites. For this, we incorporated a carboxy-terminal Tap tag at the genomic locus of these kar9 mutants (Moore et al., 2006). In asynchronous cultures, replacement of serine 197 with an alanine residue (S197A) had a minimal effect on the three bands (lane 3). However with hydroxyurea treatment, S197A decreased the intensity of the slowest migrating band and moderately enriched the middle band (lane 4). This moderate effect suggests that only a small fraction of the total Kar9p in asynchronous cultures is phosphorylated at serine 197 and that this residue may be transiently phosphorylated during S phase.
Figure 2.
Clb4p may promote the phosphorylation of Kar9p at serine 197. Asynchronous cultures were grown to mid-log phase and lysed by vortexing with glass beads. For S-phase arrest, 100 mM HU was added to early log phase cells for 2 h before cell lysis. Anti-HA was used to probe for the HA epitope within the tap tag. (A) Samples prepared from asynchronous or hydroxyurea-arrested cultures expressing Kar9p-tap from the chromosomal KAR9 locus (yRM4366), Kar9p-S197A-tap (yRM6168), Kar9p-S496A-tap (yRM6015), and Kar9p-AA-tap (yRM6170) in the wild-type background. Anti-actin was used as a loading control. (B) Samples prepared from cultures expressing Kar9p-tap in the wild-type background (yRM4366), Kar9p-tap in clb4Δ (yRM6094), Kar9p-S197A-tap in clb4Δ (yRM6211), Kar9p-S496A-tap in clb4Δ (yRM6142), and Kar9p-AA-tap in clb4Δ (yRM6235). async, asynchronous; WT, wild type.
In contrast to S197A, inhibiting phosphorylation at serine 496 (S496A) eliminated the slowest migrating band in both asynchronous and hydroxyurea-arrested cells (Figure 2A, lanes 5 and 6; Moore et al., 2006), suggesting that phosphorylation at serine 496 is required for the formation of the slowest migrating band. Therefore, the majority of Kar9p is phosphorylated at serine 496 during S phase. Furthermore, we observed a faint intermediate band in the S197A S496A double mutant in both asynchronous and S-phase–arrested cells. This indicates that at least one additional phosphorylation site exists on Kar9p (lanes 7 and 8) as postulated previously (Maekawa and Schiebel, 2004; Moore et al., 2006).
To determine how Clb4p contributes to these phosphorylation bands, we examined extracts from clb4Δ cells. In clb4Δ, Kar9p ran as a doublet and lacked the slowest migrating band in both asynchronous and HU-treated cells (Figure 2B, lanes 10 and 14). This suggests that at least one phosphorylation event is impeded in the clb4Δ mutant. To determine whether serine 197 or 496 is the target of Clb4p-dependent phosphorylation, we tested whether phospho-inhibiting mutations at either site would further diminish the Kar9p-tap isoforms observed in the clb4Δ strain. For both asynchronous and HU-treated cultures, Kar9-S197A-tap in a clb4Δ background ran as doublet and seemed identical to wild-type Kar9p-tap seen in this background (Figure 2B, lanes 11 and 15). Thus, S197A did not confer an additive defect to that seen in clb4Δ extracts. This is consistent with the premise that Clb4p phosphorylates serine 197. When S496A was combined with clb4Δ, a single Kar9p-tap band resulted that corresponded to the fastest migrating band of the Kar9p-tap triplet (lane 12, also compare lane 16). Thus, S496A did confer an additive defect in combination with clb4Δ. This suggests that the phosphorylation of serine 496 is not dependent on Clb4p.
Although S197A did not diminish the isoforms of Kar9p-tap observed in clb4Δ (Figure 2B, lane 11), the single clb4Δ and the single S197A mutations did not produce an equivalent banding pattern in either asynchronous (compare lanes 3 and 10) or hydroxyurea-treated cultures (compare lanes 4 and 14). In both cases, the deletion of CLB4 seemed to have a more pronounced effect on Kar9p phosphorylation than the S197A mutation alone. The treatment with hydroxyurea argues against this being due to a difference in the cell cycle position of clb4Δ. Thus, it is likely that Clb4p targets serine 197 and an additional residue on Kar9p. Consistent with this, we did observe a faint slower migrating Kar9p-tap band in the S197A S496A mutant arrested with hydroxyurea (Figure 2A, lane 8). However a similar, albeit less intense band, for this mutant is present in the absence of Clb4p (Figure 2B, lane 16), perhaps indicating the presence of a Clb4p-independent site. The nature of this modification and whether it represents additional means of regulating Kar9p function remains an intriguing question.
Phosphorylation at Serine 197, but Not 496, Is Required in the Absence of Dynein
In the absence of Kar9p, spindle positioning is accomplished through the compensatory function of the dynein pathway that draws the spindle across the bud neck by exerting pulling forces on cMTs, presumably from the bud cortex (Kahana et al., 1995; Yeh et al., 1995; Adames and Cooper, 2000). Simultaneous mutations in both the Kar9p and dynein pathways result in synthetic lethality (Miller and Rose, 1998). In a recent a genome-wide screen, Tong et al. (2004) demonstrated a synthetic lethal interaction between clb4Δ and mutants in the dynein pathway (Tong et al., 2004). This suggests that Clb4p may contribute to an important function of the Kar9p pathway. If the phosphorylation of Kar9p at serine 197 is the essential role that Clb4p plays in the Kar9p pathway, then kar9-S197A should also be synthetically lethal with mutants in the dynein pathway. Indeed, we found this to be the case. In haploid cells generated from meiotic crosses, double mutants containing kar9-S197A-tap in combination with mutations in the dynein pathway (either dyn1Δ, jnm1Δ, or bik1Δ) either exhibited no growth or formed microcolonies with a severe growth impairment (Table 2 and Supplemental Figure S1, A). In contrast, kar9-S496A-tap produced no obvious growth defects when combined with mutations in the dynein pathway (Supplemental Figure S1, B). Thus, the function of the two phosphorylation sites can be separated genetically. This suggests that phosphorylation at serine 197, but not serine 496, contributes to the essential function of Kar9p that is revealed by the absence of dynein. These results are consistent with the observation that the kar9-S197A S496A mutant does not suppress the inviability of the kar9Δdyn1Δ double mutant (Liakopoulos et al., 2003).
Table 2.
