Abstract
Lymphocytic choriomeningitis virus (LCMV), strain WE, is a non-cytopathic RNA virus that is highly adapted to its natural host, the mouse. Acute infection of adult mice leads to generalized virus spread, followed by cytotoxic T lymphocyte-mediated virus clearance below the detection levels of conventional assays within 2–3 weeks. Indirect evidence had suggested that virus or viral antigen might persist in the immune mouse. Here we demonstrate LCMV-WE persistence at low levels after infection with 102 or 106 plaque-forming units, shown as viral genome, viral antigen, and replicative virus using sensitive in vitro and in vivo assays. The finding that LCMV-WE persists in the face of apparently intact immune responses resembles the situation in some viral (hepatitis B and C, HIV) and bacterial (tuberculosis, leprosy) infections in humans; the results are relevant to the understanding not only of other murine and human persistent viral infections but also of protective immunological memory by “infection immunity.”
There are a number of features that suggest that, after apparent clearance of lymphocytic choriomeningitis virus (LCMV) infection (1, 2), virus or viral antigen may persist to variable extent dependent on the LCMV strain (3–7). First, unlike vaccination using antigen in other forms, the cytotoxic T lymphocyte (CTL) response against LCMV remains demonstrable after restimulation or as precursor CTLs over many months (8, 9) and bears hallmarks of acute effector status—rapid killing ability in vivo and ex vivo and homing to the peripheral tissues accompanied by immediate in vivo protective capacity (10–12). Second, neutralizing antibodies (nAb) responses usually arise late after infection (e.g., LCMV-WE) or only marginally [e.g., LCMV-Armstrong (13)], when virus appears to have been cleared for several weeks (13, 14); a situation similar to bona fide persistent viruses such as hepatitis B (15, 16), hepatitis C (17), and HIV (18), but also tuberculosis (19) and leprosy (20) in humans. Third, mice deficient in MHC class II, CD4+ T cells or B cells (and consequently unable to produce T cell help or nAb) initially appear to clear virus from all tissues below the detection level of conventional assays, but succumb to renewed viremia after weeks or months (21, 22); so, under these conditions there is certainly low level persistence of replication-competent virus. Finally, treatment with antilymphocytic serum 30–50 days after LCMV infection provoked recrudescence of viremia in mice infected with and immune to LCMV-Traub (6).
We previously demonstrated low levels of LCMV reverse transcripts (i.e., viral cDNA) in spleens of LCMV-WE immune mice or after in vitro infection (23). However, a direct role for this DNA in persistence of viral antigen has not yet been shown. Several attempts to isolate infectious LCMV or influenza virus or to detect reverse transcriptase–PCR (RT-PCR) signals from mice acutely infected with 102–106 plaque-forming units (pfu) of LCMV or other RNA viruses, after they apparently had cleared the virus, have failed so far (23–26). We find in this study that it is possible to detect viral RNA consistently in the majority of mice infected 40–80 days previously with LCMV-WE by using nested RT-PCR, that virus in replicative form also may be isolated by using sensitive in vivo readouts, and that viral antigen may be detected in a few cells in tissues (particularly spleen, lung, and kidney) by immunohistochemistry.
Materials and Methods
Mice and Viruses.
C57BL/6 (B6), BALB/c, and IFN-α/β/γ receptor−/− (AG129) mice were obtained from the breeding colony at the Institut für Labortierkunde (University of Zurich, Zurich) and were kept under specific pathogen-free conditions. They were infected i.v. with low (200 pfu) or high (2 × 106 pfu) doses of LCMV-WE (27) originally obtained from F. Lehmann-Grube (Pette Institut, Hamburg, Germany). The same virus stock was used for all of the described experiments. The viral titer of the stock was measured by focus-forming assay.
Virus Determination in Vitro.
Focus-forming assay.
The focus-forming assay was used as described (28). Values of virus titers are expressed as log10 pfu per organ or per milliliter of blood.
Molecular analysis.
