Abstract
Plant flowering is a crucial developmental transition from the vegetative to reproductive phase and is properly timed by a number of intrinsic and environmental cues. Genetic studies have identified that chromatin modification influences the expression of FLOWERING LOCUS C (FLC), a MADS-box transcription factor that controls flowering time. Histone deacetylation and methylation at H3K9 and H3K27 are associated with repression of FLC; in contrast, methylation at H3K4 and H3K36 activates FLC expression. However, little is known about the functions of histone arginine methylation in plants. Here, we report that Arabidopsis Shk1 binding protein 1 (SKB1) catalyzes histone H4R3 symmetric dimethylation (H4R3sme2). SKB1 lesion results in upregulation of FLC and late flowering under both long and short days, but late flowering is reversed by vernalization and gibberellin treatments. An skb1-1flc-3 double mutant blocks late-flowering phenotype, which suggests that SKB1 promotes flowering by suppressing FLC transcription. SKB1 binds to the FLC promoter, and disruption of SKB1 results in reduced H4R3sme2, especially in the promoter of FLC chromatin. Thus, SKB1-mediated H4R3sme2 is a novel histone mark required for repression of FLC expression and flowering time control.
Keywords: FLC, flowering time, histone arginine methylation, PRMT5/SKB1
Introduction
The lifespan in flowering plants is highly controlled by a developmental switch for the vegetative to reproductive transition. Investigations in Arabidopsis have identified four major pathways (photoperiod, vernalization, gibberellin (GA) and autonomous) involved in regulating flowering time (Komeda, 2004; He and Amasino, 2005). The photoperiod and vernalization pathways render Arabidopsis able to flower in response to external cues. The vernalization pathway accelerates flowering, and at least three genes (VRN1, VRN2 and VIN3) are involved (Gendall et al, 2001; Levy et al, 2002; Sung and Amasino, 2004). vrn1, vrn2 and vin3 mutants reduce the vernalization response. VRN1 and VRN2 are expressed constitutively, but VIN3 expression is cold induced. The photoperiod flowering pathway is controlled by CONSTANS, CRY2, FHA, GIGANTEA, FT and FWA, and mutants for all are weakly or not at all sensitive to cold treatment (Martinez-Zapater and Somerville, 1990; Koornneef et al, 1991; Guo et al, 1998; Searle and Coupland, 2004).
The autonomous pathway is defined by a group of late-flowering mutants fca, fpa, fve, fld, ld, fy and flk, whose expression does not depend on photoperiod (Koornneef et al, 1991; Lee et al, 1994; Macknight et al, 1997; Schomburg et al, 2001; He et al, 2003; Simpson et al, 2003; Ausin et al, 2004; Lim et al, 2004; Mockler et al, 2004). In contrast to photoperiod mutant plants, the autonomous mutant plants exhibit a marked reduction in flowering time under vernalization treatment (Koornneef et al, 1991). Thus, vernalization can overcome the requirement for the autonomous pathway.
In promoting flowering, GA increases the expression of SOC1 and LEAFY, whereas photoperiod controls flowering time through regulation of CO (Komeda, 2004; He and Amasino, 2005). Autonomous and vernalization pathways both control flowering by decreasing of FLC mRNA level (Komeda, 2004; He and Amasino, 2005). FLC is a MADS-box transcription factor that plays a central role in blocking developmental transition from the vegetative to flowering stage (Michaels and Amasino, 1999a; Sheldon et al, 1999). Loss of FLC function elevates the expression of SOC1 and FT, two flowering-time integrator genes, and leads to earlier flowering (Hepworth et al, 2002). The late-flowering phenotype in autonomous pathway mutant plants was blocked by introducing flc (Michaels and Amasino, 2001; He et al, 2003). Vernalization accelerates flowering by a permanent epigenetic repression of FLC expression despite the presence of autonomous pathway mutations (Komeda, 2004).
Recent studies have revealed that FLC-mediated transition of flowering time is associated with histone covalent modification, including acetylation and methylation (He and Amasino, 2005). The autonomous pathway repressors FLD and FVE, as components of an HDAC complex, inhibit the expression of FLC by increasing deacetylation of FLC chromatin (He et al, 2003; Ausin et al, 2004). Vernalization also elevates deacetylation of histone tails of FLC chromatin and at the same time, increases methylation in H3K27 and H3K9, and decreases methylation of H3K4 (Bastow et al, 2004; He et al, 2004; Sung and Amasino, 2004; Mylne et al, 2006; Greb et al, 2007). Recently, methylation of H3K36 was found to be a histone mark required for increasing FLC expression in Arabidopsis (Zhao et al, 2005).
