Abstract
Expansion of polyglutamine repeats in several unrelated proteins causes neurodegenerative diseases with distinct but related pathologies. To provide a model system for investigating common pathogenic features, we have examined the behavior of polyglutamine expansions expressed in Caenorhabditis elegans. The expression of polyglutamine repeats as green fluorescent protein (GFP)-fusion proteins in body wall muscle cells causes discrete cytoplasmic aggregates that appear early in embryogenesis and correlates with a delay in larval to adult development. The heat shock response is activated idiosyncratically in individual cells in a polyglutamine length-dependent fashion. The toxic effect of polyglutamine expression and the formation of aggregates can be reversed by coexpression of the yeast chaperone Hsp104. The altered homeostasis associated with polyglutamine aggregates causes both the sequestration of an otherwise soluble protein with shorter arrays of glutamine repeats and the relocalization of a nuclear glutamine-rich protein. These observations of induced aggregation and relocalization have implications for disorders involving protein aggregation.
The folding of a protein to its native state is a complex process that occurs against significant odds and often involves the transient appearance of conformational intermediates that are susceptible to misfolding (1). The efficiency of protein folding can be affected by several factors, including primary sequence mutations or alterations in the cellular environment. As a result of these factors, abnormal protein conformations can arise that may be prone to aggregation. This mechanism operates in a number of human diseases including transmissible spongiform encephalopathy, Alzheimer's disease, and the CAG repeat diseases (2, 3). A common theme is the initial appearance of an altered protein conformation, which can apparently act as a seed for misfolded proteins, initiating a cascade of events that culminates in the deposition of proteinaceous aggregates inside or outside the cell (4). The presence of misfolded proteins itself can be a potent force to initiate and sustain protein aggregation leading to pathology and disease.
The CAG repeat disorders are caused by expansions of polyglutamine tracts in unrelated proteins and include Huntington's disease, dentatorubral pallidoluysian atrophy, spinal bulbar muscular atrophy, and several types of spinocerebellar ataxias (5). Huntington's disease, the most frequent of these autosomal dominant diseases, is characterized by insoluble granular and fibrous deposits of huntingtin protein in neurons (6, 7). Pathology is typically associated with polyglutamine expansions of greater than 40 residues, and the longer the length of the expansion, the earlier the onset of disease (8).
Several in vitro and in vivo approaches have demonstrated that the expansion of glutamine repeats is the root cause of pathogenesis. It also produces an aggregation of the proteins containing the expansion. The expression of polyglutamine repeats containing proteins in different model animal systems reproduces features of the diseased state, such as the age-dependent appearance of aggregates and neuronal death (9–13). Huntingtin aggregates in vivo contain a truncated fragment retaining the glutamine repeats, presumably due to proteolysis (7, 14, 15). The formation of aggregates in vitro depends on the appearance of fragments of huntingtin containing polyglutamine repeats, consistent with the proposal that protein aggregate formation is generally preceded by the appearance of an initial seed of misfolded protein (16). Studies of a model polyglutamine peptide in vitro suggest a model in which the polyglutamine strands self-associate in β-sheets strongly held together by hydrogen bonds (17).
The appearance of misfolded protein has been proposed to be the conserved signal for activation of the heat shock response. Specific examples include the expression of a mutant λ repressor in Escherichia coli and an aberrant flight muscle actin in Drosophila (18, 19). The elevated levels of heat shock proteins, also termed molecular chaperones because of their involvement in various steps of protein folding, assist the cell in restoring the protein-folding homeostasis after stress exposure (20, 21). Recent evidence has also pointed to a direct role for the chaperones HDJ-1 (22), HDJ-2 (22–24), and Hsp104 (25) in preventing polyglutamine aggregate formation and a role for Hsp70 and HDJ-1 in eliminating the toxicity of polyglutamine expression (12, 26). The effect of the yeast chaperone Hsp104 on polyglutamine aggregates correlates with its ability to resolubilize and reactivate aggregated proteins in vitro and its ability to eliminate the [PSI+] prion-like element when overexpressed. Although a direct ortholog for Hsp104 has not been identified in humans or Caenorhabditis elegans, there are several paralogs in the C. elegans genome database (27).
The forces that govern the misfolding of proteins with glutamine expansions, the nature of the toxic species, the mechanism by which these species produce diverse pathologies, and the manner in which polyglutamine–protein misfolding and pathologies interface with homeostatic physiological responses are problems of enormous difficulty that would benefit from the establishment of simpler, genetically tractable model systems. Here we set aside the complex issue of the differential toxicity caused by individual polyglutamine proteins in neurons, and focus instead on an underlying common thread: polyglutamine length-dependent protein misfolding. Using the nematode C. elegans, we examine the ability of an otherwise innocuous protein with a polyglutamine expansion to induce toxicity and a homeostatic response, to affect the folding of other proteins, and to be influenced by molecular chaperones.
Methods
Plasmid Constructs.