Organs (a quarter of spleen, lung, or kidney) were harvested from immunized mice 40–80 days after infection, immediately were snap frozen by immersing the tubes with long forceps in liquid nitrogen, and were stored at −80°C. The frozen tissues were disrupted and homogenized in the presence of Qiagen (Valencia, CA) lysis buffer by using a rotor-stator homogenizer (Polytron PT 1200, Kinematica AG, Lucerne, Switzerland). The tissue lysate was additionally homogenized by using QIAshredder and RNA extracted in 40 μl of solution by using RNeasy Kit (Qiagen). After DNase-treatment (20 units per 10 μl of reaction volume; Boehringer Mannheim) performed at 37°C for at least 90–120 min, 8 μl of RNA solution was used as a template for reverse transcription by using a first strand cDNA synthesis kit (Amersham Pharmacia) and specific glycoprotein (GP) or nucleoprotein (NP) primers. The primer sequences (antisense R1 and sense 001 for GP and antisense 2818 or sense 3060 for NP) were previously published (23) and consistent with the cDNA sequences for LCMV-WE (29). Amplification of GP sequences then was performed by using nested PCR on 2 μl of template with primer pairs 001/R1 (primary reaction) followed by RC1/DM1 (secondary reaction). PCR conditions were 50 μl of total volume containing 20 pmol of each primer, 0.2 μM dNTPs, 2.5 units of Taq polymerase, and 1 × PCR buffer (Boehringer Mannheim). The cycling conditions for GP-PCR were 94°C for 1 min, 55°C for 1 min, 72°C for 2 min, 35 cycles (primary reaction), 94°C for 1 min, 42°C for 1 min, 72°C for 1 min, 35 cycles (secondary reaction). The primers for amplification of NP sequences were 2818/3060 and NP5′I/NP3′I for primary and secondary reactions, respectively (23). The conditions for NP-PCR were 94°C for 30 s, 50°C for 1 min, 72°C for 30 s, 35 cycles for both amplifications. In the secondary reaction, 2 μl of each primary product was used. The final product (10 μl) was run on 1% agarose gels and was visualized with ethidium bromide. PCR sequencing of these products also was performed after purification by using the QIAquick gel extraction kit (Qiagen). We were careful to minimize the potential for cross contamination and PCR artifacts. PCR mixes were made up in a separate laboratory with a dedicated set of instruments; cDNA template was added to PCR mixes in a third laboratory in which LCMV culture was not performed; aerosol resistant tips were used. The results were obtained independently by three separate operators. Control uninfected mouse tissue was processed in parallel and was uniformly negative; water controls in both primary and secondary reactions were negative throughout; material from cDNA reactions in the absence of reverse transcriptase was also negative.
Immunohistology.
Samples of freshly removed organs either were immersed in Hank’s balanced salt solution and snap frozen in liquid nitrogen or were fixed with 4% formalin solution and embedded in Paraplast. Frozen tissue sections were cut in a cryostat, were placed on siliconized glass slides, were air dried, were fixed with acetone, and were stored at −80°C. To stain for LCMV, sections were incubated with rat anti-LCMV-NP monoclonal Ab (VL-4) (28). Affinity-purified goat anti-rat-IgG Ab, followed by alkaline phosphatase-labeled donkey anti-goat-IgG Ab (Jackson ImmunoResearch), were used as secondary reagents. Paraffin sections were pretreated with 0.1% pronase for 25 min. After incubation with rabbit anti-LCMV antiserum, affinity-purified biotinylated swine anti-rabbit Ab were applied, followed by ABC/AP (Dako). Alkaline phosphatase was detected by a red color reaction product using naphthol AS-BI phosphate/New Fuchsin as substrates. Sections were counterstained with hemalum.
Virus Determination in Vivo.
Organs were harvested from immunized mice, were snap frozen exactly as for molecular analysis, and then were stored at −80°C. After thawing, organs were kept on ice. Spleens were gently pressed through a fine stainless steel grid, and other organs were homogenized before intraperitoneal adoptive transfer into individual AG 129 mice, which are extremely susceptible to LCMV-WE (30). The AG 129 mice were kept individually in separate cages because of very efficient spread of virus from infected to uninfected AG 129 mice. Recipient mice were killed on day 4 after transfer, and viral titers in blood and solid tissues (spleen, kidney, lung) were measured in a focus-forming assay.
Results
Evaluation of the Different Methods To Detect LCMV-WE.
We evaluated the sensitivity of three different techniques to detect LCMV or LCMV sequences in solid organs: (i) infectious focus-forming assay on MC57 cells, (ii) nested RT-PCR, and (iii) injection of tissue homogenates into AG 129 mice. It should be emphasized that it is very difficult to validate the various methods with respect to absolute values. The reasons that virus isolated from organs of infected mice yield a value that cannot be reliably related to the real value in the animal are as follows. First, there are theoretical lower limits of detectable units when the different tests were used to determine virus or RNA in biological samples; the factors that influence these are presented in Table 1. Second, there are limitations in the sensitivity of what can be detected (Table 2). Attempts at mixing known amounts of virus or infected cells to uninfected tissues are the best approximations to imitate actual values in infected organs. Two test materials were analyzed: first, about 1.5 × 107 splenocytes (a quarter of a naïve spleen) were mixed with decreasing numbers of LCMV-WE-infected MC57 cells, starting at 103 cells (2 days after infection, multiplicity of infection 0.01). This procedure leads to >90% cells becoming infected as determined by flow cytometry (data not shown). The MC57 cells were washed four times at 4°C to remove free virus particles. Alternatively, the equivalent of a quarter of a naïve spleen was mixed with increasing dilutions of the LCMV-WE stock, starting at 105 pfu; the virus titer of the stock had been determined by focus-forming assay. For nested GP- or NP-specific RT-PCR, mixtures were snap frozen in liquid nitrogen and then were processed as detailed in Materials and Methods. Snap-frozen mixed materials also were used for infection of AG 129 mice by intraperitoneal injection after thawing. For the focus-forming assay in vitro, the virus or cells were added to a quarter of spleen, all were homogenized with a teflon pestle in a tight fitting glass tube, were frozen at −80°C, were thawed, were kept from then on strictly on ice, and were analyzed.
Table 1.