In addition to acetylation and methylation at lysines, methylation at histone arginine is a covalent modification that results in monomethylarginines, asymmetric dimethylarginines or symmetric dimethylarginines (Bedford and Richard, 2005; Wysocka et al, 2006). In humans, of nine protein arginine methyltransferases (PRMT1–9), three (PRMT1, PRMT4 and PRMT5) have been identified as methylating arginines of histones H2A, H3 and H4 (Bedford and Richard, 2005; Cook et al, 2006). PRMT5, also called JBP1 or Skb1, is the only enzyme that methylates histone arginines in symmetric dimethylation (Gilbreth et al, 1996; Pollack et al, 1999; Bao et al, 2001; Bedford and Richard, 2005; Wysocka et al, 2006). Histone arginine methylation has been shown to regulate chromatin remodelling, gene transcription, and cell proliferation and differentiation (Bedford and Richard, 2005; Ancelin et al, 2006; Wysocka et al, 2006; Dacwag et al, 2007; Liu et al, 2007). In plants, however, the biological functions of histone arginine methylation have never been elucidated.
In this report, we present the identification and characterization of a protein arginine methyltransferase SKB1 involved in controlling flowering time. The late-flowering phenotype of T-DNA insertional skb1 mutant plants is due to upregulation of FLC expression. SKB1 binds to FLC chromatin and catalyzes H4R3 symmetric dimethylation (H4R3sme2). H4R3sme2 is a novel histone mark associated with FLC expression.
Results and discussion
Loss or gain of SKB1 function alters flowering time in Arabidopsis
The Arabidopsis genome contains a single-copy SKB1 gene (At4g31120; see www.arabidopsis.org) of 23 exons (Figure 1A). Arabidopsis SKB1, encoding a 642-amino acid-protein of approximately 72 kDa (Figure 1B and C), shows high homology with the human PRMT5 (Pollack et al, 1999). SKB1 is about 58% identical to PRMT5 over a motif for the S-adenosylmethionine binding site and the C-terminal region of 338 amino acids, and the similarity is about 74% over the same region (see Supplementary Figure S1). SKB1 contains consensus methyltransferase regions I, post I, II, III, and a THW loop in the C-terminus (Pollack et al, 1999; Zhang et al, 2000; Zhang and Reinberg, 2001; Bedford and Richard, 2005) (Figure 1A), which is an evolutionarily highly conserved core region found in homologous proteins of diverse organisms (see Supplementary Figure S2).
Figure 1.
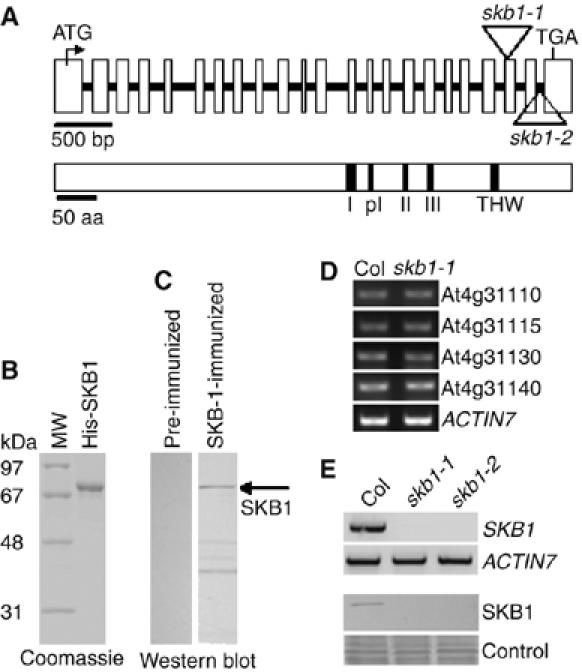
Structure of the SKB1 gene and identification of skb1 mutants. (A) Structure of the SKB1 gene and a diagram of the SKB1 protein. Exons are indicated as boxes and introns as lines. T-DNA insertions in skb1 mutants are indicated by arrowheads. The SKB1 methyltransferase regions I, post I, II, III, and THW loop are shown in black. (B) A 72-kDa 6 × His-tagged full-length SKB1 protein purified from Escherichia coli. (C) Western blot detection of Arabidopsis endogenous SKB1 protein with the use of polyclonal anti-SKB1 antiserum. (D) RT–PCR analysis of the expression of genes upstream and downstream of skb1-1. (E) RT–PCR (upper panel) and Western blot (lower panel) analyses of SKB1 expression in wild-type Col and skb1-1 and skb1-2 plants 20 days after sowing; ACTIN7 serves as an internal control; total proteins stained with Coomassie blue showed equal loading.