Phosphorylation of Ser 197 but not Ser 496 is essential for Kar9p function
| Cross | Viability | Tetrads (PD:TT:NPD) | Double mutants (viable/predicted) |
|---|---|---|---|
| kar9-AA × dyn1Δ | Not viable | 19 (5:10:4) | 5/18 |
| S197A × dyn1Δ | Not viable | 19 (7:10:2) | 1/14 |
| S496A × dyn1Δ | Viable | 18 (1:15:2) | 19/19 |
| S197A × jnm1Δ | Not viable | 20 (2:13:5) | 1/23 |
| S496A × jnm1Δ | Viable | 19 (2:16:1) | 17/18 |
| S197A × bik1Δ | Not viable | 14 (3:9:2) | 0/13 |
| S496A × bik1Δ | Viable | 15 (3:10:2) | 13/14 |
| S197A × bim1Δ | Viable | 17 (3:11:3) | 15/17 |
| S496A × bim1Δ | Viable | 20 (2:14:4) | 19/22 |
| S197A ×clb4Δ | Viable | 15 (3:9:3) | 14/15 |
| S496A ×clb4Δ | Viable | 15 (2:10:2) | 13/14 |
| kar9-AA ×clb4Δ | Viable | 18 (2:11:5) | 20/21 |
| kar9Δ ×clb4Δ | Viable | 19 (5:12:2) | 16/16 |
| dyn1Δ ×clb4Δ | Sick | 18 (3:10:5) | 11/20 |
| jnm1Δ ×clb4Δ | Sick | 19 (1:15:3) | 3/21 |
Double mutants were generated by meiotic crosses and scored for growth after 2-3 d. Progeny exhibiting no growth or forming microcolonies ∼10-fold smaller than single mutant or wild-type cells (see Supplemental Figure S1) were scored as “inviable.” Microcolonies also exhibited poor growth upon streaking to new media. The dyn1Δclb4Δ and jnm1Δclb4Δ double mutants were scored as “sick” as these colonies were smaller than those produced by single mutants but did not exhibit phenotypes as severe as those observed for other double mutants (see Supplemental Figure S1). Of the 20 predicted dyn1Δ × clb4Δ double mutants, 11 produced colonies equivalent to the size of those formed by single mutants. Six produced small colonies that displayed severely retarded growth, and three exhibited no growth at all. For the 21 predicted jnm1Δ × clb4Δ double mutants, three formed colonies similar to the size of single mutants. Eight produced small colonies that exhibited severely retarded growth, and 10 exhibited no growth. The following parental strains were used in these crosses: Kar9p-AA-tap (yRM6170), Kar9p-S197A-tap (yRM6168), Kar9p-S496A-tap (yRM6015), dyn1Δ (yRM1094), jnm1Δ (yRM469), bik1Δ (yRM565), bim1Δ (yRM2060), clb4Δ (yRM6152), and kar9Δ (yRM369). The clb4Δ strain yRM6026 was used for crosses with dyn1Δ, jnm1Δ, and kar9Δ.
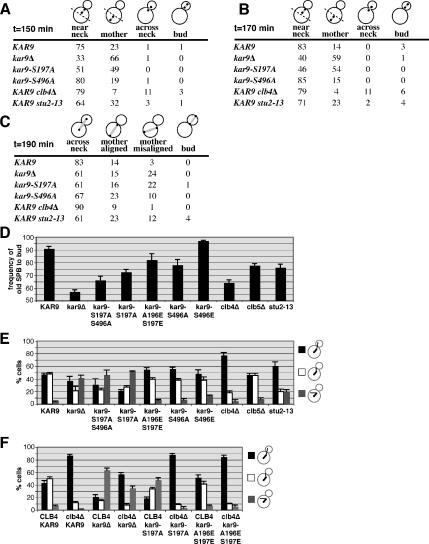
To gain further understanding of the functional differences of these two phosphorylation sites, we examined the position of short bipolar spindles marked with Spc110p-CFP in both of these mutants. Cells were synchronized in stationary phase as described and then released into G1 phase (Allen et al., 2006). In wild-type cells at 150 min after release, 75% of preanaphase bipolar spindles were located within the mother cell and adjacent to the bud neck. In kar9Δ mutants, only 33% were positioned at the neck. Instead, 66% of the spindles were located at the distal end of the mother. In kar9-S197A, 49% of the spindles were located distal to the bud neck, a phenotype similar to kar9Δ (Figure 3A). In contrast, the placement of the spindle in kar9-S496A seemed similar to wild type in this assay at both 150 and 170 min post-release (Figure 3, A and B). By anaphase, both kar9-S197A and kar9-S496A displayed an increase in the number of spindles elongating within the mother cell and misaligned with respect to the long axis of division (Figure 3C). For all time points, the severity of the S197A phenotype is greater than the S496A phenotype, correlating with the strength of the genetic interactions seen in the absence of dynein. Similar results were seen in asynchronous cultures examining preanaphase spindles (Supplemental Figure S2).
Figure 3.
Mutations altering the phosphorylation status of Kar9p produce defects in spindle positioning and/or microtubule orientation. (A) Spindle position shown as the percentage of total cells with short bipolar spindles. Spindle position was scored in WT (yRM6374), kar9Δ (yRM6900), kar9-S197A (yRM6144), kar9-S496A (yRM6007), clb4Δ (yRM6099), and stu2-13 (yRM6601) cells with SPBs labeled with SPC110-CFP. Quiescent populations of cells were isolated using the methods of Allen et al. (2006) and released into fresh media to initiate synchronous reentry into the cell cycle and then briefly fixed in formaldehyde. At 150 min after introduction into fresh media, the majority of cells in each strain contained short bipolar spindles. Cells in which both poles were located in the bud proximal half of the mother cell were scored as “near neck.” If at least one pole was located in the distal half of the mother cell, the cell was scored at “mother.” Cells displaying one pole on either side of the bud neck were scored as “across neck.” Cells in which both poles were located in the bud were scored as “bud.” At least 100 cells were scored for each strain. The stu2-13 strain (yRM6601) was derived from CUY1143 (Kosco et al., 2001) and is not isogenic with other strains used in this experiment. (B) At 170 min after release cells were scored as described in A. (C) The position of anaphase spindles at 190 min after release. Anaphase spindles were identified by Spc110p- labeled poles that were separated by more than 0.5 μm. Cells in which the two poles were located on opposite sides of the bud neck were score as “across neck.” Cells displaying both poles within the mother cell but aligned parallel to the long axis of division were scored as “mother aligned.” If both poles were contained within the mother cell but the alignment of the spindle deviated from the axis of division, the cells were scored as “mother misaligned.” Cells in which both poles were located in the bud were scored as “bud.” At least 100 cells were scored for each strain. (D) Kar9p phosphorylation is important for directing SPB inheritance. Quantification of cells with the older SPB proximal to the bud in wild type (yRM6902; n = 500), kar9Δ (yRM5031; n = 200), kar9-S197A S496A (yRM6904; n = 200), kar9-S197A (yRM6905; n = 200), kar9-A196E S197E (yRM6903; n = 200), kar9-S496A (yRM6906; n = 255), kar9-S496E (yRM6901; n = 200), clb4Δ (yRM6246; n = 250), clb5Δ (yRM5509; n = 250), and stu2-13 (yRM6601; n = 200). Only cells with a bipolar spindle oriented along the long axis of the mother-bud were scored. The origin was set at 50% because this represents random inheritance. Error bars represent the SEM of the percentages obtained from at least four experiments. (E) Microtubule orientation was scored in the following preanaphase cells, WT (yRM3886), kar9Δ (yRM6907), kar9-S197A S496A (yRM6908), kar9-S197A (yRM6110), kar9-A196E S197E (yRM6275), kar9-S496A (yRM6602), kar9-S496E (yRM6909), clb4Δ (yRM6910), clb5Δ (yRM6911), and stu2-13 (yRM6518). Cells with at least one cMT entering the bud were scored as “bud.” Cells with no cMTs entering the bud, but at least one terminating at the bud neck were scored as “neck.” Cells with no cMTs entering the bud or reaching the bud neck, but with cMTs visible in the mother cell were scored as “mother.” Error bars represent the SE of the mean. At least 200 cells were scored for each strain. The stu2-13 strain (yRM6518) was derived from CUY1143 (Kosco et al., 2001) and is not isogenic with other strains used in this experiment. (F) The microtubule orientation defect of clb4Δ is only partially dependent on KAR9. Microtubule orientation was scored as described for E in the following preanaphase cells, CLB4 KAR9-3GFP (yRM3886), clb4Δ KAR9-3GFP (yRM6912), CLB4 kar9Δ (yRM6907), clb4Δ kar9Δ (yRM6914), CLB4 kar9-S197A-3GFP (yRM6110), clb4Δ kar9-S197A-3GFP (yRM6915), CLB4 kar9-A196E S197E-3GFP (yRM6275), and clb4Δ kar9-A196E S197E-3GFP (yRM6916). At least 200 cells were scored for each strain.
