Abstract
Pathway-specific therapy is the future of cancer management. The oncogenic phosphatidylinositol 3-kinase (PI3K) pathway is frequently activated in solid tumors; however, currently, no reliable test for PI3K pathway activation exists for human tumors. Taking advantage of the observation that loss of PTEN, the negative regulator of PI3K, results in robust activation of this pathway, we developed and validated a microarray gene expression signature for immunohistochemistry (IHC)-detectable PTEN loss in breast cancer (BC). The most significant signature gene was PTEN itself, indicating that PTEN mRNA levels are the primary determinant of PTEN protein levels in BC. Some PTEN IHC-positive BCs exhibited the signature of PTEN loss, which was associated to moderately reduced PTEN mRNA levels cooperating with specific types of PIK3CA mutations and/or amplification of HER2. This demonstrates that the signature is more sensitive than PTEN IHC for identifying tumors with pathway activation. In independent data sets of breast, prostate, and bladder carcinoma, prediction of pathway activity by the signature correlated significantly to poor patient outcome. Stathmin, encoded by the signature gene STMN1, was an accurate IHC marker of the signature and had prognostic significance in BC. Stathmin was also pathway-pharmacodynamic in vitro and in vivo. Thus, the signature or its components such as stathmin may be clinically useful tests for stratification of patients for anti-PI3K pathway therapy and monitoring therapeutic efficacy. This study indicates that aberrant PI3K pathway signaling is strongly associated with metastasis and poor survival across carcinoma types, highlighting the enormous potential impact on patient survival that pathway inhibition could achieve.
Keywords: breast cancer, metastasis, stathmin, microarray
Using high-throughput technologies, a number of gene expression profiling studies have found tumor signatures capable of discriminating cancer patients with good vs. poor outcomes (1–4). Despite these promising results, deciphering the biological basis of why these signatures are predictive and how the identified risk groups relate to activation of oncogenic pathways and sensitivity to molecularly targeted therapies remains a significant challenge (5). Moreover, optimal efficacy of targeted therapies will be possible only when the appropriate subgroups of patients with target and/or pathway-activated tumors can be identified and used to guide treatment (6–8).
The oncogenic phosphatidylinositol 3-kinase (PI3K) signaling pathway has been implicated in nearly all aspects of tumor biology: cell transformation, growth, proliferation, migration, protection from apoptosis, genomic instability, angiogenesis, and metastasis, as well as cancer stem cell maintenance (7, 9). Aberrant PI3K pathway signaling is estimated to be present in >30% of human cancers (7) and occurs by two nonexclusive mechanisms: by oncogenic alterations such as of HER2 and PIK3CA or by inactivation of the tumor suppressor PTEN (phosphatase and tensin homolog deleted on chromosome 10), which catalyzes the precise opposite reaction to PI3K and is the pathway's most important regulatory brake.
The degree of PI3K pathway activation in a tumor varies depending on the type of pathway lesion(s) that are present, with inactivation of PTEN being perhaps the most common lesion (7) as well as the most potent (10). Despite evidence suggesting activated PI3K signaling would confer an aggressive tumor phenotype, there has been a lack of consensus among studies associating pathway lesions to markers of pathway activation and of these variables to actual cancer patient outcome (e.g., refs. 11–14). Moreover, although this pathway contains many attractive therapeutic targets, recent clinical trials of pathway-targeted drugs have in most cases not been as promising as hoped. These issues are likely because of the heterogeneity of PTEN/PI3K pathway lesions, resistance mediated by cooperation with other oncogenic pathways, the lack of markers for therapy response, and the lack of a reliable quantitative assay that integrates the multiple mechanisms of pathway activation for appropriate patient stratification. These issues are apparent even for targeted-therapy successes such as trastuzumab for HER2-positive breast cancer (BC), where recent evidence suggests that resistance may be mediated by PTEN (6).