Theoretical detection levels of the various LCMV detection methods in organs of immune mice
| Detection limit
| ||
|---|---|---|
| Focus-forming assay | Nested NPRT-PCR | Transfer in AG 129 mice |
| 40–400 pfu* | 100–200 copies† | 2–4 infective units‡ |
The indicated detection limits for three different LCMV detection methods can be calculated from the following dilution factors and optimal sample volumes used for each technique:
As only a quarter of spleen (1/4) was harvested for the focus forming assay, yielding 2 ml of tissue homogenate from which only 200 μl (1/10) were used in the assay, the theoretical detection level of the focus-forming assay is 40 pfu if one plaque is detected in the first dilution step and 400 pfu if the cell layer in the first dilution well is destroyed by the toxic effects of the homogenate.
† Only 8 μl of the total 40-μl RNA solution extracted from a quarter spleen were used for the reverse transcription reaction (1/5), and 2 μl of the resulting total of 16 μl cDNA (1/8) were used in the PCR reaction. The theoretical detection limit if one copy gives a positive signal would therefore be 160 copies per spleen.
‡ We transferred half or a quarter of the whole amount of tissue homogenate in AG 129 mice. If one infective unit of LCMV leads to viremia in these mice, a minimum of 2–4 units would be expected in the whole organ.
Table 2.
Methodological sensitivities of different LCMV detection methods evaluated with diluted virus stock and infected MC57 cells
| Material tested | Detectable units | Detection limit per spleen
|
||
|---|---|---|---|---|
| Focus-forming assay | Nested NPRT-PCR | Transfer in AG 129 mice | ||
| Spleen + Dilutions of LCMV stock | pfu | 104 | 102 | 0.2-1 |
| Spleen + LCMV-infected MC57 cells | No. of infected MC57 cells | 102 | 10-40 | 1-4 |
Increasing dilutions of a LCMV-WE stock (viral titer measured by focus-forming assay) or decreasing numbers of LCMV-infected MC57 cells were mixed with the equivalent of a quarter of spleen and the indicated three LCMV-detection methods (as detailed in Materials and Methods) were performed on these samples to evaluate the respective detection limits.
The results are summarized in Table 2. The detection limit of the conventional focus forming assay was ≈102 infected MC57 cells or 104 pfu of titrated virus per total spleen. In contrast, nested NP RT-PCR detected 10–40 infected MC57 cells and ≈102 pfu of LCMV per spleen. Similar results were obtained for nested GP RT-PCR. Interestingly, RT-PCR after tissue homogenization using a teflon pestle [as we had used in earlier studies (23) and still use for preparing homogenates for focus-forming assay] instead of a rotor-stator homogenizer was 10- to 100-fold less sensitive. To determine the sensitivity of the PCR, cDNA was synthesized from RNA extracted from spleen of naive C57BL/6 mice and was mixed with decreasing numbers of LCMV-plasmid copies. Nested PCR specific for GP was able to detect 3–6 plasmid copies (data not shown).
Infection of AG 129 mice detected 1–4 infected MC57 cells and 0.2–1 pfu of titrated LCMV per spleen. Similarly, i.v. infection of AG 129 mice revealed viremia after inoculation with 0.1 but not fewer pfu LCMV of a titrated virus stock (data not shown). As outlined above, a comparison of the various methods in the sensitivity assay as shown in Table 2 offers a baseline but may not apply readily to the comparison of units detected in organs of infected mice by the same methods (see below; Table 4) because the status of virus infection and ratio of infectious units versus viral RNA in vivo is generally unknown.
Table 4.
Summary of experiments that detect LCMV in the same groups of immune mice by various methods
| Experimental group
|
Ratio of LCMV positive mice in different detection systems
|
||||
|---|---|---|---|---|---|
| Infection with LCMV-WE | Time after infection | Focus-forming assay | Transfer in AG129 mice | Nested RT-PCR | Immunohistology |
| 200 pfu i.v. | Day 40 | 0/3 | 1/3 | 3/3 | 3/3 |
| Day 80 | 0/4 | 2/4 | 4/4 | 4/4 | |
| 2 × 106pfu i.v. | Day 40 | 0/3 | 3/3 | 3/3 | 3/3 |
| Day 80 | 0/3 | 3/3 | 3/3 | 3/3 | |
Three to four B6 mice per group and time point were immunized with 200 or 2 × 106 pfu of LCMV i.v. After 40 or 80 days, organs were harvested, and four different methods were used to detect either LCMV genome (nested RT-PCR), LCMV antigen (immunohistology with rat anti-LCMV-NP antibody; VL-4) or replication-competent LCMV (focus-forming assay and transfer of tissue homogenates into AG 129 mice), as detailed in Materials and Methods. The ratio of LCMV-positive mice by the respective technique is shown for each group.
Detection of Persistent Viral RNA in Immune Mice by RT-PCR.
We used nested RT-PCR with GP- and NP-specific primers to identify LCMV GP and NP RNA in different organs of LCMV-WE-immune mice. The term “RNA detection” in the following sections implies detection of reverse transcribed RNA by nested RT-PCR, as described in Material and Methods. The primers and strategy are shown in Fig. 1A.
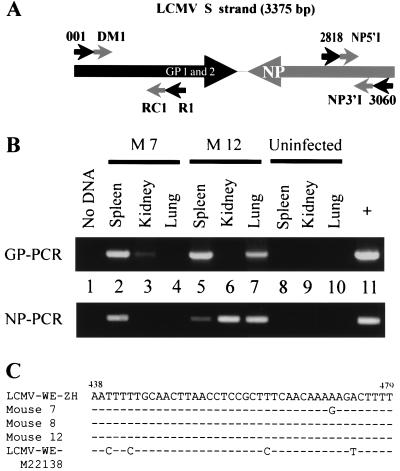
Figure 1.