Kar9p plays an important role in directing only one SPB, usually the old SPB, into the bud. To characterize the role that phosphorylation at these sites plays in SPB inheritance, we assayed the frequency at which the old SPB was transferred into the bud. For this assay, SPBs were marked with Spc110p conjugated with DsRed.T1.N1. Using this fluore, the older SPB appears brighter than the newly synthesized SPB (Pereira et al., 2001; Yoder et al., 2003; Moore et al., 2006). Consistent with previous reports, only 57% of kar9Δ mutants faithfully transferred the old SPB into the bud, indicating that SPB inheritance is nearly random in the absence of Kar9p (Figure 3D). In kar9-S197A, 72% of cells placed the old SPB into the bud. In contrast, kar9-S496A showed a less severe SPB inheritance defect. Both of these inheritance defects can be suppressed by mimicking phosphorylation at each site by using glutamic acid substitutions. clb4Δ shows a defect that is worse than that of kar9-S197A. The defect seen in clb5Δ is similar to that seen in kar9-S496A. These data are consistent with the model that the phosphorylation of Kar9p is important for ensuring the fidelity of SPB inheritance.
To explore the basis of the spindle displacement defects in these phosphoinhibited mutants, we examined the orientation of cMTs in cells with short bipolar spindles. CFP-Tub1p–labeled microtubules were scored as either extending into the bud, interacting with the bud neck, or misoriented in the mother cell. In wild-type cells, >94% of microtubules either entered the bud or contacted the bud neck (Figure 3E). In contrast for kar9-S197A, only 49% of microtubules were oriented in this manner. This defect was rescued by the phosphomimetic mutation kar9-A196E S197E in which nearly 94% of cells display microtubules oriented either into the bud or to the bud neck. This suggests that the phosphorylation of serine 197 is required for Kar9p to guide cMT ends to the bud. For both the kar9-S496A mutant and the clb5Δ strains, we did not observe a microtubule orientation defect. As reported previously by Maekawa and Schiebel (2004), we found that the clb4Δ strain exhibited a significant increase in the number of microtubules extending into the bud, confirming their findings (p < 0.0001 compared with wild-type cells).
The role of Clb4p in antagonizing the interaction of microtubules with the bud cortex is thought to be dependent upon the function of Kar9p in guiding microtubule ends toward the bud and delivering Clb4p to those microtubule ends. In this model, the microtubule orientation defect seen in kar9Δ would be predicted to be epistatic to the persistent bud-directed microtubules seen in clb4Δ. To test this prediction, microtubule orientation was scored in the clb4Δkar9Δ double mutant. The double mutant displayed a microtubule orientation defect that was intermediate between the two single mutants (Figure 3F). These data suggest that Clb4p has both Kar9p-dependent and -independent functions for microtubule orientation and that Clb4p may influence microtubule–cortex interactions through factors other than Kar9p. We also observed the kar9-S197A clb4Δ mutant displays more bud-oriented microtubules than the kar9Δ clb4Δ double, consistent with the idea that the S197A allele maintains some degree of Kar9p function that enhances the guidance of microtubule ends into the bud.
Phosphorylation at Serine 197 Modulates the Kar9p–Stu2p Two-Hybrid Interaction
The phosphorylation of Kar9p is likely to be translated into its asymmetric localization to one SPB by modulating an interaction between Kar9p and an SPB-associated factor. We considered two candidates for controlling the loading of Kar9p onto the SPBs, Bim1p and Stu2p. Both localize to the SPBs, interact with Kar9p physically, and function in the Kar9p genetic pathway (Lee et al., 2000; Miller et al., 2000; Kosco et al., 2001; Wolyniak et al., 2006). In bim1Δ cells, endogenous levels of Kar9p are not detected on SPBs or cMTs (Liakopoulos et al., 2003; our unpublished observations). The role of Stu2p in Kar9p localization has remained unclear (Miller et al., 2000).
To test whether the interactions of Kar9p with either Bim1p or Stu2p were sensitive to the phosphorylation status of Kar9p, we used a two-hybrid approach. BIM1-AD and STU2-AD fusions were scored for interaction with kar9 mutants that either inhibited or mimicked phosphorylation at serines 197 and 496. Because we observed that Bim1p enhanced the two-hybrid interaction between Stu2p and the carboxy-terminal region Kar9p (Figure 5), we carried these assays out in a reporter strain deleted for BIM1. As shown in Figure 4A, the Kar9p–Stu2p interaction was decreased by the phosphomimetic S197E mutation and abrogated by the A196E S197E mutation, with its greater negative charge. Conversely, the interaction was enhanced by preventing phosphorylation at serine 197 by using the S197A mutation. At the 496 site, preventing phosphorylation had little apparent effect on the interaction, whereas the S496E mutation enhanced the interaction. We then investigated whether combinations of these residues produced an additive effect on the Kar9p–Stu2p interaction. The A196E–S197E S496A combination did not interact with Stu2p, similar to the A196E–S197E mutation. The S197A S496E double mutant displayed a slight enhancement of the interaction compared with wild-type Kar9p, like the 197A single mutation. Surprisingly, the S197A S496A mutation displayed the greatest enhancement of the Kar9p–Stu2p interaction. This enhancement was most obvious on −Ade plates (Figure 4A). Thus, the enhancement of the interaction by the S197A and S496E mutations did not act in a simple additive manner. These effects were not as readily pronounced in the wild-type two-hybrid reporter strain (data not shown). We also tried to investigate the Stu2p–Kar9p interaction at endogenous protein levels by using immunoprecipitation techniques. These results, however, were inconclusive in part due to the high background level of epitope-tagged Stu2p on the negative control beads (data not shown). Nevertheless, the two-hybrid data suggest that the interaction of Stu2p with Kar9p may be regulated by the phosphorylation of serine 197.
Figure 5.