To characterize the function of SKB1, we identified two T-DNA insertional mutants (skb1-1 and skb1-2) at the At4g31120 region of Arabidopsis in a Columbia (Col) genetic background from the Salk T-DNA collection (Alonso et al, 2003). Polymerase chain reaction (PCR) confirmed that the T-DNA in skb1-1 and skb1-2 was inserted in exon 21 and intron 22, respectively (Figure 1A) and appeared not to affect the expression of the four genes flanking SKB1 (Figure 1D). The full-length SKB1 mRNA was undetectable in the skb1 mutants, and neither mutants expressed the SKB1 protein (Figure 1E). Both skb1-1 and skb1-2 plants flowered much later than wild-type plants under a long-day photoperiod (Figure 2A and B). In addition, as compared with wild-type plants, skb1 plants had an increased number of rosette leaves (26 leaves as compared with 12 for wild-type plants at bolting) (Figure 2B and C), and displayed severe developmental retardation 14 days after germination (DAG) (Figure 2D), which is often associated with late-flowering mutants (Lee et al, 1994; Schomburg et al, 2001; He et al, 2003; Simpson et al, 2003; Ausin et al, 2004; Kim et al, 2004; Lim et al, 2004; Mockler et al, 2004; Henderson et al, 2005). The skb1 mutant plants also formed leaves slightly more curled and darker than wild-type plants and showed slightly reduced fertility (∼85% of wild-type seed set when self-pollinated), although floral organs were normal (data not shown). The late-flowering phenotype observed only in homozygous plants revealed that the skb1 mutation is recessive.
Figure 2.
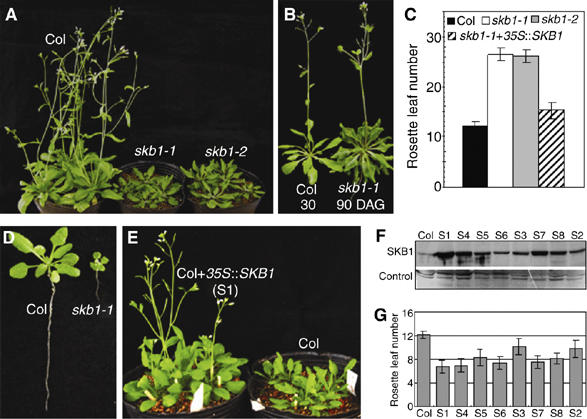
Phenotype of skb1 mutants and SKB1 overexpression. (A) Wild-type Col (Col) and skb1 mutations grown under long-day (LD) conditions for 35 days after germination (DAG). (B) Col and skb1-1 mutant at the flowering stage grown under LD. (C) Flowering times of Col, skb1 mutants and transgenic plants under LD. (D) Col and skb1-1 mutant at 14 DAG under LD. (E) Col- and SKB1-overexpressing (S1) plants at 24 DAG under LD. (F) Abundance of SKB1 protein levels in homozygous transgenic seedlings (12 DAG under LD) determined by Western blot with anti SKB1 antibody. (G) Flowering times of the transgenic plants in (F) under LD. Bars represent means±s.d. of rosette leaf number at bolting. For each line, 20 plants were scored.
We introduced a 35S∷SKB1 construct that expressed SKB1 cDNA constitutively into skb1-1 mutant plants. 35S∷SKB1 could rescue the skb1-1 mutant, resulting in transgenic plants with a wild-type phenotype (Figure 2C). Further, by introducing 35S∷SKB1 into wild-type plants, plants overexpressing SKB1 had early-flowering features (Figure 2E and G), and SKB1-promoted flowering time showed a stoichiometric relation with SKB1 protein level (Figure 2F and G). As loss of SKB1 function induced late flowering and overexpression of SKB1 resulted in early flowering, we concluded that SKB1 is a positive regulator of floral initiation. Gain-of-function 35S∷SKB1 transgenic plants flowering early resembled those from a previous report showing that overexpression of FLK, a gene of the autonomous pathway, accelerates the flowering process (Mockler et al, 2004), but differed from FVE-overexpressing plants, which did not show altered flowering time (Ausin et al, 2004).
SKB1 is a new member of the autonomous pathway repressing FLC expression
As four major pathways (photoperiod, vernalization, GA and autonomous) have been found to regulate flowering time in Arabidopsis thaliana (Komeda, 2004), we sought to identify the pathway that SKB1 was involved in by treating skb1 mutant plants with the different physiologic conditions. Under long- or short-day photoperiods, both skb1-1 and skb1-2 mutant plants displayed later flowering than wild-type plants, but flowered later in short day than in long day (Figure 3A and C), which indicates that skb1 mutants were sensitive to photoperiod. After exposure to vernalization, skb1 mutant plants flowered rapidly (Figure 3A and B), which is similar to results for several autonomous pathway mutants (Schomburg et al, 2001; He et al, 2003; Simpson et al, 2003; Ausin et al, 2004; Lim et al, 2004; Mockler et al, 2004; Henderson et al, 2005). GA treatment reversed in part the delayed flowering of skb1 mutant plants (Figure 3A and D), an observation consistent with that of the autonomous pathway mutants flk and fca, which respond to both vernalization and GA (Michaels and Amasino, 1999b; Mouradov et al, 2002; Lim et al, 2004). Taken together, these results suggest that SKB1 is a new member of the autonomous pathway and prevents late flowering.
Figure 3.