Detection of LCMV-WE RNA in immune mice. (A) PCR amplification strategy with schematic representation of the S strand of LCMV, which encodes the glycoproteins (GP1 and GP2) and the nucleoprotein (NP) in opposite sense. The primers used are indicated. (B) RT-PCR on reverse-transcribed DNase-treated RNA extracted from spleen, kidney, and lung of two B6 mice (M7 and M12 in Table 3) infected 80 days previously with LCMV-WE (200 pfu i.v. for M7 and 2 × 106 pfu i.v. for M12) using GP- and NP-specific primers as detailed in Material and Methods. Products were run on 1% agarose gels and were visualized with ethidium bromide. Product sizes are 907 bp for GP and 207 bp for NP. Controls in this experiment were no cDNA (lane 1) and RNA extracted from organs of an uninfected mouse (lanes 8–10). The positive control was cDNA template from MC57 cells infected with LCMV (multiplicity of infection 0.02) and was cultured for 48 h. (C) Partial sequence alignment (nucleotides 438–479) from amplified GP products (spleen isolates). Products were sequenced by using inner PCR primers (RC1/DM1) and were compared with LCMV-WE sequence derived from our viral stock (LCMV-WE-ZH) and from a plasmid frequently used in our laboratory, containing GP cDNA (GenBank accession number M22138). Positions of difference in nucleotides are indicated.
In the first set of experiments, we analyzed organs (spleen, kidney, and lung) of B6 mice 40 and 80 days after i.v. infection with low dose (200 pfu) or high dose (2 × 106 pfu) LCMV-WE for the presence of GP and NP RNA (3–4 mice per group). These organs were chosen because they have often been found to harbor persistent viruses in humans (e.g., CMV). The results are summarized in Table 3. RT-PCR data are shown for two mice in Fig. 1B. The agarose gel bands in Fig. 1B are representative of the RT-PCR-positive raw data used for Table 3. GP-specific RNA could be detected at least in one of the mentioned organs in all 13 mice tested in this experiment: 40 days after low dose (200 pfu) LCMV-WE infection 2/3 spleens and 3/3 lungs were positive for GP RT-PCR whereas kidneys were negative. The proportion of detectable GP-specific RNA was 2/4 for spleen, lung, or kidney on day 80 after infection. As expected, the percentage of positive organs by detection of LCMV-GP RNA was higher after high dose (2 × 106 pfu) LCMV-WE immunization: With one exception, all organs of three mice tested were PCR-positive on day 40 after infection. GP-specific RNA could still be detected 80 days after infection in 1/3 spleens, 2/3 kidneys, and 2/3 lungs, each mouse being LCMV positive in one or two of the three tested organs.
Table 3.
LCMV detection by nested RT-PCR specific for GP and NP and by immunohistochemistry in different organs of individual LCMV immune mice
| Group | Time after infection | Mouse | LCMV-WE detection
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spleen
|
Kidney
|
Lung
|
|||||||||
| PCR*
|
Histo† | PCR*
|
Histo† | PCR*
|
Histo† | ||||||
| GP | NP | GP | NP | GP | NP | ||||||
| 200 pfu of LCMV i.v. | Day 40 | M1 | + | − | + | − | − | − | + | + | − |
| M2 | − | − | − | − | − | − | + | + | − | ||
| M3 | + | − | + | − | − | − | + | + | − | ||
| Day 80 | M4 | − | ND | + | + | ND | − | − | + | − | |
| M5 | − | − | − | − | − | − | + | + | − | ||
| M6 | + | − | ND | − | − | ND | + | − | ND | ||
| M7 | + | + | ND | + | − | ND | − | − | ND | ||
| 2 × 106pfu of | Day 40 | M8 | + | + | + | + | + | + | + | + | + |
| LCMV i.v. | M9 | − | + | + | + | − | + | + | + | + | |
| M10 | + | + | + | + | + | + | + | + | + | ||
| Day 80 | M11 | − | − | − | + | + | + | + | + | + | |
| M12 | + | + | + | − | + | + | + | + | + | ||
| M13 | − | + | + | + | + | + | − | − | − | ||
Spleens, kidneys, and lungs were harvested from individual mice (M 1-13) infected 40 or 80 days previously with 200 pfu of LCMV-WE i.v. or 2 × 106 pfu of LCMV-WE i.v. ND, not determined.
Nested PCR specific for GP and NP on reverse-transcribed RNA extracted from indicated organs.
† Immunohistological analysis of tissue sections with anti-LCMV-NP antibody (VL-4). Positive and negative samples are indicated.
Eight PCR products sequenced from eight individual B6 LCMV-WE immune mice confirmed LCMV-GP-specific amplification (Fig. 1C). The GP of our LCMV-WE strain (LCMV-WE-ZH) has some characteristic nucleotide changes in comparison to the most frequently used plasmid (containing LCMV-WE cDNA) in our laboratory [LCMV-WE-M22138 (29)]. The sequenced region in Fig. 1C is diagnostic for these mutations and indicates that contamination by plasmids is unlikely. The region of GP encoding the GP33 Db-restricted epitope, which represents the immunodominant CTL target in B6 mice after infection with LCMV-WE, was not mutated in these sequences, rendering it unlikely that the persistent sequences were derived from CTL escape mutant viruses (31).