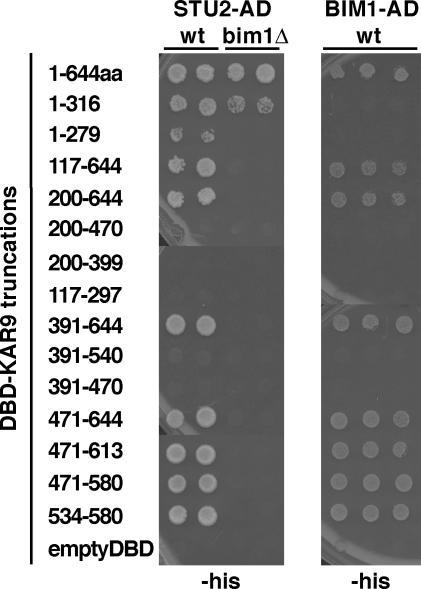
Two regions of Kar9p are sufficient for Stu2p binding, whereas one region is sufficient for Bim1p binding. Truncations of KAR9 were fused to the GAL4-DNA binding domain and assayed for interaction with STU2Δ649-AD (pRM1916) and BIM1187-276-AD (pRM2023). Two independent colonies were analyzed for each STU2-AD set. Three independent colonies were analyzed for each BIM1-AD set. The empty DBD and AD plasmids were used as controls. WT, wild type two-hybrid reporter strain. bim1Δ, two-hybrid reporter strain deleted for BIM1.
Figure 4.
Phosphorylation at serine 197 inhibits the two-hybrid interaction of KAR9 with STU2, but not BIM1. (A) Full-length KAR9 fused to the GAL4-DBD was assayed for interaction with STU2Δ649 (pRM1916) and BIM1187-276 (pRM2023) fused to the GAL4-activation domain in a two-hybrid reporter strain in which BIM1 was disrupted (yRM2057). Three independent colonies were spotted onto SC plates lacking histidine (−His) or adenine (−Ade). Cells were scored after 3 d of growth at 30°C. Growth was equivalent for all spots on plates lacking uracil and leucine to demonstrate the presence of both plasmids (data not shown). Mimicking phosphorylation at serine 197 through DBD-kar9-S197E (pRM5617) and DBD-kar9-A196E S197E (pRM6255) disrupted the two-hybrid interaction with STU2. The interaction with BIM1 did not seem to be affected by the phosphorylation status of either residue S197 or S496. AD, activation domain; DBD, DNA binding domain. (B) The Kar9p–Bim1p interaction is not dependent on the phosphorylation status of Kar9p. Kar9p-tap (yRM6920), Kar9p-S197A-tap (yRM6919), Kar9p-A196E S197E-tap (yRM6918), Kar9p-S496A-tap (yRM6921), Kar9p-S496E-tap (yRM6923), and Kar9p-S197A S496A-tap (yRM6922) were affinity purified from exponentially growing cultures by using IgG-Sepharose. A parallel experiment was carried out in a wild-type strain in which chromosomal KAR9 was not tap-tagged (yRM6917). This shows that Bim1p did not interact significantly with the IgG-Sepharose. The precipitates were analyzed by immunoblotting with anti-HA to detect Kar9p-tap and rabbit anti-Bim1p.
Phosphorylation has been postulated to regulate the interaction between Kar9p and Bim1p (Liakopoulos et al., 2003). Serine 496 of Kar9p is conserved in mammalian APC, and phosphorylation of this region of APC attenuates its binding to the Bim1p homologue EB1 (Askham et al., 2000; Honnappa et al., 2005). Therefore, we tested whether the phosphorylation status of Kar9p at serine 496 or serine 197 might regulate the interaction between Kar9p and Bim1p. As seen in Figure 4A, Bim1p interacted equally well with each kar9 phosphomutant by two-hybrid analysis. Consistent with this, similar amounts of Bim1p copurified with Kar9p-tap regardless of its phosphorylation status (Figure 4B), albeit less Kar9p-A196E S197E-tap was precipitated. Although it is possible that the effect of Kar9p phosphorylation on Bim1p binding is beyond the sensitivity of these assays, these results suggest that Kar9p phosphorylation does not significantly affect its interaction with Bim1p.
Stu2p Interacts with Two Regions of Kar9p
The observation that the interaction with Stu2p is inhibited by phosphorylation at serine 197 suggested the possibility that Stu2p interacts with this region of Kar9p. Therefore, we mapped the regions of Kar9p that interact with Stu2p by two-hybrid analysis as described previously (Moore et al., 2006). As shown in Figure 5, two regions of Kar9p were sufficient for interaction with Stu2p. The first region lay in the amino-terminal half of Kar9p, from amino acids 1 to 316. The second region was a 47-amino acid region located in the carboxy-terminal third of the protein from amino acid 534 to 580. This 47-amino acid region was also sufficient for interaction with Bim1p (Figure 5). This region contains a portion of the EB1 binding site that is conserved in APC (Bienz, 2001; Honnappa et al., 2005). Because Bim1p interacts with Stu2p (Chen et al., 1998), we tested whether BIM1 was required for either of these interactions (Miller et al., 2000). The 534-580 amino acid (aa) region of Kar9p failed to interact with STU2-AD when BIM1 was deleted from the two-hybrid reporter strain, whereas the interaction with the 1-316 aa region was retained. These data suggest that Stu2p may interact with Kar9p through Bim1p-dependent and Bim1p-independent mechanisms.
Stu2p Is Required for the Accumulation of Kar9p at SPBs
If phosphorylation of Kar9p at serine 197 modulates the stability of a Kar9p–Stu2p interaction at the SPB, then stu2 mutants would be expected to also affect the localization of Kar9p to the SPBs. Because STU2 is an essential gene, we used the temperature-sensitive stu2-13 allele generated by Kosco et al. (2001). Strains bearing this allele are inviable at 37°C. However, at 30°C the stu2-13 single mutant is viable, but it exhibits synthetic lethality with mutants of the dynein pathway (Kosco et al., 2001). To confirm that stu2-13 exhibits defects in Kar9p-dependent processes, we analyzed the position of preanaphase spindles and spindle pole body inheritance in the stu2-13 mutant at the permissive temperature (Figure 3, A–E). For spindle positioning, the stu2-13 mutant displayed a slightly increased percentage of short bipolar spindles located distal to the bud (Figure 3, A and B, p < 0.0001 compared with wild type), similar to kar9Δ albeit less severe. We also found that the combined rate of anaphase occurring within the mother was similar between kar9Δ and stu2-13. However, the rate at which anaphase spindles were misaligned in the mother cell was less severe in stu2-13 than in kar9Δ (Figure 3C). We also characterized the rate of old versus new SPB inheritance in stu2-13 by using Spc110p marked with the DsRed fluore. Approximately 75% of stu-13 cells directed the older pole to the daughter cell. These data together with the genetic data of Kosco et al. (2001) suggest that the function of the stu2-13 mutant in the Kar9p pathway is compromised even at the permissive temperature.