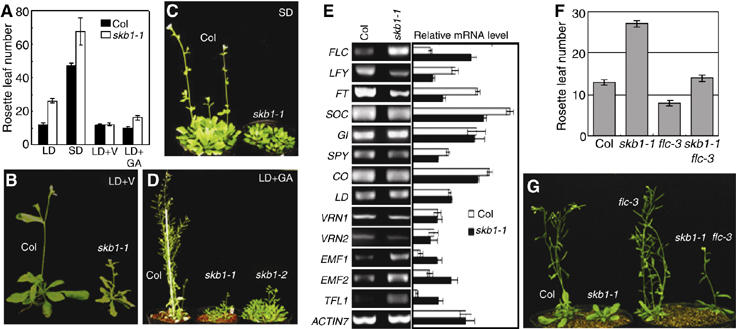
Analysis of skb1 mutant response to environmental cues and target genes. (A) Flowering times of skb1-1 under LD, short-day (SD), vernalization (V) and GA treatment. Bars represent means±s.d. For each line, 30–50 plants were scored. (B) Col and skb1-1 mutant plants germinated and grown at 2–4°C for 6 weeks and moved to LD conditions for 26 days. (C) Plants grown at 98 DAG in SD. (D) Plants treated with GA3 and grown at 37 DAG under LD. (E) RT–PCR analysis of flowering control genes expression in wild-type and skb1-1 mutant plants. RNA was isolated from 20-day-old seedlings grown under LD. ACTIN7 was used as a loading control. Signal intensities were normalized relative to ACTIN7 with the use of ImageQuant and shown by bar graphs at the right from three independent experiments. (F) Flowering time of Col, skb1-1, flc-3 and skb1-1 flc-3. Ten plants were scored and grown under LD. (G) The phenotypes of wild-type Col, skb1-1, flc-3 and skb1-1 flc-3 mutant grown under LD.
To investigate the molecular mechanism underlying the late-flowering phenotype of skb1, we examined the transcript levels of critical genes in the different flowering pathways (Komeda, 2004; He and Amasino, 2005). Consistent with their showing a normal photoperiod-responsive phenotype, skb1 mutants and wild-type plants did not show a significant difference in transcript levels of GI and CO genes, two key components of the photoperiod pathway (Samach et al, 2000; Suarez-Lopez et al, 2001); SPY in the GA pathway (Tseng et al, 2004); VRN1 and VRN2 in the vernalization pathway (Gendall et al, 2001; Levy et al, 2002) (Figure 3E). The expression of LD, in the autonomous pathway upstream of FLC (He and Amasino, 2005), did not differ between skb1-1 and wild-type plants. The expression of the flowering repressor gene FLC, however, was significantly upregulated in skb1-1 mutant plants (Figure 3E). Increased FLC expression is a characteristic of other autonomous mutants such as fca, ld, fy, fld, flk, fpa and fve (Schomburg et al, 2001; He et al, 2003; Simpson et al, 2003; Ausin et al, 2004; Mockler et al, 2004; Henderson et al, 2005). Loss of SKB1 downregulated LFY, FT and SOC1, and upregulated EMF1, EMF2 and TFL1 (Figure 3E). LFY, FT and SOC1 are all downstream of FLC and are expressed in an FLC-dependent manner (He and Amasino, 2005). The function of EMF1, EMF2 and TFL1 genes may not influence the four flowering pathways, but their reduced expression induces early flowering (Shannon and Meeks-Wagner, 1991; Moon et al, 2003; Komeda, 2004).
To examine whether accumulation of FLC mRNA directly results in late flowering in skb1 mutant plants, we introduced skb1-1 into an FLC-null mutant, flc-3 (Michaels and Amasino, 1999a). The late-flowering phenotype of skb1-1 was suppressed by flc-3 (Figure 3F and G). Thus, the flowering time control by SKB1 is targeted towards FLC expression. The skb1-1 flc-3 double-mutant plants (14.8 rosette leaves) flowered later than flc-3 (9 rosette leaves) or wild-type Col plants (12.7 rosette leaves) but much earlier than skb1-1 mutant plants (27 rosette leaves). It is likely that FLC-independent factors also contribute to the later flowering phenotype of the skb1 mutants. This assumption is consistent with the previous observation of upregulated expression of EMF1, EMF2 and TFL1 (Figure 3E).
Expression pattern of SKB1 protein and mRNA
The temporal and spatial expression patterns of the SKB1 protein was analyzed with the use of a specific antibody against SKB1 (Figure 1C). Western blotting results showed that SKB1 was highly expressed in flowers, roots and siliques, but less so in stems and mature leaves (Figure 4A). The SKB1 protein was maintained at high levels during vegetative development (from 5 to 20 days after germination) (Figure 4B). Consistent with the protein expression pattern, SKB1 RNA was more abundant in shoot apex, young leaves and leaf primordia, floral and inflorescence meristems (Figure 4C–E), and was also expressed in gynoecium, stamens, sepals and young siliques (especially ovules), but not as much in older leaves, petals and vascular tissues (Supplementary Figure S3). This expression pattern was similar to that of genes affecting flowering transition (Lee et al, 1994; Michaels and Amasino, 1999a; Schomburg et al, 2001; He et al, 2003, 2004; Simpson et al, 2003; Ausin et al, 2004; Bastow et al, 2004; Mockler et al, 2004; Henderson et al, 2005; Zhao et al, 2005).