To address these problems, we generated a gene expression signature of PTEN protein loss from human BC biopsies with the hope that a signature of this PI3K pathway-activated state would be pathway-integrative and would identify tumors that have a similar signature because of other activating lesions. Bearing out this hypothesis, our signature identifies tumors with complete PTEN loss as well as those with cooperating secondary alterations to the pathway and is predictive of clinical outcome in multiple independent data sets of varied carcinoma types. Furthermore, the signature gene stathmin is a robust surrogate marker for the signature, its protein levels are directly related to risk for BC recurrence, and it exhibits anti-PI3K pathway pharmacodynamic properties in vitro and in vivo. Our results indicate that PTEN/PI3K pathway activation is a strong biological correlate of metastasis and poor prognosis in carcinoma, implying an enormous potential advantage for new drugs that target this pathway. The signature or components such as stathmin may be clinically useful for patient selection for PI3K pathway-targeted therapy as well as monitoring therapeutic efficacy.
Results and Discussion
A Gene Expression Signature of PTEN Loss.
Because PTEN is rarely mutated in BC but protein levels are diminished in up to 48% (11, 12, 15, 16), we evaluated PTEN by immunohistochemistry (IHC) for 351 stage II breast tumors. Frozen samples for 35 PTEN-deficient (PTENIHC−) and 70 PTEN-normal (PTENIHC+) cases, matched on estrogen receptor (ER) status where possible [supporting information (SI) Table 1], were selected for gene expression profiling. A 3-fold cross-validation strategy (SI Fig. 5 and SI Methods) was applied to the microarray data to identify genes most significantly associated to PTEN IHC status, and a consensus-ranked gene list was generated by sorting on the average P value (APV) from each cross-validation analysis. There were 246 signature genes with an APV ≤0.02 (hereinafter “the signature”; see SI Table 2), a >15-fold overabundance compared with permutation simulations (SI Methods). Notably, the PTEN gene was the top discriminator (APV = 5.6 × 10−5), demonstrating that PTEN transcript levels, and not posttranslational events, are the primary determinant of protein levels in BC.
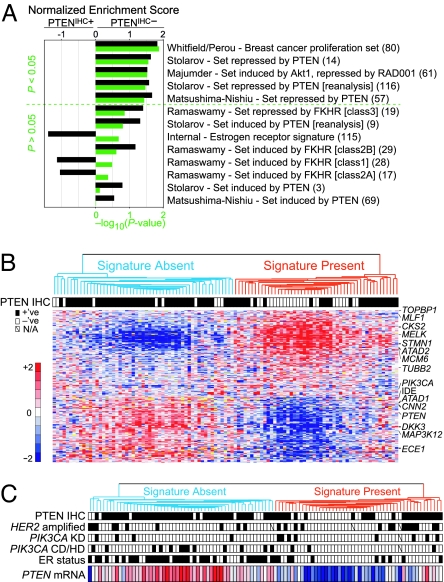
We applied gene set enrichment analysis (17) to further verify the biological consistency of our PTEN-associated signal to the literature. We found two in vitro PTEN-regulated gene cassettes (refs. 18 and 19; P = 0.027 and P = 0.037, respectively), an in vivo Akt/mTOR/RAD001-regulated cassette (ref. 20; P = 0.030), and three of four sets regulated by the downstream pathway component FOXO1A (21), to be enriched toward the expected PTEN status group (Fig. 1A and SI Notes). Moreover, a proliferation-associated cassette (22) was significantly enriched in PTENIHC− tumors (P = 0.014), consistent with the known potent stimulatory effect of PI3K signaling on the cell cycle. Similar biological themes were identified by Gene Ontology analysis (SI Notes and SI Tables 3 and 4).
Fig. 1.
The PTEN/PI3K microarray gene expression signature in breast carcinoma. (A) Gene set enrichment analysis (17) verifies that the PTEN signature faithfully captures known biological outputs of the pathway. For each gene set tested (see SI Methods), the normalized enrichment score (black bars; positive and negative values indicate higher expression in the PTENIHC− or PTENIHC+ group, respectively) and the corresponding nominal P value (green bars) are plotted. The number of matched genes per set is within parentheses. (B) Hierarchical clustering of the 105 breast tumor samples (columns) by using the top 246 signature genes (rows) with an APV < 0.02. The two major tumor dendrogram clusters, “Signature Absent and Signature Present,” are indicated by blue- and red-colored branches, respectively. PTEN IHC status is indicated by filled (positive) or white (negative) boxes. In the heat map, fold change is relative to the median for each gene according to the color scale shown (red, overexpression; blue, underexpression; yellow, missing values), and selected gene symbols are displayed to the right. Clustering was performed by using the 1-Pearson correlation metric and centroid linkage. (C) Microarray PTEN mRNA expression levels from B illustrate PTEN to be a better marker of the SP group than PTEN IHC status. HER2 amplification, PIK3CA KD or CD/HD mutation, and the ER status for each case are indicated by filled (positive) boxes, open (negative) boxes, or a diagonal line (missing data).