The plasmids pEGFP-N1-Q19 and pEGFP-N1-Q82 (28) contain the polyglutamine-encoding sequences cloned as a cassette, into the EcoRI and BamHI sites of pEGFP-N1, or pEYFP-N1 and pECFP-N1 (CLONTECH), respectively, resulting in yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) constructs. The cDNAs encoding the relevant polyglutamine-fluorescent fusion proteins or untagged YFP were subcloned into the NheI and KpnI sites [for the Q19/Q82-green fluorescent protein (GFP) fusions], NheI and SalI sites (for Q19-YFP/Q82-CFP fusions), or the KpnI site (for YFP) of plasmid pPD30.38 (29) for expression in body wall muscle cells. The DNA fragment corresponding to Q82-GFP was subcloned into pPD118.33 (30), creating myo-2∷Q82-GFP for expression in pharyngeal cells.
The DNA-encoding residues 294 to 534 of heterogeneous ribonuclear protein (HRP) 1 was subcloned into BglII and AgeI sites of pEYFP-NI and subcloned into pPD30.38, resulting in unc-54∷HRP1-YFP. PCR primers were used to amplify DNA sequences, encoding wild-type and mutant Hsp104 from the constructs pYS104 (31) and pKT218 + KT620 (32), respectively, and Ydj, which were subcloned into the SalI and NcoI sites of pPD30.38.
C. elegans Protocols.
Nematodes were maintained by using standard procedures (33). Subsequent to staining for β-galactosidase activity (34), the animals were processed for immunostaining with anti-GFP Abs (CLONTECH) by using 0.1% Triton X-100 in solutions for Ab incubation and washing (35). Alternatively, fixed animals were treated with 1 mg/ml RNase A overnight at 37°C, followed by the addition of propidium iodide (30 μg/ml) to stain the nuclei, washed three times in PBS, and mounted. For heat shock treatments, the animals were incubated at 33°C for 1 h, collected by centrifugation, and treated with formaldehyde.
A mixture of the plasmids encoding the GFP fusions, and chaperones were coinjected with the rol-6(su1006) dominant marker plasmid pRF4, at a total concentration of 200 μg/ml, into early-adult hermaphrodites (36, 37). The injection mixtures contained 50 μg/ml each of pRF4, pPD30.38-Q82-GFP, pPD118.33-Q82-GFP, and pPD30.38-Hsp104 or pPD30.38-mutant Hsp104 or pPD30.38-Ydj1. For each combination of plasmid DNAs, the injections yielded multiple lines that expressed the fusion protein and chaperones from extrachromosomal arrays (37). pPD30.38-Q82-CFP and pPD118.33-Q82-GFP or pPD30.38-Q19-YFP were coinjected, along with a plasmid containing sqt-1(sc13), into PC72 ubIn5 animals that contain an integrated copy of the hsp16∷lacZ reporter. The sqt-1(sc13) mutation causes a left-roller phenotype, which can suppress the rol-6(su1006) right-roller phenotype (38).
Fluorescence Microscopy.
Animals were examined with Zeiss Axioscope and Nikon Optiphot epifluorescence microscopes equipped with MTI 3CCD charge-coupled device camera and IP Lab (Fairfax, VA) spectrum imaging software. The GFP fusion proteins were visualized by using XF-114-E (CFP), XF116-E (GFP), and XF-104-E (YFP) filter sets (Omega Optical, Brattleboro, VT). Propidium iodide staining was visualized by using filters used for Texas red. Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA) was used for pseudocoloration of images, and image overlays.
Immunoblotting Analysis.
Extracts were prepared by resuspending frozen worm pellets in buffer C (20 mM Hepes, pH 7.9/25% glycerol/0.42 M NaCl/1.5 mM MgCl2/0.2 mM EDTA/0.5 mM PMSF/0.5 mM DTT) and sonicating intermittently for a total of 30 s (Branson Sonifier 450 Output: 6). The sonicates were centrifuged at 40,000 × g for 5 min, and the supernatants were analyzed on 12% native PAGE and SDS/PAGE. Alternatively, the worm pellets were resuspended in 1× Laemmli sample buffer containing 0.4 or 2% SDS, boiled for 10 min, and analyzed by SDS/PAGE. Blots were probed with a 1:200 dilution of anti-GFP peptide polyclonal antiserum (CLONTECH), 1:10,000 or 1:2,500 dilution of anti-Hsp104 or anti-Ydj1 polyclonal antiserum, respectively, and visualized by chemiluminescence (ECL, Amersham).
Results
Expansion of Polyglutamine Repeats Results in the Appearance of Protein Aggregates in C. elegans.