Figure 4.
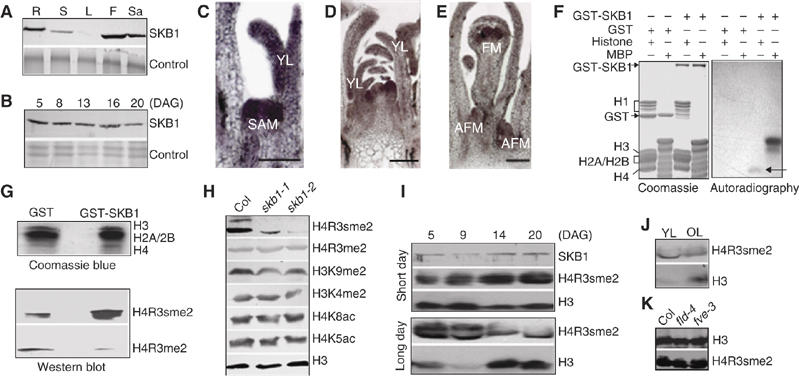
Analysis of SKB1 expression pattern and methyltransferase activity. (A, B) Immunoblot analyses of SKB1 with the use of polyclonal anti-SKB1 antiserum. (A) Total protein extracts from different tissues of the wild type at 38 days. R, root; S, stem; L, leaf; F, flower; Sa, silique. Control shows equal loading. (B) Expression of SKB1 protein at different developmental stages. Col was grown under LD and whole parts were harvested at the indicated growth stages for total protein isolation. (C–E) RNA in situ hybridization analysis of SKB1 expression in Col grown under LD for 10 (C), 21 (D) and 26 (E) days. AFM: axillary flower meristem; SAM, shoot apical meristem; FM, floral meristem; YL, young leaf. Scale bar, 100 μm. (F) In vitro methylation of histones (10 μg) and myelin basic protein (MBP, 10 μg) by GST-SKB1 (2 μg) purified from E. coli. GST (3 μg) was a negative control; left, Coomassie blue-stained gel; right, autoradiograph of 3H-labelled proteins produced by in vitro methylation; methylated H4 is indicated by an arrow. (G) SKB1-mediated H4R3sme2 was analyzed by Western blot with specific antibodies. (H) Western blot analysis of modification of histone H3 and H4. Histone-enriched protein extracts from 20-day-old Col and skb1 mutant plants grown under LD were probed with antibodies that specifically recognize the indicated forms of histone H3 and H4. (I) Levels of SKB1 and H4R3sme2 at different developmental stages in Col. Total soluble proteins isolated from seedlings grown in SD were exposed to antibody against SKB1. Histone-enriched proteins were extracted from plants grown under SD or LD and immunoblotted with antibodies against H4R3sme2 and H3. (J) The level of H4R3sme2 in specific tissues of Col grown under LD. YL: young leaves plus apex from seedlings at 5 DAG; OL: old leaves from plants at 35 DAG. (K) Abundance of H4R3sme2 in Col, fld-4 and fve-3. Proteins were isolated from seedlings grown at 20 DAG under LD.
Arginine methyltransferase SKB1 methylates H4R3sme2 both in vitro and in vivo
PRMT5, the human homologue of SKB1, methylates histones H2A, H3 and H4 (Pollack et al, 1999; Ancelin et al, 2006; Dacwag et al, 2007). To examine whether Arabidopsis SKB1 methylates histone arginines, we assayed in vitro a GST-SKB1 fusion protein, purified by affinity chromatography from Escherichia coli, for methylation activity, with histones and myelin basic protein (MBP) used as substrates. Both histone and MBP were methylated, but of the five histones (H1, H2A, H2B, H3 and H4), only H4 was methylated (Figure 4F). We then used specific antibodies to examine whether SKB1 methylates H4R3. GST-SKB1 catalyzed H4R3 symmetric dimethylation (H4R3sme2) but not asymmetric dimethylation (Figure 4G), which suggests that SKB1 can symmetrically methylate H4R3 as does PRMT5 (Pollack et al, 1999; Ancelin et al, 2006).
We then examined whether SKB1 methylates H4R3 in vivo. Figure 4H shows the skb1 mutants, with no difference from wild type in the level of asymmetric dimethylated H4R3, but markedly reduced level of symmetric dimethylated H4R3, which indicates that H4R3 contains symmetric and asymmetric forms of dimethylation, and that SKB1 is a major enzyme controlling H4R3 symmetric dimethylation in Arabidopsis. Further investigation revealed that impaired H4R3 symmetric dimethylation had little effect on dimethylated H3K4, H3K9 and acetylated H4K5 and H4K8 (Figure 4H).