We next assessed the localization of Kar9p-3GFP in stu2-13 mutants. These cells exhibited bright foci of Kar9p-3GFP that were not associated with the SPBs marked with Spc110p-DsRed. These Kar9p foci moved dynamically through the cytoplasm, often contacting the cortex of the bud (data not shown; see Supplemental Movies). We then confirmed that these foci were localized at microtubules (MT) plus ends by using stu2-13 cells labeled with CFP-Tub1p (Figure 6A).
Figure 6.
Stu2p is required for the accumulation of Kar9p at the SPB. (A) Examples of Kar9p-3GFP (yRM3886), Kar9p-S197A-3GFP (yRM6110), and Kar9p-A196E S197E-3GFP (yRM6275) in a wild-type background, Kar9p-3GFP in the stu2-13 mutant (yRM6518), all containing CFP-Tub1p labeled MTs. (B) Quantification of the fluorescence intensity of Kar9p-3GFP at SPBs and the plus ends of cMTs. At least 70 cells were scored for each strain. Cells were grown in SC media −Trp at 30°C. Error bars denote the SEM within each data set. (C) Quantification of the fluorescence intensity of Kar9p-3GFP at the SPBs of unbudded cells from the same captured images used in B.
In wild-type cells, Kar9p is often present simultaneously at the SPBs and plus ends of cMTs. The distribution of Kar9p between these two sites is dynamic, with Kar9p accumulating at the SPBs and eventually moving out from the SPB toward the plus end (Liakopoulos et al., 2003; Maekawa et al., 2003). To compare the amount of plus end localization of Kar9p versus its SPB localization in stu2-13, we quantified the intensity of the Kar9p-GFP signal at both the SPB and the plus end in cells containing short bipolar spindles and cyan fluorescent protein (CFP)-labeled microtubules. In wild-type cells, the average fluorescence intensity of Kar9p foci at cMT plus ends was less intense than at the SPB (Figure 6B). In contrast, stu2-13 cells exhibited an average fluorescence intensity at cMT plus ends that was approximately threefold greater than that at the SPBs. Furthermore, the Kar9p foci at the plus ends of stu2-13 cells were nearly twofold more intense than the average plus end foci observed in wild-type cells (Figure 6B). The stu2-13 mutants also displayed a less intense Kar9p–3GFP signal at the SPB during the G1 phase of the cell cycle than wild type (Figure 6C). Thus, Kar9p is enriched at plus ends and diminished at the SPBs in the stu2-13 mutant. These data support a role for Stu2p in binding Kar9p at the SPB.
Because the pseudophosphorylation of Kar9p at serine 197 disrupts the two-hybrid interaction of Kar9p with Stu2p, we tested whether Kar9p-A196E S197E-3GFP would also display an increased accumulation of Kar9p at the plus ends of cMTs. Indeed, in comparison to preanaphase cells expressing wild-type Kar9p, this phospho-mimic exhibited substantially more GFP signal at plus ends than SPBs (Figure 6B). In contrast, the phospho-inhibited S197A mutant did not display this increase. Thus, the phosphorylation of serine 197 is important for regulating the accumulation of Kar9p at the SPB.
DISCUSSION
The restriction of Kar9p to one SPB and set of cMTs is important for directing a specific spindle pole toward the nascent daughter cell during cell division. Two phosphorylation sites were previously identified by Liakopoulos et al. (2003) for directing the localization of Kar9p to a specific set of cMTs. Although both sites are phosphorylated by Cdc28p (Liakopoulos et al., 2003; Maekawa et al., 2003), the results presented in this work suggest that the two modifications act by different mechanisms to regulate the localization and function of Kar9p. We tested the hypothesis that specificity is conferred by distinct cyclins and target residues on Kar9p. Although additional regulation may exist, we propose a working model in which Clb5p promotes the phosphorylation of serine 496, and Clb4p targets serine 197 for phosphorylation. These modifications enable Kar9p to recognize inherent differences between the two SPBs. We present evidence that phosphorylation at serine 197 also disrupts the interaction between Kar9p and the SPB-associated Stu2p, a novel role for this MAP. In addition, our results indicate that the phosphorylation of serine 197 is required for Kar9p to direct cMTs into the bud. These findings provide new insight into how centrosomes may influence the establishment of asymmetric microtubule arrays during cell division.
Our findings reported here extend several observations made previously and reconcile some apparent points of difference in the literature. Work from both the Barral and Schiebel groups showed that Clb4p was important for the asymmetric localization of Kar9p on SPBs (Liakopoulos et al., 2003; Maekawa et al., 2004). Results from the Barral group indicated that the effect of clb4Δ mutations on Kar9p asymmetry is somewhat stronger than those seen by the Schiebel group, although their scoring parameters were different. Both groups agreed that the Kar9p–S197A S496A mutant mislocalizes to both poles. However, the two groups differed on whether Clb5p has an effect on Kar9p asymmetry. The Schiebel group showed that Kar9p and Clb5p interact by two-hybrid analysis (Maekawa et al., 2003). Their work also suggested that clb5Δ mutations have a modest effect in increasing the amount of Kar9p at the mother-directed SPB (Maekawa et al., 2004). In contrast, the Barral group concluded that Clb5p has no effect (Liakopoulos et al., 2003).
From our data comparing Kar9p localization in both cyclin mutants (Figure 1, B and D), we conclude that each has an effect on Kar9p asymmetry but to differing extents. Clb4p has the stronger effect, with Clb5p making a modest contribution to Kar9p asymmetry. Consistent with this, alanine mutations preventing phosphorylation at serine 197 produce a stronger disruption of Kar9p asymmetry than do similar mutations at serine 496.
Several differences between the experimental protocols may explain some of the apparently discrepant observations. A portion of the work from the Schiebel laboratory (Maekawa et al., 2003) was carried out in strains containing double mutations between cdc28-4 and cyclin deletions. Even at the permissive temperature, the cdc28-4 mutation limits the overall activity of the kinase. Our work and the Liakopoulos work used strains deleted for cyclins in a wild-type CDC28 background. Another difference is the fluore used in the fusion to visualize Kar9p. The Barral laboratory used one yellow fluorescent protein fused to Kar9p and the Schiebel laboratory used one GFP, whereas our fusion was made with three tandem GFPs. Our construct may result in an increased level of sensitivity in detecting smaller amounts of Kar9p, thus increasing the level of partial asymmetry detected. Furthermore, it is also possible that strain differences may account for a portion of the observed differences.
An important contribution of our study is the analysis of the impact of individual phosphorylation events on spindle positioning and Kar9p function. This reveals that CDK-phosphorylation of distinct sites produces different effects not only on Kar9p localization but also on the Kar9p-dependent linkage of cMTs to cortical polarity. Although the inhibition of phosphorylation at serine 496 disrupts Kar9p asymmetry at the SPBs, genetic and cell biological assays indicate that Kar9p-S496A retains a significant degree of functionality. In contrast, preventing phosphorylation of serine 197 disrupts the delivery of cMT plus ends into the bud. This result is likely to account for the aberrant placement of preanaphase spindles away the bud neck (Figure 3, A–C) and the impaired inheritance of the older SPB into the daughter cell (Figure 3D).