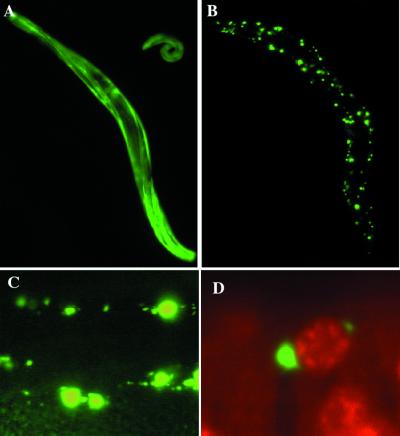
The effect of polyglutamine expansions in C. elegans was examined by expressing GFP fusion proteins with 19 or 82 glutamine residues (Q19-GFP or Q82-GFP) in body wall muscle cells under the control of the unc-54 promoter. Whereas Q19-GFP was distributed evenly throughout the body wall muscle cells (Fig. 1A), Q82-GFP formed discrete intracellular aggregates, generally more than two per cell (Fig. 1, B–D). These aggregates were detected in early embryos and increased in number and size during development from the larval to the adult stage. At higher magnification, these foci of Q82-GFP were found to be irregular in size (Fig. 1C) and cytoplasmic (Fig. 1D).
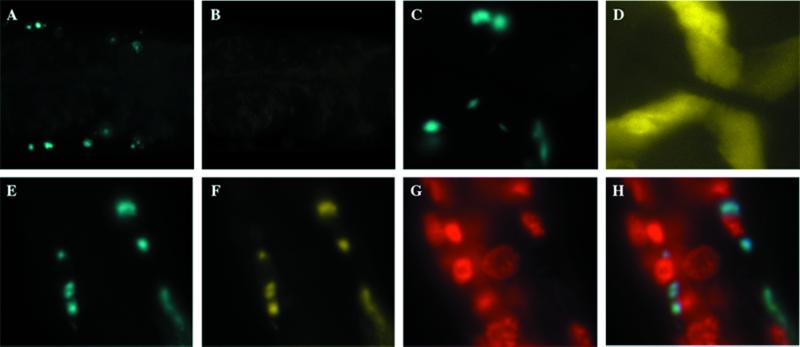
Figure 1.
Expression of polyglutamine expansions results in protein aggregates in C. elegans. The localization of Q19-GFP (A) and Q82-GFP (B) expressed in body wall muscle cells in adult C. elegans detected by fluorescence microscopy. (C) Q82-GFP protein aggregates in body wall muscle cells of adults observed at high magnification (×40) reveals that the aggregates varied widely in size. (D) The cytoplasmic localization of Q82-GFP aggregates was determined by propidium iodide staining (in red) and presented as a merged image (×100).
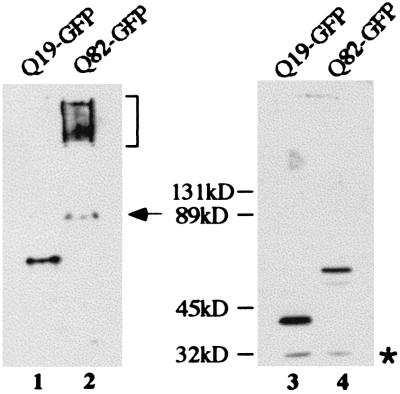
The biochemical properties of Q19-GFP and Q82-GFP were investigated by native and denaturing PAGE (Fig. 2 A–C). Under native conditions, Q82-GFP exhibited the properties of a high molecular weight complex that was largely retained in the stacking gel (Fig. 2A). In contrast, Q19-GFP was soluble, with a mobility corresponding to its molecular size (Fig. 2A). Under mild denaturing conditions, the majority of Q82-GFP remained insoluble and was trapped in the well of the gel (Fig. 2B). However, the protein was solubilized by higher concentrations of denaturant, indicating that the aggregates are not formed by covalent linkage (Fig. 2C).
Figure 2.
Biochemical properties of Q19-GFP and Q82-GFP. Extracts of animals expressing Q19-GFP and Q82-GFP were separated by native (lanes 1 and 2) and denaturing (lanes 3 and 4) PAGE and immunoblotted with anti-GFP Ab. Q19-GFP exhibits a mobility that corresponds to the size expected for a soluble monomer on native (lane 1) and denaturing (lane 1) gels, whereas Q82-GFP runs as a high molecular mass complex at the top of the native gel (lane 2) and as a monomer on the denaturing gel (lane 4). The arrow indicates a band (lane 2) with the mobility of the Q82-GFP monomer and the asterisk (lane 4) corresponds to GFP alone.
Expression of Polyglutamine Expansions Constitutively Activates the Heat Shock Response.
