Abstract
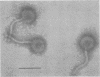
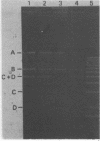
The temperate bacteriophage phiLC3, isolated from Lactococcus lactis subsp. cremoris, has an isometric head and a flexible tail containing 1 major protein and 8 minor proteins. Infection of a permissive L. lactis host strain yields a burst of about 50 phages per infected cell with a latent period of 60 min. A detailed restriction map of the phage chromosome was constructed by using 12 different restriction enzymes. The phage chromosome is a 33-kb linear double-stranded DNA molecule with unique cohesive ends and with a G + C content of 36.5%. Chemical sequencing of the DNA ends revealed 13-base 3' extended complementary single strands with a relatively high percentage of G + C. Pulsed-field gel electrophoretic analysis of DNA from a strain lysogenized with phiLC3 was used to localize the prophage to a 320-kb BamHI restriction endonuclease fragment from the host chromosomal DNA. This result indicates that lysogeny involves integration of the phage into the host chromosome. A spontaneous phiLC3 clear plaque mutant that was unable to give rise to lysogens was isolated.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Becker A., Murialdo H. Bacteriophage lambda DNA: the beginning of the end. J Bacteriol. 1990 Jun;172(6):2819–2824. doi: 10.1128/jb.172.6.2819-2824.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967 Dec;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coveney J. A., Fitzgerald G. F., Daly C. Detailed characterization and comparison of four lactic streptococcal bacteriophages based on morphology, restriction mapping, DNA homology, and structural protein analysis. Appl Environ Microbiol. 1987 Jul;53(7):1439–1447. doi: 10.1128/aem.53.7.1439-1447.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. E., Powell I. B., Hillier A. J. Temperate bacteriophages and lysogeny in lactic acid bacteria. FEMS Microbiol Rev. 1990 Sep;7(1-2):79–90. doi: 10.1111/j.1574-6968.1990.tb04880.x. [DOI] [PubMed] [Google Scholar]
- Ellis D. M., Dean D. H. Nucleotide sequence of the cohesive single-stranded ends of Bacillus subtilis temperate bacteriophage phi 105. J Virol. 1985 Aug;55(2):513–515. doi: 10.1128/jvi.55.2.513-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Waldman A. S., Benjamin R. C., Huang P. C., Scocca J. J. Nucleotide sequence and properties of the cohesive DNA termini from bacteriophage HP1c1 of Haemophilus influenzae Rd. Gene. 1984 Nov;31(1-3):197–203. doi: 10.1016/0378-1119(84)90210-5. [DOI] [PubMed] [Google Scholar]
- Huggins A. R., Sandine W. E. Incidence and properties of temperate bacteriophages induced from lactic streptococci. Appl Environ Microbiol. 1977 Jan;33(1):184–191. doi: 10.1128/aem.33.1.184-191.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Meyer J. Electron microscopic heteroduplex study and restriction endonuclease cleavage analysis of the DNA genomes of three lactic streptococcal bacteriophages. Appl Environ Microbiol. 1986 Mar;51(3):566–571. doi: 10.1128/aem.51.3.566-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lakshmidevi G., Davidson B. E., Hillier A. J. Circular Permutation of the Genome of a Temperate Bacteriophage from Streptococcus cremoris BK5. Appl Environ Microbiol. 1988 Apr;54(4):1039–1045. doi: 10.1128/aem.54.4.1039-1045.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmidevi G., Davidson B. E., Hillier A. J. Molecular characterization of promoters of the Lactococcus lactis subsp. cremoris temperate bacteriophage BK5-T and identification of a phage gene implicated in the regulation of promoter activity. Appl Environ Microbiol. 1990 Apr;56(4):934–942. doi: 10.1128/aem.56.4.934-942.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist B. H. Recombination between satellite phage P4 and its helper P2. I. In vivo and in vitro construction of P4: :P2 hybrid satellite phage. Gene. 1981 Sep;14(4):231–241. doi: 10.1016/0378-1119(81)90156-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Murialdo H., Becker A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol Rev. 1978 Sep;42(3):529–576. doi: 10.1128/mr.42.3.529-576.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray K., Isaksson-Forsen A. G., Challberg M., Englund P. T. Symmetrical nucleotide sequences in the recognition sites for the ter function of bacteriophages P2, 299 and 186. J Mol Biol. 1977 May 25;112(3):471–489. doi: 10.1016/s0022-2836(77)80193-9. [DOI] [PubMed] [Google Scholar]
- Potter H., Dressler D. A 'Southern Cross' method for the analysis of genome organization and the localization of transcription units. Gene. 1986;48(2-3):229–239. doi: 10.1016/0378-1119(86)90081-8. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Characterization of streptococcal bacteriophage c6A. J Gen Virol. 1985 Dec;66(Pt 12):2737–2741. doi: 10.1099/0022-1317-66-12-2737. [DOI] [PubMed] [Google Scholar]
- Prevots F., Mata M., Ritzenthaler P. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl Environ Microbiol. 1990 Jul;56(7):2180–2185. doi: 10.1128/aem.56.7.2180-2185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relano P., Mata M., Bonneau M., Ritzenthaler P. Molecular characterization and comparison of 38 virulent and temperate bacteriophages of Streptococcus lactis. J Gen Microbiol. 1987 Nov;133(11):3053–3063. doi: 10.1099/00221287-133-11-3053. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snook R. J., McKay L. L., Ahlstrand G. G. Transduction of Lactose Metabolism by Streptococcus cremoris C3 Temperate Phage. Appl Environ Microbiol. 1981 Nov;42(5):897–903. doi: 10.1128/aem.42.5.897-903.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]