Phosphorylation at Serine 496 Enables Kar9p to Act on a Preexisting Asymmetry at the SPBs
During S phase, the majority of Kar9p is phosphorylated at serine 496 and mimicking this phosphorylation directs Kar9p to one SPB. Although this does not preclude the possibility that serine 496 could be phosphorylated at a specific subcellular site, the fact that Kar9p-S496E localizes to one SPB demonstrates that the phosphorylation event itself need not be restricted to a specific location. Furthermore, the viability of Kar9p-S496E in combination with the bik1Δ and kip2Δ mutations indicates that this phosphomimic is accomplishing the functions of the wild-type protein. These findings argue against a model in which an asymmetrically localized kinase phosphorylates serine 496 to generate Kar9p asymmetry and are instead consistent with a model in which this phosphorylation alters the interaction of Kar9p with a selective factor that is present at one of the two poles. This factor could either repel phosphorylated Kar9p from the mother bound SPB or recruit it to the daughter-bound SPB.
Although the identity of this factor is not yet clear, our data do not indicate that Bim1p acts as this selective factor. Although Bim1p functions in loading Kar9p onto the SPB (Liakopoulos et al., 2003), our results suggest that the Kar9p–Bim1p interaction is not noticeably affected by the phosphorylation status of either serine 496 or 197 (Figure 4). Furthermore, Bim1p is present at both SPBs (Liakopoulos et al., 2003), whereas the selective factor is likely to be found at only one pole. In mammalian cells, APC has been shown to preferentially localize to one centrosome (Louie et al., 2004). APC contains a phosphorylation site that is conserved with serine 496 and is thought to regulate its interaction with EB1, the Bim1p homologue (Askham et al., 2000; Honnappa et al., 2005). Thus, these findings suggest potential differences between the yeast and mammalian systems. Interestingly, the interaction with EB1 is not required for the localization of APC to centrosomes, supporting the idea that additional factor(s) may mediate this localization (Louie et al., 2004).
We also considered the possibility that phosphorylation at serine 496 could affect Kar9p localization by regulating its stability. An enhancement of Kar9p levels is observed when the S496A mutant is arrested in S phase; however, this is not seen in the S197A S496A mutant (Figure 2A, lanes 6 and 8). Similar levels of Kar9p are also detected in asynchronous cultures expressing the S496E or S496A mutants (Figure 4B). Further work will be necessary to determine whether the phosphorylation of serine 496 plays a role in Kar9p degradation. However, the A196E S197E phosphomimetic mutant did seem to decrease Kar9p levels in cellular extracts and precipitated fractions (data not shown; Figure 4B).
Our model that Cdc28p-Clb5p phosphorylates S496 is supported by several pieces of evidence. Work from the Morgan laboratory has shown that Clb5p specifically targets Kar9p for phosphorylation (Loog and Morgan, 2005). Clb5p interacts with a region on Kar9p that contains serine 496. Inhibiting phosphorylation on this site with an alanine mutation produces a phenotype similar to that seen in clb5Δ for both Kar9p localization (Moore et al., 2006) and SPB inheritance (Figure 3D). Furthermore, a phosphomimetic residue at position 496 suppresses the Kar9p mislocalization defect seen in clb5Δ (Figure 1B). However, in analyzing Kar9p-tap in the clb5Δ strain, a detectable alteration in Kar9p phospho-isoforms was not apparent by Western blot analysis (data not shown). Although this finding is not consistent with our hypothesis, we speculate that this may be due to clb5Δ cells being substantially delayed in S phase, when Cdc28p activity is at its peak. This delay may allow alternative cyclin–Cdk complexes to phosphorylate Kar9p and mask the putative clb5Δ defect.
Does Clb4p-dependent Phosphorylation Regulate Kar9p?
Several pieces of data support the idea that Clb4p targets serine 197 for phosphorylation. First, a mutant form of Clb4p interacts with Kar9p (Moore et al., 2006). Second, phosphomimetic mutations at serine 197 partially suppress the mislocalization of Kar9p to both SPBs in clb4Δ. Third, the serine 197A mutation does not result in an additive effect on the banding pattern of Kar9p when combined with clb4Δ. Fourth, both Clb4p and phosphorylation at serine 197 are required for viability in the absence of dynein function. Fifth, both S197A and clb4Δ display a defect in SPB inheritance.
We find that preventing phosphorylation at serine 197 does not severely alter the banding pattern of Kar9p visualized by Western blot analysis from asynchronous cultures (Figure 2A). This suggests that in comparison with the 496 site, which does significantly change the banding pattern, only a small pool of Kar9p is phosphorylated at serine 197. It is possible that this phosphorylation event may be restricted to specific regions of the cell and/or short periods during the cell cycle.
Overexpression studies show that Clb4p localizes to the mother-bound SPB (Liakopoulos et al., 2003), whereas endogenously expressed Clb4p localizes to the daughter-bound SPB in a Kar9p-dependent manner (Maekawa and Schiebel, 2004). These data might be reconciled if Clb4p were to execute temporally distinct functions at both poles. During spindle assembly, Clb4p–Cdc28p activity could disrupt the Kar9p association with the incipient mother-bound pole, releasing Kar9p from that pole. Using time-lapse microscopy, Huisman et al. (2004) observed that Kar9p is indeed cleared from the mother-bound SPB during SPB separation (Huisman et al., 2004). Because the localization of Clb4p to the SPB is dependent on Kar9p (Maekawa and Schiebel, 2004), it is possible that Clb4p may initially associate with the mother-bound SPB but be released from that pole along with Kar9p during spindle assembly. Subsequently, Clb4p-Cdc28p could then disengage Kar9p from the bud-directed SPB for deployment to the plus end.
There are several reasons that might explain the synthetic lethality of the phospho-inhibited kar9-S197A mutant in combination with mutations in the dynein pathway. First, it is possible that the synthetic lethality results from the association of Kar9p with both SPBs, which might allow both poles to be directed into the bud. However, this does not seem like the most likely explanation, because the S496A mutation also disrupts Kar9p asymmetry, albeit to a lesser extent, and it is viable in the absence of the dynein pathway. Furthermore, we did not observe a hypermigration of preanaphase spindles into the bud for the kar9-S197A mutant (Figure 3, A–C), despite the presence of Kar9p-S197A on both SPBs and sets of cMTs. The ability of a Bim1p-Myo2p chimera to rescue the synthetic lethality of a kar9Δdyn1Δ double mutant also argues that a loss of Kar9p asymmetry does not compromise the viability of dynein mutants (Hwang et al., 2003). The Bim1p–Myo2p fusion does not display a selective localization to one set of cMTs, yet it sufficiently accomplishes the function of Kar9p by capturing cMTs from either SPB or orienting them toward the bud (Hwang et al., 2003). Interestingly, this chimera increases the frequency of hypermigrated spindles in kar9Δ cells, suggesting that this may be negatively regulated through Kar9p modulation.