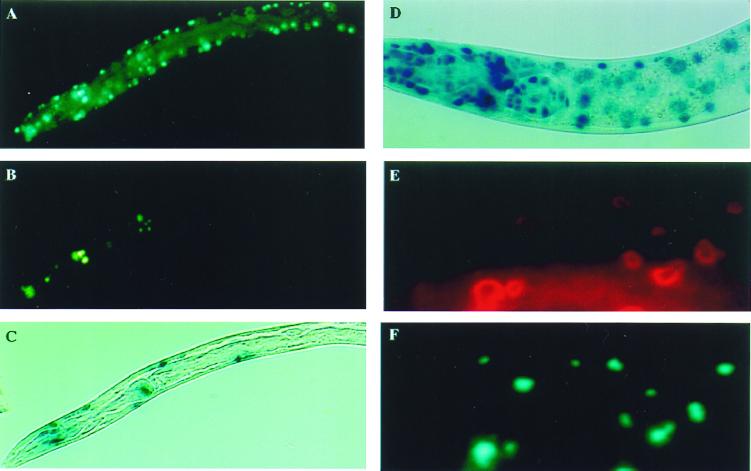
To examine the induction of the stress response, the polyglutamine–GFP fusions were expressed in PC72 animals that contain integrated copies of a hsp16∷lacZ reporter (39). Expression of Q82-CFP in body wall and Q82-GFP in pharyngeal muscle cells, regulated by the unc-54 and myo-2 promoters, respectively, resulted in protein aggregates in the respective tissues (Fig. 3 A and B). Expression of these proteins selectively activated the stress response as observed by elevated expression of β-galactosidase in some cells (Fig. 3C). The analysis of hundreds of Q82 animals ruled out any consistent pattern of a subset of cells exhibiting particular pattern of stress responsiveness. By comparison, exposure to heat stress, whether in the presence or absence of polyglutamine aggregates, resulted in a uniform and robust heat shock response (Fig. 3D). The hsp16∷lacZ reporter was not induced in Q19-GFP-expressing animals, indicating that the Q82 motif itself initiated biochemical stress.
Figure 3.
Q82-GFP constitutively activates the heat shock response. Q82-CFP or Q82-GFP was coexpressed in body wall muscle cells (A) and pharyngeal cells (B), respectively, of PC72 animals containing the heat shock reporter (hsp16∷lacZ). The animals were fixed and stained for β-galactosidase activity. Dual color epifluorescence microscopy was used to visualize Q82-CFP (A) and Q82-GFP (B). (C) Q82 aggregates caused constitutively activated β-galactosidase activity in a subset of the body wall and pharyngeal muscle cells. (D) Exposure of PC72 animals to heat shock-activated β-galactosidase activity in all cells. (E) Animals expressing Q82-CFP were stained with anti-GFP Ab, detected by indirect immunofluorescence by using rhodamine-labeled secondary Ab, and compared with the pattern of Q82-CFP fluorescence (F).
One possible cause for the lack of correlation between activation of the stress response and accumulation of Q82-GFP aggregates could be that some of the Q82-GFP protein is nonfluorescent but still capable of initiating the stress response. We used an Ab to assess the accumulation of GFP, rather than relying solely on its autofluorescence. The patterns of Q82-GFP activity and protein distribution were identical (Fig. 3 E and F). Thus, there is no appreciable accumulation of cryptic misfolded GFP in body wall muscle cells, and polyglutamine expansion proteins initiate a stress signal leading to the idiosyncratic and variable activation of the heat shock response.
The Expression of Polyglutamine Protein Inhibits Development.
Animals expressing aggregation-prone polyglutamine fusions exhibited a retardation of growth rates compared with animals expressing soluble forms of polyglutamine. This effect was established by measuring the time for progression from embryo to adult. Whereas 100% of wild-type (N2) embryos progressed to adult in 48 h, only 43% of Q82-GFP-expressing embryos progressed to the L4/young adult stage (Table 1), with the slower growing animals often requiring up to 96 h to attain adulthood (data not shown). In contrast, expression of Q19-GFP had no effect on developmental timing (data not shown).
Table 1.
Expression of Hsp104 restores the growth rates of animals expressing polyglutamine aggregates
| Line | % at stage
|
Sample, n | ||
|---|---|---|---|---|
| L1/L2 | L3 | L4/adult | ||
| N2 | 0 | 0 | 100 | 177 |
| Q82-GFP | 32 | 25 | 43 | 282 |
| Hsp104 no. 1 | 5 | 6 | 89 | 219 |
| Hsp104 no. 2 | 14 | 21 | 65 | 270 |
| Hsp104 no. 3 | 8 | 24 | 68 | 187 |
| Hsp104 no. 4 | 6 | 24 | 70 | 238 |
| Mut Hsp104 no. 1 | 21 | 22 | 57 | 171 |
| Mut Hsp104 no. 2 | 35 | 27 | 38 | 197 |
| Mut Hsp104 no. 3 | 37 | 23 | 40 | 262 |
| Mut Hsp104 no. 5 | 41 | 23 | 36 | 222 |
| Ydj1 no. 1 | 37 | 29 | 34 | 155 |
| Ydj1 no. 2 | 20 | 38 | 42 | 179 |
Transgenic lines expressing Q82-GFP fusion protein alone or coexpressing Q82-GFP and wild-type (four lines) or mutant (Mut) Hsp104 (four lines) or Ydj1 (two lines) in body wall muscle cells were established. Gravid adults were allowed to lay embryos for 1 h. The developmental stage of the animals after 48 h was noted, and the percentage of animals at the various stages was determined. The experiment was performed at least three times for each of the lines, of which one is shown. Consistently faster rates of growth were observed in animals expressing wild-type Hsp104 than the mutant protein.