SKB1 and the histone mark H4R3sme2 are associated with FLC expression
The level of H4R3sme2 was also examined during developmental stages of the wild-type Col line. Plants grown under short- and long-day conditions at 5, 9, 14 and 20 days after germination were collected for protein isolation. SKB1 protein levels were weakly increased with plant age under short-day condition (Figure 4I) and slightly decreased with age under long-day conditions (Figure 4B). Consistent with SKB1 expression, H4R3sme2 was maintained at high levels during vegetative development under both short- and long-day photoperiods, but showed a weak increase under short-day and a slight decrease under long-day condition with plant age (Figure 4I). As FLC is also highly expressed at these states (Michaels and Amasino, 1999a), SKB1 activity and H4R3sme2 could be associated with FLC expression. This association was further confirmed by tissue-specific differences. Both FLC and SKB1 are highly expressed in young leaves and apical meristems, with low levels in old leaves (Michaels and Amasino, 1999a) (Figure 4A–E). Indeed, H4R3sme2 was also detected at high levels in young leaves and apical meristems, but at low levels in old leaves (Figure 4J).
We then examined H4R3sme2 in other late-flowering mutants: fld-4 and fve-3, two autonomous pathway mutants upregulating FLC expression by increasing histone acetylation (He et al, 2003; Ausin et al, 2004; Kim et al, 2004). Histone-enriched proteins were isolated from plants grown for 20 days after germination under long-day conditions. The level of H4R3sme2 in both fld-4 and fve-3 mutant plants did not significantly differ from that in wild-type plants (Figure 4K), which suggests that increased histone acetylation resulting from loss of FLD or FVE function may not affect the H4R3sme2 mark on global level.
SKB1 associates with FLC chromatin and is required for symmetric dimethylation of H4R3 in FLC chromatin
The transcription of FLC regulated through chromatin remodelling has been well studied (He et al, 2003, 2004; Bastow et al, 2004; Zhao et al, 2005). In addition, the mammalian SKB1 homologue PRMT5 interacts with chromatin and suppresses gene transcription (Bedford and Richard, 2005). We thus asked whether SKB1 binds to the five FLC chromatin regions covering its promoter (region A), the first exon (region B) and the first intron (regions C–E), as was previously reported (Bastow et al, 2004) (Figure 5A). Chromatin immunoprecipitation (ChIP) assay revealed that SKB1 antibody could specifically pull down the FLC promoter (region A) (Figure 5B), a chromatin region that is critical for FLC transcription (He et al, 2003; Bastow et al, 2004). SKB1 association with other regions (B–E) was undetectable (Figure 5B). By contrast, SKB1 antibody could not pull down DNA in regions A–E in the skb1-1 mutant (Figure 5B). Consistent with SKB1 strongly binding to the FLC promoter, the FLC promoter also showed a high level of H4R3sme2 in the wild type; in contrast, the skb1 mutant showed greatly decreased H4R3sme2 (Figure 5B and Supplementary Figure S4).
Figure 5.
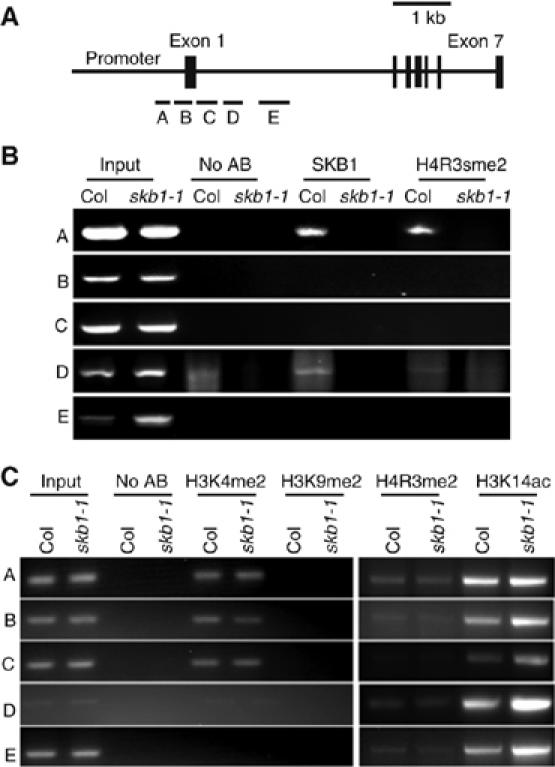
ChIP assays of wild type and skb1-1 mutant at the FLC locus. (A) A diagram of the FLC gene structure, with bars representing the A–E regions examined by ChIP. (B, C) ChIP results with antibodies against SKB1, H4R3sme2, H3K4me2, H3K9me2, H4R3me2 and acetylated H3K14 (H3K14ac). Samples were from wild-type and skb1-1 plants in LD for 20 days. The input is chromatin before immunoprecipitation. ‘No AB': the control sample lacks an antibody. ChIP assays involved at least three independent experiments; data represent results of one experiment.