Affirming that our PTEN-associated signal is independent of ER, an ER signature of genes up-regulated in our ER-positive (ER+) cases (see SI Methods) was not significantly enriched in our PTEN-associated signal (Fig. 1A).
PTENIHC+ Tumors with PTENIHC−-Like Profiles.
In the same 3-fold cross-validated approach, support vector machines were used to classify each tumor as either PTENIHC− or PTENIHC+ based on their gene expression pattern (SI Fig. 5 and SI Methods). Good classification performance was achieved [receiver operating characteristic (ROC) area, a measure of the tradeoff between sensitivity and specificity of a test, of 0.758, P = 0.0001]. This performance was not dependent on the information from the PTEN gene (ROC area = 0.758, P = 0.0005, when excluding PTEN; see also SI Notes). Regardless of the genes used, a perfect classification could not be attained, with up to 44% of the tumors classified as PTENIHC− being in fact PTENIHC+, suggesting the likelihood that other PI3K pathway lesions could also induce the transcriptional program of PTEN loss.
Fig. 1B illustrates the pattern of signature gene expression across the 105 tumor samples. As expected, the pattern of expression was biphasic, with most of PTENIHC+ tumors clustered together in one main branch [denoted Signature Absent (SA)], and the majority of PTENIHC− tumors clustering in the other main branch [Signature Present (SP)]. Consistent with the support vector machines result, exceptions were observed (Fig. 1B).
Interestingly, ATAD1, which is the nearest neighbor to PTEN at 10q23.31 in a head-to-head orientation, was significantly down-regulated in PTENIHC− tumors (APV <0.004) and the absolute expression levels of ATAD1 closely tracked PTEN levels (Pearson r = 0.676, P < 0.0001; SI Fig. 6 and SI Notes). Moreover, irrespective of PTEN IHC status, PTEN mRNA levels were below the median for 86% of SP compared with 20% of SA tumors (P < 0.0001). Thus, presence of the signature in 19/25 PTENIHC+ cases may be in part because of reduced PTEN message levels below the sensitivity of our IHC analysis (Fig. 1C).
Influence of Other PI3K Pathway Lesions.
To test whether other PI3K pathway abnormalities are associated to the activated signature, we analyzed the tumors for p110α (PIK3CA) mutations and for amplification of HER2 (Fig. 1C). Notably, 67% (8 of 12) of the PIK3CA kinase domain (KD) mutants clustered with the SP tumors, whereas only 19% of non-KD mutants [3 of 16: 2/11 helical domain (HD), and 1/5 C2 domain (CD)] were in this main branch (P = 0.019). Consistent with this, downstream pathway signaling differences between PIK3CA domain mutants were also seen in vitro (SI Fig. 7 and SI Notes). Moreover, suggestive of a dose-responsive inhibition of the PI3K pathway by PTEN (independent of PTEN IHC status), in the SP cluster 82% (9/11) of PIK3CA mutants had PTEN mRNA levels below the median compared with only 18% (3/17) of PIK3CA mutants in the SA cluster (P = 0.0001). HER2 amplification (HER2+) did not appear to correlate with the presence of the signature (Fig. 1C), with 58% of HER2+ cases (14/24) clustering among the PTENIHC− branch (SI Notes). However, among these 14 cases exhibiting the signature, 11 had PTEN mRNA levels below the median, in contrast to 3/10 HER2+ SA cases (P = 0.035). In total, 75% (15/20) of lesions to PIK3CA or HER2 in the SP group had low PTEN message levels, in contrast to 17% (4/24) in the SA group (P = 0.0002).