Hsp104 Reduces the Appearance of Polyglutamine Aggregates and Associated Developmental Delay.
To address whether molecular chaperones affect the formation of polyglutamine aggregates or growth retardation in C. elegans, genes encoding different yeast chaperones were coexpressed along with Q82-GFP in body wall muscle cells. Coexpression with wild-type Hsp104 dampened the growth inhibitory effects of Q82-GFP by nearly 2-fold, such that an average of 73% of the embryos from separate transgenic lines attained the adult stage (Table 1). This effect was due to the activity of Hsp104 because coexpression with a mutant of Hsp104, defective in protein remodeling activity (40), had no effect. Coexpression of another yeast chaperone, Ydj1, had no noticeable effect on Q82-associated developmental delay (Table 1).
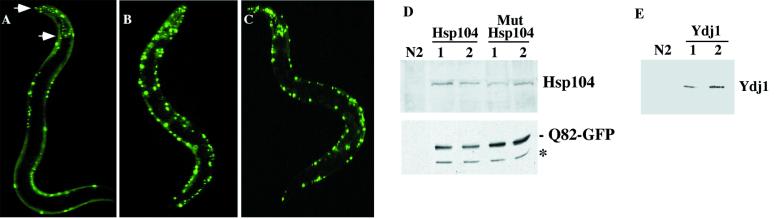
The restoration of normal growth rates in animals coexpressing Q82-GFP and Hsp104 correlated with a striking reduction in aggregate formation in body wall muscle cells. This effect was cell autonomous, because there was no reduction in aggregate formation in pharyngeal muscle cells in which the Hsp104 transgene was not expressed (Fig. 4A). In contrast, expression of mutant Hsp104 and other chaperones, including Ydj1 (DnaJ family), Ssa1 (Hsp70 family), and Hsp82 (Hsp90 family) did not have a marked effect on the expression of polyglutamine aggregates (Fig. 4 B and C, and data not shown). The levels of wild-type or mutant Hsp104, Ydj1, and Q82-GFP in transgenic animals were shown to be similar among the transgenic lines (Fig. 4 D and E). The ability of Hsp104 to ameliorate growth defects (Table 1) correlated with its effect on aggregate formation.
Figure 4.
Expression of wild-type Hsp104 diminishes aggregation of Q82-GFP. To assess the effects of chaperone expression on polyglutamine aggregates, wild-type or mutant Hsp104 or Ydj1 (dnaJ chaperone) was coexpressed with Q82-GFP in body wall muscle cells (unc-54 promoter), whereas only Q82-GFP was expressed in the pharyngeal cells (myo-2 promoter). (A) Overexpression of Hsp104 reduced Q82-GFP aggregates in body wall muscle cells but not in the pharyngeal cells (between the arrows). In animals expressing mutant Hsp104 (B) or Ydj1 (C), Q82 aggregates were unaffected. (D and E) Immunoblot analysis of extracts of wild-type N2 or chaperone-expressing animals for (D) two lines, each expressing Hsp104 and mutant Hsp104 by using anti-Hsp104 and anti-GFP Ab (asterisk corresponds to mobility of free GFP); and (E) Ydj-1 levels in N2 animals and two lines expressing Ydj1.
Polyglutamine Aggregates Alter the Folding Environment: Effects on Induced Aggregation.
Having demonstrated that Q19-GFP is expressed as a soluble protein (Figs. 1A and 2A), we asked whether it would coassociate with Q82. Q82-CFP and YFP or Q19-YFP were expressed in body wall muscle cells. The distinct spectral properties of these GFP variants ensured unambiguous detection of each (Fig. 5 A and B). Q82-CFP did not affect the localization of YFP (Fig. 5 C and D) but profoundly affected Q19-YFP through coaggregation (Fig. 5 E–H).
Figure 5.
Polyglutamine aggregates sequester proteins containing shorter polyglutamine arrays. (A) Expression of Q82-CFP in body wall muscle cells resulted in the formation of aggregates whose fluorescence did not bleed into the YFP channel (B). The ability of Q82-CFP aggregates to sequester other proteins was tested by coexpressing Q82-CFP with YFP. In animals coexpressing YFP and Q82-CFP (C and D), YFP alone was distributed diffusely throughout the muscle cells, whereas Q19-YFP (E and F) formed coaggregates. These aggregates were cytosolic, adjacent to the nuclei in red (G), as shown in the merge (H) of E, F, and G.