Second, the synthetic lethality could be explained by our observation that Kar9-197A lacks bud-oriented cMTs. This suggests that the phosphorylation of serine 197 affects the ability of Kar9p to function at the cMT plus end. It is possible that the accumulation of Kar9p-S197A at the SPB prevents Kar9p from traveling to the cMT plus end, in which case there would be inadequate amounts of Kar9p-S197A there for it to orient microtubules to the bud. However, the fact that small amounts of Kar9p localized at the plus end of microtubules in kip2Δ mutants are sufficient for it to carry out its essential function (Moore et al., 2006) indicates that this may not be the most likely explanation. Moreover, recent work by Cuschieri et al. (2006) describes a γ-tubulin mutant that is defective for Kar9p function, despite displaying an enrichment of Kar9p at the plus end. In that study, the authors posit that the SPB could serve as a platform for the assembly of functional Kar9p complexes and that the precocious release of Kar9p from the SPB leads to the accumulation of nonfunctional Kar9p at the plus end. Thus, the SPB-asymmetry and the amount of Kar9p at the plus end may be less important for cell viability than the functionality of Kar9p at the plus end. It is also possible that phosphorylation at serine 197 is necessary for plus end-associated Kar9p to properly interact with the cortical myosin Myo2p. This notion is supported by our observation that cMTs are not oriented toward the bud in the presence of Kar9p-S197A, which is reminiscent of phenotypes reported for myo2 mutants that are deficient for interaction with Kar9p (Yin et al., 2000). More work is needed to test this possibility.
Maekawa and Schiebel (2004) showed that the localization of Clb4p to cMT plus ends is dependent on Kar9p. Therefore, it is also possible that the S197A mutant could impede the translocation of Clb4p-Cdc28p to the plus end. This might occur either because the release of kar9-S197A from the SPB is inhibited or because S197A might not interact as well with Clb4p. Arguing against this, however, is our finding that the S197A single mutant displays less microtubule orientation into the bud than does the clb4Δ kar9-S197A double mutant (Figure 3F). This suggests that Clb4p is still regulating microtubule orientation in the presence of S197A. Furthermore, the finding that clb4Δ partially suppresses the microtubule misorientation of kar9Δ, enhancing the budward orientation of microtubules, suggests that the function of Clb4p in releasing microtubules from the cortex is not completely dependent upon Kar9p. Therefore, one might speculate that Clb4p is acting on components in addition to Kar9p to release microtubules from the bud cortex, consistent with suggestions made previously by the Schiebel laboratory.
If phosphorylation of serine 197 is the only important function of Clb4p in the Kar9p pathway, then the kar9-A196E S197E mutation should suppress the growth defect seen in double mutants between clb4Δ and the dynein pathway. We tested this by using a deletion of the dynactin complex component, JNM1, the yeast homologue of dynamitin. We compared the growth of clb4Δ jnm1Δ KAR9+ and clb4Δ jnm1Δ kar9-A196E S197E colonies obtained from meiotic crosses. However, we did not observe an obvious suppression of the growth defect (n = 40 tetrads; data not shown). This finding does not support the hypothesis that Clb4p directs phosphorylation to serine 197.
The observation that the A196E S197E mutant was viable in combination with mutants of the dynein pathway (data not shown) suggests that Kar9p function requires the phosphorylated, but perhaps not the unphosphorylated, state of serine 197. This also implies that the “cycling” of the phosphate on and off serine 197 is not required for the essential function of Kar9p seen in the absence of dynein.
The model that Clb4p targets serine 197 also predicts that clb4Δ and S197A mutants would share a number of similar phenotypes. However, in contrast to their similar phenotypes for Kar9p localization, genetic interactions, and SPB inheritance (Figure 3D), the phenotypes of these mutants differed in the spindle positioning (Figure 3, A–C) and microtubule orientation assays (Figure 3, E and F). These discrepancies with the model could be based on the ability of Clb4p to phosphorylate other elements that are important for spindle positioning and microtubule stability, as postulated previously (Maekawa et al., 2004). The identification of these other elements should prove interesting for future research. Based on our Western blot analysis, it is also likely that Clb4p promotes the phosphorylation of additional sites on Kar9p (Figure 2B). It is not known whether these modifications also contribute to the regulation of Kar9p function.
Stu2p as a Regulator of Kar9p Association with the SPB
Our results suggest a model in which the phosphorylation of Kar9p at serine 197 is responsible for attenuating its interaction with Stu2p at the SPB, allowing Kar9p to be released from the SPB. At the newly synthesized SPB, a decreased interaction may act to simply clear Kar9p from this pole, whereas at the older bud-directed SPB, release from Stu2p may promote its subsequent travel to the microtubule plus end. In support of this model, phosphomimetic mutations at serine 197 disrupted the two-hybrid interaction with Stu2p and also displayed an enriched localization of Kar9p at the plus end. Similarly, the stu2-13 allele diminished the localization of Kar9p at SPBs and greatly enriched its localization at the plus ends of cMTs (Figure 6). In contrast, preventing phosphorylation at serine 197 enriched the pool of Kar9p at the SPB. Because Stu2p is at both poles, the mislocalization of Kar9p-S197A to both poles could be explained by its increased interaction with Stu2p. Together, these results suggest a novel role for Stu2p in maintaining the association of Kar9p at the SPB. We cannot, however, eliminate the possibility that the small pool of Stu2p localized at cMT plus ends (Kosco et al., 2001; Wolyniak et al., 2006) might also be a regulator of the accumulation of Kar9p there.
Our model is consistent with several other recent observations. In addition to Kar9p, Stu2p also interacts with the CLIP-170 homologue Bik1p, and it is required for the localization of Bik1p to the SPBs (Carvalho et al., 2004; Wolyniak et al., 2006). Furthermore, both Stu2p and the γ-tubulin Tub4p at the SPB can influence the dynamic behavior of cMT plus ends (Vogel and Snyder, 2000; Usui et al., 2003). Like stu2-13, the tub4Δdsyl mutant containing a deletion in its carboxy-terminal tail also results in increased amounts of Kar9p localized at the plus end with a concomitant decrease of Kar9p at the SPB (Cuschieri et al., 2006). Because the spindle pole body component Spc72p binds to Stu2p and interacts indirectly with Tub4p, (Chen et al., 1998; Knop and Schiebel, 1998; for review, see Helfant, 2002; Usui et al., 2003), it will be interesting for future studies to determine whether Stu2p and this complex act as a general regulator for the release of other microtubule plus end binding proteins from the SPB. Given that the Stu2p/XMAP215/TOGp family is conserved throughout eukaryota, it is possible that analogous mechanisms may control the trafficking of MAPs in many organisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank John Cooper, Bruce Goode, Tim Huffaker, Eric Phizicky, and the Yeast Resource Center at the University of Washington for providing yeast strains, plasmids, and reagents. We especially thank John Cooper for unique contributions. We thank the reviewers for insightful comments. This work was supported by grants to R.K.M. from the Ruth Estrin Goldberg Memorial for Cancer Research, the Basil O'Connor Starter Scholar Research Award from the March of Dimes Birth Defects Foundation 5-FY01-523, and the National Science Foundation MCB-0414768.