Together, these results suggest that our identified signature integrates various PI3K pathway lesions, some of which we have discerned but others not yet uncovered, that singly or, particularly in the setting of moderately reduced PTEN message levels, collaboratively activate PI3K signaling to a similar extent to that seen with IHC-detectable loss of PTEN. Conversely, cases with HER2 amplification or PIK3CA CD/HD mutations rarely exhibit the activated signature in the context of high PTEN expression. We hypothesize that the differential clustering of HER2+ cases by the signature could relate to trastuzumab sensitivity; however, this remains to be tested. These data demonstrate an important relationship between PTEN dosage and tumor phenotype in humans and imply potential clinical utility of therapies that reactivate PTEN.
Despite controlling for ER status in our sample selection and verifying independence of our PTEN signal from an ER signal, 78% of ER+ tumors clustered in the SA group, whereas 66% of ER– tumors clustered in the SP group (Fig. 1C). This suggests that frequent activation of the PI3K pathway is part of the natural history of ER− BC and is consistent with our observations in an independent large population-based BC cohort in which most ER− tumors harbor ≥1 PI3K pathway activating aberrations, in stark contrast to ER+ tumors (data not shown; SI Notes).
Clinical Implications of the PTEN/PI3K Pathway Signature.
We next investigated whether our signature could predict patient outcome. Although the selection of our samples was not designed for survival analysis, we found that the SP group had a significantly higher proportion of distant metastases [P = 0.025; SI Fig. 8; consistent with the pathway-integrative nature of the full signature, we note that PTEN IHC was not prognostic (P = 0.705; data not shown)].
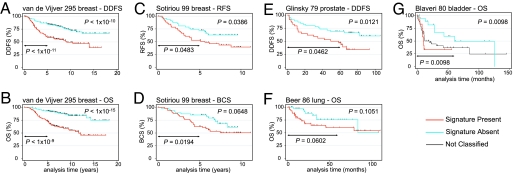
To ascertain whether this result was reproducible, we queried the prognostic association of the PTEN signature in two independent breast tumor data sets. The Dutch 295 BC series (3) serves as a gold standard data set for independent validation (23). We mapped (24) our 246-gene PTEN signature genes to the Dutch data set and generated a nearest centroid classifier (NCC; ref. 2) trained on our data set. Each independent tumor was then predicted by the NCC as SP or SA, and a continuous signature score proportional to the degree of correlation to these classes was calculated. As illustrated by Kaplan–Meier (KM) survival estimates, our signature-based NCC separated the 295 tumors into the SP and SA groups, with SP cases expressing the signature of PTEN loss having a significantly worse distant disease-free survival [DDFS; hazard ratio (HR), 2.48, P = 1.15 × 10−5] and overall survival (OS; HR 3.69, P = 2.21 × 10−7; Fig. 2 A and B and SI Table 5). Multivariate analysis showed the classification to be independent of and the most significant variable after adjustment for lymph node metastasis and ER status (SI Table 6). Indicating an underlying mechanistic explanation for most poor prognosis BCs, our signature had a sensitivity of 74% and specificity of 94% in identifying the same poor prognosis (76 of 114 ER+ and 58 of 66 ER−) and good prognosis (105 of 112 ER+ and three of three ER−) tumors as the Dutch 70-gene classifier (3).
Fig. 2.
KM survival estimates for tumors classified as SP (red curves) or SA (blue curves) indicates the PTEN/PI3K signature to predict patient survival across independent carcinoma microarray data sets. Log-rank P value comparing these classes at complete follow-up and at 5-year follow-up are given. Cox regression survival analyses are presented in SI Table 5. (A) DDFS and (B) OS for the classification of 295 Dutch BC (3). (C) Relapse-free survival and (D) BCS for the classification of 99 BCs (4). (E) DDFS for the classification of 79 prostate cancers (25). (F) OS for 86 lung cancers (1). (G) OS for 80 bladder cancers (26). Here, a three-group classification was used (see SI Methods), with a black curve for unclassified cases. For all KM graphs, the corresponding three- or two-group analyses are presented in SI Fig. 9.
We next tested whether the PTEN signature score was predictive of outcome on a cutoff-independent continuous scale in the Dutch data set. The signature score ranged from −1.44 to +1.45 (positive values indicating greater presence of the signature) and was highly predictive of DDFS (HR 1.80, P = 4.10 × 10−6) and OS (HR 2.33, P = 1.98 × 10−8; SI Table 5).