Polyglutamine aggregates also influenced the solubility and subcellular localization of glutamine-rich proteins that did not contain long homopolymeric stretches of glutamine. HRP1, a yeast protein involved in mRNA processing (41), was identified in a search of sequence databases for proteins with high asparagine and glutamine content, one of the common features of prion-like elements in yeast (42). Although 25% of the C-terminal 241 residues of HRP1 are asparagine or glutamine, neither residue is present as a consecutive stretch longer than three residues. HRP1-YFP expressed from the unc-54 promoter was localized to the nucleus of body wall muscle cells in 1 to 20 distinct foci (Fig. 6A). When coexpressed with Q82-CFP, however, the proteins coaggregated (Fig. 6 B and C) in a pattern similar to Q82-CFP alone. Inspection at higher magnification also revealed the presence of foci exclusively of HRP1-YFP (Fig. 6C). Examination of these animals revealed that the foci containing HRP1-YFP alone were restricted to the nucleus, and coaggregates of HRP1-YFP and Q82-CFP were cytoplasmic (Fig. 6 D–G). These results show that Q82-CFP has a dominant effect on the localization of other glutamine-rich proteins.
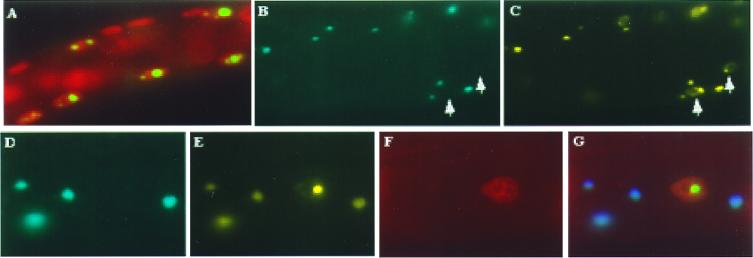
Figure 6.
Polyglutamine aggregates cause mislocalization of a nuclear protein to the cytoplasm. (A) HRP1-YFP fusion protein expressed in the body wall muscle cells coalesces in nuclei as determined by propidium iodide staining (red). (B and C) Animals coexpressing Q82-CFP (B) and HRP1-YFP (C) in body wall muscle cells were analyzed for coaggregates containing both Q82-CFP and HRP1-YFP. Aggregates containing only nuclear HRP1-YFP are indicated with arrows. Shown in D through G are images from a representative animal where the formation of coaggregates of Q82-CFP (D) and HRP1-YFP (E) were detected. Aggregates composed solely of HRP1 were restricted to the nucleus (E), whereas the coaggregates of HRP1-YFP and Q82-CFP were confined to the cytoplasmic compartment, shown by a merge of the images of the coaggregates and nuclear staining (D–G).
Discussion
We show that the aggregation of a GFP reporter protein in C. elegans depends on the length of a fused polyglutamine tract. These observations create a model system for dissecting the interactions of polyglutamine-expanded proteins in polyglutamine repeat disorders. The appearance and accumulation of polyglutamine aggregates perturbs protein-folding homeostasis, inducing a variable heat shock response and altering the solubility and localization of nonpolyglutamine proteins. Remarkably, these imbalances can be reversed by expression of the yeast chaperone Hsp104. The developmental defect induced by the polyglutamine expression, and the reversion of that defect by Hsp104, provide a tractable system for testing models of pathogenesis and therapeutic strategies. Finally, the alternate conformational states adopted by the polyglutamine proteins present an opportunity to study general aspects of protein misfolding and aggregation in C. elegans.
Deregulation of the Heat Shock Response by the Chronic Biochemical Stress of Polyglutamine Expansions.
The expression of a polymer of glutamine residues causes activation of the stress-sensing machinery of C. elegans. This constitutive activation of the heat shock response may have dire consequences for the organism, because the accumulation of molecular chaperones may interfere with events critical to cell growth and cell death (43, 44). The deregulation of an otherwise transient cytoprotective response to acute stress may become deleterious as a chronic stress response. Deregulation of the heat shock response by the expression of an aggregation-prone protein was also observed during infection of cultured human cells by the scrapie agent (45). Alteration in the cellular stress surveillance machinery due to proteins in altered conformational states may be an important element in the toxicity of protein misfolding diseases.
C. elegans cell types do not show a uniform response to the stress of polyglutamine expression. Several factors may lead to this mosaicism. It is possible that several different types of aggregates may be formed, and that only certain structures lead to heat shock activation. It is also possible that small stochastic differences in the stress responses of individual cells are amplified by a positive feedback mechanism that is initiated by the response to the polyglutamine protein. This varied response may parallel the mosaic response of individual neurons in the polyglutamine disorders. Further studies in this system may help to uncover why the progress of disease is so variable from cell to cell in groups of affected neurons.
Molecular Chaperones Reestablish Homeostasis After Expression of Polyglutamine Expansions.