These immune B6 mice also were analyzed for LCMV nucleoprotein RNA. NP RT-PCR was positive in a similar proportion of mice as for GP RT-PCR in low and high dose infection and at both time points analyzed (Table 3 and Fig. 1B). In the majority of cases, NP RNA was detected in the same organs that were positive for GP RNA. The few discrepancies observed are probably due either to different copy numbers of LCMV GP and NP RNA, slightly different sensitivities, or to the fact that RNA levels in immune mice are just at the detection level, as assessed by nested RT-PCR performed at limiting dilution conditions on reverse-transcribed RNA from immune mice (data not shown).
The approximate number of RNA copies present in organs of immune mice cannot be readily extrapolated from the plasmid titration experiment presented in the first section because neither the efficiency of RNA extraction nor that of the reverse transcription step are known. Nevertheless, a relative estimation of viral RNA copies can be obtained by the comparison of acutely infected mice (day 4) with immune mice (after day 60) after i.v. infection with 200 pfu of LCMV-WE. Nested PCR performed at limiting dilution demonstrated that the relative value of RNA copies in an acute infection is 106× in excess compared with the value during the memory phase; this confirms that levels of infection are very low indeed.
We next performed NP-specific RT-PCR on spleens taken from immune BALB/c (H-2d) mice. In accordance with the results from B6 (H-2b) mice, 3/5 spleens were PCR positive on day 55 after low dose (200 pfu) infection and sequencing confirmed LCMV-WE NP amplification (data not shown). As controls, spleens from 3/3 LCMV-WE-carrier BALB/c mice and 0/3 spleens from uninfected mice were positive in this series of experiments. The region of NP encoding the NP118 Ld restricted epitope, which represents the immunodominant CTL target in BALB/c mice after infection with LCMV-WE, was not mutated in a total of five sequences obtained from five individual BALB/c memory mice.
In all of the experiments presented in this section, spleens, lungs, and kidneys from uninfected control mice, as well as no-DNA-controls, were always negative. A single round PCR performed on some positive organs of the nested RT-PCR-tested immune mice either with conditions and primers of primary reaction (R1/001 for GP and 2818/3060 for NP) or of secondary reaction (RC1/DM1 for GP and NP5′I/NP3′I for NP) was negative, even though these protocols will amplify GP or NP from cDNA prepared from acutely infected MC57 cells as a positive control (data not shown). This is consistent with previous observations using LCMV-Armstrong strain (24).
It is important to emphasize that spleens, lungs, and kidneys from all of the immune mice tested were uniformly negative for LCMV by conventional focus-forming assay. Because of methodological limitations, the detection limit was ≈40 pfu per spleen and 20 pfu per lung or kidney (data summarized in Table 4).
Virus Persists in Replicative Form.
We next addressed the question of whether the viral genome detectable by PCR was associated with replication-competent virus. As previously described (23), attempts to isolate virus directly from tissue homogenates were unsuccessful. In a more sensitive in vivo assay, AG 129 mice were used as recipients for adoptively transferred splenocytes from immune mice. Such IFN-α/β/γ receptor-deficient mice are extremely susceptible to LCMV infection, and virus grows rapidly in all tissues (30). It is important to note that these mice were housed individually because virus secreted by a LCMV-positive AG 129 mouse infects AG 129 mice in the same cage within 1–2 days (data not shown). Such mice are susceptible to viremia after inoculation with 0.1 pfu LCMV, as stated before.
We injected 107 intact spleen cells from immune mice i.v. into AG 129 mice and determined LCMV titers in blood and organs of the recipients by focus-forming assays 4 days after transfer. Two of five B6 mice infected for >60 days previously with LCMV-WE (200 pfu i.v.) were positive by this technique, as were three of four mice infected with 2 × 106 pfu. Viral titers in the blood were between 5 × 104 and 5 × 105 pfu per ml. Transfer of splenocytes from uninfected control mice, uninfected MC57 cells, or medium alone did not lead to detectable viremia in the AG129 mice.
In a separate set of experiments, lung homogenates from individual LCMV immune mice (which were used in the RT-PCR experiments above) were injected i.p. into AG 129 mice. Lung homogenates from mice infected 40 and 80 days previously with 200 pfu LCMV-WE caused viremia in 1/3 and 2/4 AG129 mice, respectively (Table 4). All AG129 mice (three per group) injected with lung homogenates isolated 40 and 80 days after infection with 2 × 106 pfu LCMV-WE exhibited viremia (Table 4). The results in Table 4 indicate that it was relatively more difficult to find infectious virus that could replicate in AG 129 mice than to detect viral RNA by RT-PCR in organs of immune mice. This does not conflict with the findings of the sensitivity assays using acutely infected material (Table 2) in which AG 129 mice proved to be very sensitive because of the different biological starting material used and the limitations stated in the first section.
Detection of Viral Antigen in Tissues of Immune Mice by Immunohistology.