In addition, consistent with a previous report (Bastow et al, 2004), dimethylation in H3K9 was undetectable in all regions in both wild-type and mutant plants (Figure 5C). The asymmetric dimethylated H4R3 was weakly detected at FLC chromatin regions A, B, D and E, with no difference between the wild type and skb1 mutant (Figure 5C). Compared with the wild type, the skb1 plants showed no change in the levels of dimethylated H3K4 in all regions (Figure 5C). The acetylation in H3K14 was found in all regions (A–E) both in the wild type and skb1 mutant, with increased levels in skb1 mutant in regions A–E (Figure 5C and Supplementary Figure S4), which is consistent with a previous report of hyperacetylation activating FLC expression (He et al, 2003; Ausin et al, 2004). These findings suggest that SKB1 affects flowering development by alterating FLC expression via symmetric dimethylation of H4R3 in its promoter.
Based on our findings, we propose that SKB1 methylates H4R3 of the FLC chromatin symmetrically, which in turn suppresses FLC expression to induce flowering. H4R3 is also asymmetrically dimethylated by PRMT1 in animal cells, which facilitates subsequent acetylation at H3K9, H3K14, H4K5 and H4K12 and is required for transcriptional activation (Huang et al, 2005). Interestingly, in plants, reduced H4R3 symmetric dimethylation resulting from SKB1 knockdown has no effect on H4R3 asymmetric dimethylation but, rather, increases H3K14 acetylation in the FLC chromatin and activates or maintains its transcription (Figure 5B and C). This finding suggests that symmetric and asymmetric dimethylations in H4R3 have distinct functions in regulation of chromatin status and gene transcription. Further exploration will define the difference in biological function between symmetric and asymmetric dimethylation in H4R3.
Materials and methods
Plant materials and growth conditions
Arabidopsis ecotype Columbia (Col) was used in this work. skb1-1 and skb1-2 alleles were isolated from the SALK T-DNA collection (Salk_065814 and Salk_095085). flc-3, fld-4 and fve-3 were described previously (Michaels and Amasino, 1999a; He et al, 2003; Ausin et al, 2004). Plants were grown at 22°C under long-day (16 h light and 8 h dark) or short-day (8 h light and 16 h dark) conditions. Vernalization treatment was as described (Bastow et al, 2004) and GA treatment was as described (Lim et al, 2004). Flowering time was measured as the number of rosette leaves at bolting.
RT–PCR analysis
RNA was isolated from 20-day-old seedlings grown under long-day conditions with use of TRI reagent as recommended by the manufacturer. cDNAs were synthesized from 2.0 μg of total RNA by use of Superscript reverse transcriptase. RT–PCR was performed with gene-specific primers (see Supplementary Table SI) and runs were 18–25 cycles depending on the linear range of products for each gene.
Western blot analysis
Histone-enriched protein extraction from 20-day-old seedlings with or without vernalization treatment was as described (Houben et al, 2003). Western blot analysis was performed with Upstate antibodies against histone H4 dimethyl Arg 3 (catalogue no. 07-213), H4 acetyl Lys 5 (catalogue no. 07-327), H4 acetyl Lys 8 (catalogue no. 07-328), H3 dimethyl Lys 4 (catalogue no. 05-790), H3 dimethyl Lys 9 (catalogue no. 05-768), H3 acetyl Lys 14 (catalogue no. 07-353), H3 (catalogue no. 07-690) and with Abcam antibodies against H4 symmetric dimethyl Arg 3 (catalogue no. 5823). Anti-SKB1 polyclonal antibody was generated with 6 × His-tagged full-length SKB1 protein purified from E. coli.
In situ hybridization
Tissue preparation of 10-, 21- and 26-day-old seedlings grown under long day, digoxigenin labelling of RNA probes and in situ hybridization were performed as described (Wu et al, 2006). DNA fragments containing the 330-base-pair (21–350) coding region of SKB1 were used as sense and antisense probes.
Chromatin immunoprecipitation
ChIP assay involved the usage of 20-day-old seedlings grown under long-day conditions as previously described (Bowler et al, 2004). Primers and PCR detection of FLC regions were as described (Bastow et al, 2004).