There is considerable evidence pointing to the destructive effects of polyglutamine aggregates leading to disease; however, recent evidence has also argued for a protective role of polyglutamine aggregates (46). Our results demonstrate that Q82 is toxic and that Hsp104 suppresses the appearance of aggregates and at least partially normalizes growth rates. That coexpression of Hsp104 can restore growth rates of animals with polyglutamine aggregates extends recent observations on the distinctive properties of this molecular chaperone. Hsp104, isolated on the basis of its potent thermotolerance properties, disaggregates thermally denatured luciferase with the aid of Hsp40 and Hsp70 (32, 40, 47). These properties are unique to Hsp104, because other chaperones function primarily to prevent protein misfolding and aggregation and assist in refolding to the native state. Overexpression of Hsp104 can also cure the [PSI+] prion-like element in yeast (48). Hsp104 interacts with a variety of protein substrates including Sup35p (the protein determinant of the [PSI+] element), mammalian prion protein (PrP) and A-β-amyloid peptide, and assists in the conversion of cellular PrP (PrPc) to the scrapie PrP (PrPSc) state in vitro (49, 50).
The association of chaperones with misfolded substrates is not exclusive to disorders involving polyglutamine aggregates. PrPSc associates with Hsp60, a member of the mitochondrial GroE family of chaperones (51), and Hsp73 (45). Additionally, in vitro, GroEL can assist in the conversion of PrPc to the protease-resistant PrPSc state (45). Cellular cofactors with chaperone-like activity may be involved in the transition and propagation of PrPc to the alternatively folded PrPSc state (52). Collectively, these observations suggest that the balance of chaperones and agents that enhance protein misfolding can affect the protein-folding environment.
Intrinsic and Induced Aggregation: Sequestration and Altered Subcellular Targeting.
Our studies indicate that Q19-YFP cannot form aggregates by itself but does coaggregate with Q82-CFP, in agreement with studies on the coaggregation of huntingtin and ataxin-1 (53, 54). Although we have not performed a comprehensive analysis of the minimal number of polyglutamine residues required for induced coaggregation, Q19-YFP (Fig. 5) and Q13-YFP (data not shown) coassociate with Q82-CFP aggregates. The coassociation of HRP1 with Q82-CFP demonstrates that induced coaggregation is not strictly dependent on the presence of a homopolymer of glutamines, but extends to other glutamine-rich proteins. The sequestration of a nuclear protein in cytoplasmic aggregates recalls the relocalization of several proteins to aggregates in human diseases (53, 55–57). Perturbation of protein homeostasis by polyglutamine protein aggregates and coassociated proteins may therefore contribute to the cytotoxicity of diverse polyglutamine expansion diseases.
Additional studies will be required to identify the determinants necessary for coassociation of cellular proteins by polyglutamine aggregates. Our results on protein sequestration and relocalization obtained in C. elegans extend to studies in human cells using the HRP1 and polyglutamine proteins, demonstrating the conserved nature of the machinery for protein homeostasis (K.K. and S. Kim, unpublished data). The ability to manipulate the aggregation state and relocalization of proteins in C. elegans offers intriguing insights into the progressive nature of polyglutamine-based diseases and an opportunity to apply genetic and biochemical tools to identify the proteins that suppress aggregate formation and cellular toxicity.
Acknowledgments
We thank Lutz Nover for his support of E.S. We also thank Robert Holmgren for assistance with microscopy, Kelly Mayo and Robert Holmgren for use of their microscopes, Shoji Tsuji for polyglutamine-containing cDNA clones, Peter Candido for the PC72 reporter strain, Andrew Fire for expression vectors, and Avrom Caplan for anti-Ydj1 antiserum. Shaoru Wang generated some of the C. elegans lines, and Susana Garcia and Jim Morley provided data on the developmental delay studies. This research was supported by a grant to R.I.M. from the National Institutes of Health (GM38109), the Daniel F. and Ada L. Rice Foundation, the Rice Institute for Biomedical Research, the Carol Gollub Foundation, and a grant to J.M.K. from the National Institute of Child Health and Human Development.