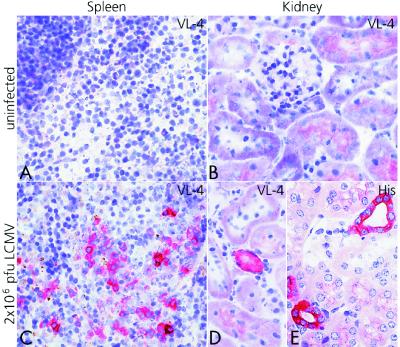
To evaluate whether viral antigen might persist in solid organs of immune mice, we next stained tissue sections from LCMV immune B6 mice with the monoclonal rat anti-LCMV-NP antibody VL-4 (32) or with a rabbit anti-LCMV hyperimmune serum, which stains all LCMV antigens. Although, in lymphoid organs, occasionally an isolated cell was found to present nonspecific staining, clusters of 3–5 stained cells (Fig. 2C) were never seen in uninfected control animals. In kidneys and lungs, epithelial cells never exhibited background staining. Multiple sections from spleens, kidneys, and lungs from >30 mice at different time points after infection (up to 120 days) with 200 pfu or 2 × 106 pfu LCMV-WE were examined. Although most of the spleen sections were positive, only rare cells or groups of cells were stained in kidney and lung sections after 200 pfu LCMV-WE; many sections were negative. However, for all mentioned conditions and time points, a couple of positive sections were found also for these organs (data not shown). Fig. 2 shows representative spleen and kidney sections from mice immunized 80 days previously with 2 × 106 pfu LCMV-WE and from uninfected control mice.
Figure 2.
Detection of LCMV-antigen in different spleens and kidneys of LCMV-immune B6 mice by immunohistological analysis. Spleens and kidneys were harvested 80 days after infection with 2 × 106 pfu LCMV-WE (C–E). Control sections from uninfected animals are shown (A and B). Frozen tissue sections were stained with VL-4, a rat anti-LCMV-NP monoclonal Ab (A–D). Paraffin sections were stained with a rabbit anti-LCMV serum [hyperimmune serum (His)] (E). (×400.)
A comparison of the different LCMV detection techniques on the set of mice immunized 40 and 80 days previously with 200 pfu LCMV-WE i.v., mentioned in the previous sections, shows that virus can be detected immunohistologically in the spleen (Table 3). After infection with 2 × 106 pfu LCMV-WE, spleens, kidneys, and lungs were regularly positive by this technique (Table 3 and Fig. 2), confirming the results obtained by RT-PCR and those obtained by infection of AG 129 mice.
Discussion
The question of whether LCMV persists in vivo in immune mice is important from both immunological and virological viewpoints. In the former, there has been ongoing debate about the role of persistent antigen [either generated by persisting infection (26, 33), antigen depots or re-exposure to cross-reactive infections (34, 35)] in maintaining protective immunological memory (36, 37). There is evidence that antigen is not required to maintain elevated CTL precursor levels as measured in vitro and CTLs, which may efficiently expand in vivo in response to viral challenge (25, 38). Moreover, there are important qualitative differences between the function of memory induced by LCMV itself (i.e., presence of “activated” effector memory T cells) and anti-LCMV memory induced by other means (recombinant vaccinia, recombinant Listeria, peptides etc.), which yields “quiescent” memory T cells, in terms of kinetics, immediate protective capacity, and ability to home to infected peripheral sites (10, 11, 39). It has been proposed that these differences were attributable to persistence of LCMV antigen in some form (40). Earlier studies had failed to demonstrate direct evidence for persistent virus (23–25). The current study has taken advantage of several improved methods and has used mice extremely sensitive to virus infection to re-evaluate this issue. The results provide evidence for persistence of small amounts of virus in the normal mouse even after low dose infection with LCMV-WE. This evidence was obtained at the level of viral genome, viral antigen, and recoverable virus, all of which were present at very low levels. The studies were not positive for all tested organs in the mice. The consequences of the differences seen after low dose (200 pfu) and high dose (2 × 106 pfu) infection should, however, not be over-interpreted. The negative results in some organs may indicate that there are probably either oscillating levels of virus production or that there is still a detection limit with the assays used. Even the most sensitive of these, PCR (which is capable of detecting 3–6 DNA copies per reaction) could only detect reverse-transcribed RNA at the level of ≈10–40 admixed LCMV-infected MC57 cells, and the amounts detected in the positive organs appeared to be just above this detection limit. The apparent discrepancy between the sensitivity assay in Table 2 and the findings summarized in Table 4, that nested RT-PCR detected more positive animals and organs than the AG 129 transfer assay, may be explained as follows: First, one may argue that cotransfer of few nAbs during injection of organ homogenates into AG 129 mice may render this in vivo readout less efficient. This is, however, unlikely, as it was shown that productive LCMV infection in the presence of excess nAb is still possible (41). Second, as pointed out, the technical and methodological limitations are obvious. In particular, the validating titrations in Table 2 cannot overcome the problem that we do not know the relative values of RNA copies versus infectious units in organs of mice infected for 40–80 days. Nevertheless, the fact that, in many immune mice, replicating virus could be isolated after transfer of organ homogenates into AG 129 mice strengthens the PCR and histological findings. When taken together, these results make a strong case for persistent LCMV-WE infection even after infection with 200 pfu.