Constructs, protein purification and methylation analysis
Full-length SKB1 cDNA was cloned into pBI121, resulting in the binary vector p35S∷SKB1, which was introduced into Agrobacterium tumefaciens to transform Arabidopsis plants. Full-length SKB1 cDNA was cloned into pGEX4T-1 in-frame. GST-SKB1 expression, purification and methylation assays were as previously described (Bao et al, 2001).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table SI
Acknowledgments
We thank Jiayang Li for his useful comments on the manuscript, Xiaofeng Cao for supplying the greenhouse and Richard Amasino, Jianru Zuo and Ligeng Ma for seeds of the flc-3, fld-4 and fve-3 mutant lines, respectively. This work was supported by National Basic Research Program of China (2005CB522400), the President's Fund of the Chinese Academy of Sciences to SB; the National Natural Sciences Foundation of China (NSFC) for distinguished Young Scholars (30525026) to KC; and NSFC (30671186) to XW.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8: 623–630 [DOI] [PubMed] [Google Scholar]
- Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM (2004) Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36: 162–166 [DOI] [PubMed] [Google Scholar]
- Bao S, Qyang Y, Yang P, Kim H, Du H, Bartholomeusz G, Henkel J, Pimental R, Verde F, Marcus S (2001) The highly conserved protein methyltransferase, Skb1, is a mediator of hyperosmotic stress response in the fission yeast Schizosaccharomyces pombe. J Biol Chem 276: 14549–14552 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S (2005) Arginine methylation an emerging regulator of protein function. Mol Cell 18: 263–272 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Cook JR, Lee JH, Yang ZH, Krause CD, Herth N, Hoffmann R, Pestka S (2006) FBXO11/PRMT9, a new protein arginine methyltransferase, symmetrically dimethylates arginine residues. Biochem Biophys Res Commun 342: 472–481 [DOI] [PubMed] [Google Scholar]
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN (2007) The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol 27: 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C (2001) The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535 [DOI] [PubMed] [Google Scholar]
- Gilbreth M, Yang P, Wang D, Frost J, Polverino A, Cobb MH, Marcus S (1996) The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc Natl Acad Sci USA 93: 13802–13807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17: 73–78 [DOI] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM (2005) Role of chromatin modification in flowering-time control. Trends Plant Sci 10: 30–35 [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Michaels SD, Amasino RM (2003) Regulation of flowering time by histone acetylation in Arabidopsis. Science 302: 1751–1754 [DOI] [PubMed] [Google Scholar]
- Henderson IR, Liu F, Drea S, Simpson GG, Dean C (2005) An allelic series reveals essential roles for FY in plant development in addition to flowering-time control. Development 132: 3597–3607 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben A, Demidov D, Gernand D, Meister A, Leach CR, Schubert I (2003) Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J 33: 967–973 [DOI] [PubMed] [Google Scholar]
- Huang S, Litt M, Felsenfeld G (2005) Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev 19: 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Hyun Y, Park JY, Park MJ, Park MK, Kim MD, Kim HJ, Lee MH, Moon J, Lee I, Kim J (2004) A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet 36: 167–171 [DOI] [PubMed] [Google Scholar]
- Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, van der Veen JH (1991) A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol Gen Genet 229: 57–66 [DOI] [PubMed] [Google Scholar]
- Lee I, Aukerman MJ, Gore SL, Lohman KN, Michaels SD, Weaver LM, John MC, Feldmann KA, Amasino RM (1994) Isolation of LUMINIDEPENDENS: a gene involved in the control of flowering time in Arabidopsis. Plant Cell 6: 75–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C (2002) Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297: 243–246 [DOI] [PubMed] [Google Scholar]
- Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhou Z, Chen G, Bao S (2007) A putative transcriptional elongation factor hIws1 is essential for mammalian cell proliferation. Biochem Biophys Res Commun 353: 47–53 [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C (1997) FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89: 737–745 [DOI] [PubMed] [Google Scholar]
- Martinez-Zapater JM, Somerville CR (1990) Effect of light quality and vernalization on late-flowering mutants of Arabidopsis thaliana. Plant Physiol 92: 770–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999a) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999b) The gibberellic acid biosynthesis mutant ga1–3 of Arabidopsis thaliana is responsive to vernalization. Dev Genet 25: 194–198 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR, Chory J, Lin C (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA 101: 12759–12764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon YH, Chen L, Pan RL, Chang HS, Zhu T, Maffeo DM, Sung ZR (2003) EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouradov A, Cremer F, Coupland G (2002) Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 (Suppl): S111–S130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem 274: 31531–31542 [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM (2001) FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, Coupland G (2004) Induction of flowering by seasonal changes in photoperiod. EMBO J 23: 1217–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR (1991) A mutation in the Arabidopsis TFL1 gene affects inflorescence meristem development. Plant Cell 3: 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120 [DOI] [PubMed] [Google Scholar]
- Sung S, Amasino RM (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Tseng TS, Salome PA, McClung CR, Olszewski NE (2004) SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Ma Q, Yam KM, Cheung MY, Xu Y, Han T, Lam HM, Chong K (2006) In situ expression of the GmNMH7 gene is photoperiod-dependent in a unique soybean (Glycine max [L.] Merr.) flowering reversion system. Planta 223: 725–735 [DOI] [PubMed] [Google Scholar]
- Wysocka J, Allis CD, Coonrod S (2006) Histone arginine methylation and its dynamic regulation. Front Biosci 11: 344–355 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou L, Cheng X (2000) Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J 19: 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7: 1256–1260 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table SI