Abbreviations used:
- APC
adenomatous polyposis coli
- CDK
cyclin-dependent kinase
- cMT
cytoplasmic microtubule
- GFP
green fluorescent protein
- MAP
microtubule-associated protein
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-04-0360) on January 24, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Adames N. R., Cooper J. A. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S. C., Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen C., et al. Isolation of quiescent and nonquiesscent cells from yeast stationary-phase cultures J. Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askham J. M., Moncur P., Markham A. F., Morrison E. E. Regulation and function of the interaction between the APC tumour suppressor protein and EB1. Oncogene. 2000;19:1950–1958. doi: 10.1038/sj.onc.1203498. [DOI] [PubMed] [Google Scholar]
- Beach D. L., Thibodeaux J., Maddox P., Yeh E., Bloom K. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 2000;10:1497–1506. doi: 10.1016/s0960-9822(00)00837-x. [DOI] [PubMed] [Google Scholar]
- Bienz M. Spindles cotton on to junctions, APC and EB1. Nat. Cell Biol. 2001;3:E67–E68. doi: 10.1038/35060140. [DOI] [PubMed] [Google Scholar]
- Byers B. Cytology of the yeast life cycle. In: Strathern J. N., Jones W. W., Broach J. R., editors. The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carminati J. L., Stearns T. Microtubules orient the mitotic spindle in yeast through dynein- dependent interactions with the cell cortex. J. Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Gupta M. L., Jr., Hoyt M. A., Pellman D. Cell cycle control of kinesin-mediated transport of Bik1(CLIP-170) regulates microtubule stability and dynein activation. Dev. Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Chen X. P., Yin H., Huffaker T. C. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuschieri L., Miller R. K., Vogel J. γ-tubulin is required for proper recruitment and assembly of Kar9-Bim1 complexes in budding yeast. Mol. Biol. Cell. 2006;17:4420–4434. doi: 10.1091/mbc.E06-03-0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfant A. H. Composition of the spindle pole body of Saccharomyces cerevisiae and the proteins involved in its duplication. Curr. Genet. 2002;40:291–310. doi: 10.1007/s00294-001-0263-x. [DOI] [PubMed] [Google Scholar]
- Honnappa S., John C. M., Kostrewa D., Winkler F. K., Steinmetz M. O. Structural insights into the EB1-APC interaction. EMBO J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman S. M., Bales O. A., Bertrand M., Smeets M. F., Reed S. I., Segal M. Differential contribution of Bud6p and Kar9p to microtubule capture and spindle orientation in S. cerevisiae. J. Cell Biol. 2004;167:231–244. doi: 10.1083/jcb.200407167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P., Halladay J., Craig E. A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahana J. A., Schnapp B. J., Silver P. A. Kinetics of spindle pole body separation in budding yeast. Proc. Natl. Acad. Sci. USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco K. A., Pearson C. G., Maddox P. S., Wang P. J., Adams I. R., Salmon E. D., Bloom K., Huffaker T. C. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L., Tirnauer J. S., Li J., Schuyler S. C., Liu J. Y., Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Liakopoulos D., Kusch J., Grava S., Vogel J., Barral Y. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 2003;112:561–574. doi: 10.1016/s0092-8674(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Loog M., Morgan D. O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Louie R. K., Bahmanyar S., Siemers K. A., Votin V., Chang P., Stearns T., Nelson W. J., Barth A. I. Adenomatous polyposis coli and EB1 localize in close proximity of the mother centriole and EB1 is a functional component of centrosomes. J. Cell Sci. 2004;117:1117–1128. doi: 10.1242/jcs.00939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Schiebel E. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 2004;18:1709–1724. doi: 10.1101/gad.298704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa H., Usui T., Knop M., Schiebel E. Yeast Cdk1 translocates to the plus end of cytoplasmic microtubules to regulate bud cortex interactions. EMBO J. 2003;22:438–449. doi: 10.1093/emboj/cdg063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., Cheng S.-C., Rose M. D. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 2000;11:2949–2959. doi: 10.1091/mbc.11.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. K., D'Silva S., Moore J. K., Goodson H. V. The CLIP-170 orthologue Bik1p and positioning the mitotic spindle in yeast. Curr. Topics Dev. Biol. 2006;76:49–87. doi: 10.1016/S0070-2153(06)76002-1. [DOI] [PubMed] [Google Scholar]
- Miller R. K., Rose M. D. Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol. 1998;140:377–390. doi: 10.1083/jcb.140.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. K., D'Silva S., Miller R. K. The CLIP-170 homologue Bik1p promotes the phosphorylation and asymmetric localization of Kar9p. Mol. Biol. Cell. 2006;17:178–191. doi: 10.1091/mbc.E05-06-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Tanaka T. U., Nasmyth K., Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Clarke D. J., Maddox P., Salmon E. D., Bloom K., Reed S. I. Coordinated spindle assembly and orientation requires Clb5p-dependent kinase in budding yeast. J. Cell Biol. 2000;148:441–452. doi: 10.1083/jcb.148.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal M., Clarke D. J., Reed S. I. Clb5-associated kinase activity is required early in the spindle pathway for correct preanaphase nuclear positioning in Saccharomyces cerevisiae. J. Cell Biol. 1998;143:135–145. doi: 10.1083/jcb.143.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D. S., Huffaker T. C. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. J. Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A. H., et al. Global mapping of the yeast interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Trzepacz C., Lowy A. M., Kordich J. J., Groden J. Phosphorylation of the tumor suppressor adenomatous polyposis coli (APC) by the cyclin-dependent kinase p34. J. Biol. Chem. 1997;272:21681–61684. doi: 10.1074/jbc.272.35.21681. [DOI] [PubMed] [Google Scholar]
- Usui T., Maekawa H., Pereira G., Schiebel E. The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 2003;22:4779–4793. doi: 10.1093/emboj/cdg459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M., Dreschsel D., Hyman A. A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J., Snyder M. The carboxy terminus of Tub4p is required for γ-tubulin function in budding yeast. J. Cell Sci. 2000;113:3871–3882. doi: 10.1242/jcs.113.21.3871. [DOI] [PubMed] [Google Scholar]
- Wang P. J., Huffaker T. C. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak M. J., Blake-Hodek K., Kosco K. A., Hwang E., You L., Huffaker T. C. The regulation of microtubule dynamics in S. cerevisiae by three interacting plus-end tracking proteins. Mol. Biol. Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R. V., Cheng J. W., Salmon E. D., Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Yang C., Chin E., Maddox P., Salmon E. D., Lew D. J., Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Pruyne D., Huffaker T. C., Bretscher A. Myosin V orientates the mitotic spindle in yeast. Nature. 2000;406:1013–1015. doi: 10.1038/35023024. [DOI] [PubMed] [Google Scholar]
- Yoder T. J., Pearson C. G., Bloom K., Davis T. N. The Saccharomyces cerevisiae spindle pole body is a dynamic structure. Mol. Biol. Cell. 2003;14:3494–3505. doi: 10.1091/mbc.E02-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.