We have previously demonstrated that LCMV-WE sequences may persist in immune mouse spleen in DNA form (23). This is attributable to reverse transcription by endogenous retroviral elements and was only seen in mouse and hamster cells, species in which LCMV is able to establish a carrier state. It is not clear whether these LCMV sequences have biological activity, as it would require illegitimate recombination and promoter action, but we have speculated that low level protein expression could occur, sufficient to contribute to immunological memory, or potentially viral persistence. The presence of RNA viral genomes demonstrated in this study represents a more conventional means for low level maintenance of infection in a “subclinical” form. The relative importance of the two potential forms of persistence remains to be determined, although the relative abundance of genomic RNA and its presence in virtually all mice tested, as seen in this study, makes viral RNA likely to be the most significant.
There is little doubt that T cell precursor frequencies depend largely on the initial burst size and then are maintained at fairly stable levels in an antigen-independent manner (25, 38). Overall, our observations do not directly answer the question of whether viral antigen is required for the maintenance of protective immunological memory, but, together with earlier experiments and a recent study by Ochsenbein et al. comparing protective memory induced by LCMV-WE versus recombinant Listeria expressing LCMV-NP (42), they provide strong supporting evidence. Therefore, viral antigen may exist during the memory phase and could contribute to protective memory mediated by effector memory T cells. These results indicate that, contrary to previous reports—but consistent with the observed immunological response to infection that includes very late emergence of nAb—LCMV-WE does persist at very low levels in normal mice after acute infection. This persistence may contribute to the maintenance of functional status of immune memory cells and to the re-emergence of virus in immune deficient mice. These findings may be relevant to other Arenaviruses and RNA viruses, where lifelong immunity is observed in the absence of known re-infection, but also so far without evidence for persistence of infection. They correlate with findings in other infections, such as HIV, hepatitis B virus, and hepatitis C virus in man (reviewed, respectively, in refs. 15–18), where virus persists in the face of apparently intact immune responses. Understanding the mechanisms involved is not only relevant to the biology of persistent virus infections in general but may also explain “infection immunity.” This term had been coined by Mackaness to describe the situation after Mycobacterium tuberculosis infection (19), which also applies to leprosy (20), where low level infections by few persisting mycobacteria keep specific and nonspecific cell-mediated immunity protective.
Acknowledgments
We thank Andrew Macpherson and Kevin Maloy for critical reading of the manuscript, Lars Hangartner and Burkhard Ludewig for helpful discussions, Niclas Rydell, Lenka Vlk, Anne Henzelin, Dieter Zimmermann, and his group for expert technical assistance, and Norbert Wey and Ida Schmieder for excellent microphotography. This work was supported by the Swiss National Science Foundation Grant 31.50900.97 to R.M.Z., the Kanton of Zürich, and the Wellcome Trust.
Abbreviations
- CTL
cytotoxic T lymphocyte
- GP
glycoprotein
- LCMV
lymphocytic choriomeningitis virus
- nAb
neutralizing antibody
- NP
nucleoprotein
- pfu
plaque-forming units
- RT-PCR
reverse transcriptase–PCR
References
- 1.Traub E. Science. 1935;81:298–299. doi: 10.1126/science.81.2099.298. [DOI] [PubMed] [Google Scholar]
- 2.Hotchin J. Cold Spring Harbor Symp Quant Biol. 1962;27:479–499. doi: 10.1101/sqb.1962.027.001.046. [DOI] [PubMed] [Google Scholar]
- 3.Rowe W P. Navy Res Rep. 1954;12:167–220. [Google Scholar]
- 4.Larsen J H. Acta Pathol Microbiol Scand. 1968;73:106–114. doi: 10.1111/j.1699-0463.1968.tb00484.x. [DOI] [PubMed] [Google Scholar]
- 5.Larsen J H. Immunology. 1969;16:15–23. [PMC free article] [PubMed] [Google Scholar]