Abbreviations
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- GFP
green fluorescent protein
- HRP
heterogeneous ribonuclear protein
- Q19 and Q82
polyglutamines with 19 and 82 residues, respectively
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.100107297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.100107297
References
- 1.Booth D R, Sunde M, Bellotti V, Robinson C V, Hutchinson W L, Fraser P E, Hawkins P N, Dobson C M, Radford S E, Blake C C, et al. Nature (London) 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 2.Kakizuka A. Trends Genet. 1998;14:396–402. doi: 10.1016/s0168-9525(98)01559-5. [DOI] [PubMed] [Google Scholar]
- 3.Perutz M F. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 4.Jarrett J T, Lansbury P T., Jr Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 5.Paulson H L, Fischbeck K H. Annu Rev Neurosci. 1997;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- 6.Roizin L, Stellar S, Liu J C. Adv Neurol. 1979;23:95–122. [Google Scholar]
- 7.DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 8.Gusella J F, MacDonald M E. Curr Opin Neurobiol. 1995;5:656–662. doi: 10.1016/0959-4388(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 9.Davies S W, Turmaine M, Cozens B A, DiFiglia M, Sharp A H, Ross C A, Scherzinger E, Wanker E E, Mangiarini L, Bates G P. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 10.Ordway J M, Tallaksen-Greene S, Gutekunst C A, Bernstein E M, Cearley J A, Wiener H W, Dure L S, 4th, Lindsey R, Hersch S M, Jope R S, et al. Cell. 1997;91:753–763. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- 11.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 12.Warrick J M, Paulson H L, Gray-Board G L, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 13.Faber P W, Alter J R, MacDonald M E, Hart A C. Proc Natl Acad Sci USA. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg Y P, Nicholson D W, Rasper D M, Kalchman M A, Koide H B, Graham R K, Bromm M, Kazemi-Esfarjani P, Thornberry N A, Vaillancourt J P, et al. Nat Genet. 1996;13:442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 15.Ona V O, Li M, Vonsattel J P, Andrews L J, Khan S Q, Chung W M, Frey A S, Menon A S, Li X J, Stieg P E, et al. Nature (London) 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 16.Scherzinger E, Sittler A, Schweiger K, Heiser V, Lurz R, Hasenbank R, Bates G P, Lehrach H, Wanker E E. Proc Natl Acad Sci USA. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perutz M F, Johnson T, Suzuki M, Finch J T. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiromi Y, Okamoto H, Gehring W J, Hotta Y. Cell. 1986;44:293–301. doi: 10.1016/0092-8674(86)90763-4. [DOI] [PubMed] [Google Scholar]
- 19.Parsell D A, Sauer R T. Genes Dev. 1989;3:1226–1232. doi: 10.1101/gad.3.8.1226. [DOI] [PubMed] [Google Scholar]
- 20.Gething M J, Sambrook J. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 21.Hartl F U. Nature (London) 1996;381:571–579. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 22.Chai Y, Koppenhafer S L, Bonini N M, Paulson H L. J Neurosci. 1999;19:10338–10347. doi: 10.1523/JNEUROSCI.19-23-10338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenoien D L, Cummings C J, Adams H P, Mancini M G, Patel K, DeMartino G N, Marcelli M, Weigel N L, Mancini M A. Hum Mol Genet. 1999;8:731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- 24.Cummings C J, Mancini M A, Antalffy B, DeFranco D B, Orr H T, Zoghbi H Y. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 25.Krobitsch S, Lindquist S. Proc Natl Acad Sci USA. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazemi-Esfarjani P, Benzer S. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- 27.Gottesman S, Wickner S, Maurizi M R. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 28.Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- 29.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 30.Okkema P G, Fire A. Development (Cambridge, UK) 1994;120:2175–2186. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez Y, Lindquist S L. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 32.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 33.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fire A. Genet Anal Tech Appl. 1992;9:151–158. doi: 10.1016/1050-3862(92)90042-4. [DOI] [PubMed] [Google Scholar]
- 35.Finney M, Ruvkun G B. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 36.Kramer J M, French R P, Park E C, Johnson J J. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kramer J M, Johnson J J. Genetics. 1993;135:1035–1045. doi: 10.1093/genetics/135.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stringham E G, Candido E P. J Exp Zool. 1993;266:227–233. doi: 10.1002/jez.1402660309. [DOI] [PubMed] [Google Scholar]
- 40.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Nature (London) 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 41.Kessler M M, Henry M F, Shen E, Zhao J, Gross S, Silver P A, Moore C L. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sondheimer N, Lindquist S. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 43.Feder J H, Rossi J M, Solomon J, Solomon N, Lindquist S. Genes Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- 44.Morimoto R I. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 45.Tatzelt J, Zuo J, Voellmy R, Scott M, Hartl U, Prusiner S B, Welch W J. Proc Natl Acad Sci USA. 1995;92:2944–2948. doi: 10.1073/pnas.92.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sisodia S S. Cell. 1998;95:1–4. doi: 10.1016/s0092-8674(00)81743-2. [DOI] [PubMed] [Google Scholar]
- 47.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 48.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 49.DebBurman S K, Raymond G J, Caughey B, Lindquist S. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schirmer E C, Lindquist S. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edenhofer F, Rieger R, Famulok M, Wendler W, Weiss S, Winnacker E L. J Virol. 1996;70:4724–4728. doi: 10.1128/jvi.70.7.4724-4728.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Telling G C, Scott M, Mastrianni J, Gabizon R, Torchia M, Cohen F E, DeArmond S J, Prusiner S B. Cell. 1995;83:79–90. doi: 10.1016/0092-8674(95)90236-8. [DOI] [PubMed] [Google Scholar]
- 53.Perez M K, Paulson H L, Pendse S J, Saionz S J, Bonini N M, Pittman R N. J Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narain Y, Wyttenbach A, Rankin J, Furlong R A, Rubinsztein D C. J Med Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matilla A, Koshy B T, Cummings C J, Isobe T, Orr H T, Zoghbi H Y. Nature (London) 1997;389:974–978. doi: 10.1038/40159. [DOI] [PubMed] [Google Scholar]
- 56.Skinner P J, Koshy B T, Cummings C J, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 57.Tran P B, Miller R J. Trends Neurosci. 1999;22:194–197. doi: 10.1016/s0166-2236(99)01409-5. [DOI] [PubMed] [Google Scholar]