- 6.Volkert M, Lundstedt C. J Exp Med. 1968;127:327–339. doi: 10.1084/jem.127.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahmed R, Jamieson B D, Porter D D. J Virol. 1987;61:3920–3929. doi: 10.1128/jvi.61.12.3920-3929.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jamieson B D, Ahmed R. J Exp Med. 1989;169:1993–2005. doi: 10.1084/jem.169.6.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murali-Krishna K, Altman J D, Suresh M, Sourdive D J, Zajac A J, Miller J D, Slansky J, Ahmed R. Immunity. 1998;8:177–187. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 10.Oehen S, Waldner H, Kündig T M, Hengartner H, Zinkernagel R M. J Exp Med. 1992;176:1273–1281. doi: 10.1084/jem.176.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kündig T M, Bachmann M F, Oehen S, Hoffmann U W, Simard J J L, Kalberer C P, Pircher H, Ohashi P S, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1996;93:9716–9723. doi: 10.1073/pnas.93.18.9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bachmann M F, Kündig T M, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1997;94:640–645. doi: 10.1073/pnas.94.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battegay M, Moskophidis D, Waldner H, Brundler M A, Fung-Leung W-P, Mak T W, Hengartner H, Zinkernagel R M. J Immunol. 1993;151:5408–5415. [PubMed] [Google Scholar]
- 14.Planz O, Seiler P, Hengartner H, Zinkernagel R M. Nature (London) 1996;382:726–729. doi: 10.1038/382726a0. [DOI] [PubMed] [Google Scholar]
- 15.Chisari F V, Ferrari C. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 16.Rehermann B, Ferrari C, Pasquinelli C, Chisari F V. Nat Med. 1996;2:1–6. doi: 10.1038/nm1096-1104. [DOI] [PubMed] [Google Scholar]
- 17.Hoofnagle J H. Hepatology. 1997;26:15S–20S. doi: 10.1002/hep.510260703. [DOI] [PubMed] [Google Scholar]
- 18.Fauci A S. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 19.Mackaness G B. Am Rev Respir Dis. 1968;97:337–344. doi: 10.1164/arrd.1968.97.3.337. [DOI] [PubMed] [Google Scholar]
- 20.Britton W J. Trans R Soc Trop Med Hyg. 1993;87:508–514. doi: 10.1016/0035-9203(93)90066-y. [DOI] [PubMed] [Google Scholar]
- 21.Thomsen A R, Johansen J, Marker O, Christensen J P. J Immunol. 1996;157:3074–3080. [PubMed] [Google Scholar]
- 22.Planz O, Ehl S, Furrer E, Horvath E, Brundler M A, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1997;94:6874–6879. doi: 10.1073/pnas.94.13.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenerman P, Hengartner H, Zinkernagel R M. Nature (London) 1997;390:298–301. doi: 10.1038/36876. [DOI] [PubMed] [Google Scholar]
- 24.Slifka M K, Matloubian M, Ahmed R. J Virol. 1995;69:1895–1902. doi: 10.1128/jvi.69.3.1895-1902.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau L L, Jamieson B D, Somasundaram T, Ahmed R. Nature (London) 1994;369:648–652. [Google Scholar]
- 26.Eichelberger M C, Wang M L, Allan W, Webster R G, Doherty P C. J Gen Virol. 1991;72:1695–1698. doi: 10.1099/0022-1317-72-7-1695. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann-Grube F. Virol Monogr. 1971;10:1–173. [Google Scholar]
- 28.Battegay M, Cooper S, Althage A, Baenziger J, Hengartner H, Zinkernagel R M. J Virol Methods. 1991;33:191–198. doi: 10.1016/0166-0934(91)90018-u. [DOI] [PubMed] [Google Scholar]
- 29.Romanowski V, Matsuura Y, Bishop D H. Virus Res. 1985;3:101–114. doi: 10.1016/0168-1702(85)90001-2. [DOI] [PubMed] [Google Scholar]
- 30.van den Broek M F, Müller U, Huang S, Aguet M, Zinkernagel R M. J Virol. 1995;69:4792–4796. doi: 10.1128/jvi.69.8.4792-4796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pircher H, Moskophidis D, Rohrer U, Bürki K, Hengartner H, Zinkernagel R M. Nature (London) 1990;346:629–633. doi: 10.1038/346629a0. [DOI] [PubMed] [Google Scholar]
- 32.Odermatt B F, Eppler M, Leist T P, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1991;88:8252–8256. doi: 10.1073/pnas.88.18.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMichael A J, Gotch F M, Dongworth D W, Clark A, Potter C W. Lancet. 1983;2:762–764. doi: 10.1016/s0140-6736(83)92297-3. [DOI] [PubMed] [Google Scholar]
- 34.Selin L K, Nahill S R, Welsh R M. J Exp Med. 1994;179:1933–1943. doi: 10.1084/jem.179.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beverley P C. Immunol Today. 1990;11:203–205. doi: 10.1016/0167-5699(90)90083-l. [DOI] [PubMed] [Google Scholar]
- 36.Gray D. Annu Rev Immunol. 1993;11:49–77. doi: 10.1146/annurev.iy.11.040193.000405. [DOI] [PubMed] [Google Scholar]
- 37.Zinkernagel R M, Bachmann M F, Kündig T M, Oehen S, Pircher H, Hengartner H. Annu Rev Immunol. 1996;14:333–367. doi: 10.1146/annurev.immunol.14.1.333. [DOI] [PubMed] [Google Scholar]
- 38.Hou S, Hyland L, Ryan K W, Portner A, Doherty P C. Nature (London) 1994;369:652–654. doi: 10.1038/369652a0. [DOI] [PubMed] [Google Scholar]
- 39.Bachmann M F, Fehr T, Freer G, Hengartner H, Zinkernagel R M. J Immunol. 1997;158:5106–5111. [PubMed] [Google Scholar]
- 40.Gray D, Matzinger P. J Exp Med. 1991;174:969–974. doi: 10.1084/jem.174.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seiler P, Brundler M A, Zimmermann C, Weibel D, Bruns M, Hengartner H, Zinkernagel R M. J Exp Med. 1998;187:649–654. doi: 10.1084/jem.187.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ochsenbein A F, Karrer U, Klenerman P, Althage A, Ciurea A, Shen H, Miller J F, Whitton J L, Hengartner H, Zinkernagel R M. Proc Natl Acad Sci USA. 1999;96:9293–9298. doi: 10.1073/pnas.96.16.9293. [DOI] [PMC free article] [PubMed] [Google Scholar]