Because the PI3K pathway is a known promoter of cell proliferation, corroborated by our gene set enrichment analysis, we removed all cell cycle-associated signature genes (SI Methods) and repeated the NCC procedure. Only 6% of tumors changed classification, and all 17 of these cases had low (r < 0.2) initial NCC correlation scores. Therefore, we conclude that the successful prognostic classification of tumors is not simply because of information from proliferation-associated gene expression but rather is associated to a complex program of transcriptional changes downstream of the PTEN/PI3K pathway. Further supporting this, we found PTEN mRNA levels to be significantly lower in the Dutch tumors classified as SP (P = 0.0004) and to be inversely correlated to the signature score as expected (r = −0.289, P < 0.0001; SI Table 7).
We obtained similar results when classifying the data set of Sotiriou et al. (4) containing 99 British breast tumors. Tumors classified as SP had a significantly worse relapse-free survival (RFS; HR 1.89, P = 0.042) and worse BC-specific survival (BCS; HR 1.87, P = 0.069) over the complete follow-up time; at 5-year follow-up analysis interval, a significant difference in BCS was evident (HR 2.70, P = 0.025; Figs. 2 C and D and SI Table 5). The signature score contained significant continuous prognostic information for both RFS (HR 2.13, P = 0.018) and BCS (HR 2.33, P = 0.020; SI Table 5).
Predicting Outcome in Other Carcinoma Types.
Prompted by the success of the PTEN/PI3K pathway signature to predict outcome in BC, we hypothesized this signature may be more generally applicable to other carcinoma types with suspected involvement of the PI3K pathway. To test this, we analyzed public microarray data sets of human prostate (25), bladder (26), and lung tumors (1). Classification by the signature could separate prostate and bladder, but not lung carcinoma, samples into groups with significant differences in survival (Fig. 2 E–G; SI Fig. 9, SI Table 5, and SI Notes). Moreover, the signature score was significantly predictive on a continuous scale for the same tumor types (SI Table 5). These results in multiple independent data sets suggest that the degree of PTEN/PI3K pathway activation is directly related to the metastatic potential of the primary tumor for several carcinoma types.
Selection of a Marker for PTEN/PI3K Pathway Activation.
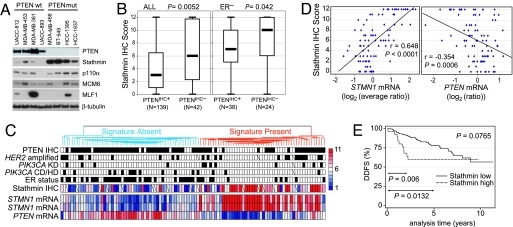
Current markers for PI3K pathway activation are inadequate. Phosphorylated Akt (p-Akt) at serine 473 is the most reliable pathway marker under highly controlled situations (e.g., cell culture); however, the available reagents do not work well in routine clinical specimens. To identify a marker of pathway activity, we selected four genes from our signature for immunoblotting experiments: p110α (PIK3CA, APV = 0.0023), stathmin (STMN1, with two significant independent reporters: APV = 0.0070 and = 0.0187), minichromosome maintenance 6 (MCM6, APV = 0.0189), and myeloid leukemia factor 1 (MLF1, APV = 0.0003). Across a panel of eight BC cell lines, strikingly, the signature gene stathmin was highly overexpressed in the PTEN mutant as compared with PTEN wild-type lines, whereas p110α had a modestly higher expression, and MCM6 and MLF1 did not show an association to PTEN (Fig. 3A).
Fig. 3.
Stathmin is a marker of the PTEN/PI3K signature. (A) Immunoblotting analysis of relative protein levels for four selected signature genes across a panel of eight BC cell lines. (B) Box plots illustrating stathmin IHC scores to be significantly higher in PTENIHC− vs. PTENIHC+ breast tumors among all 181 cases analyzed for both proteins (Left) and within the subgroup of 62 ER-negative cases (Right). P values were calculated by using the Mann–Whitney test. (C) Stathmin IHC levels are highly correlated to the presence of the PTEN/PI3K signature (ROCarea = 0.809, P < 0.0001). The hierarchical clustering tumor dendrogram, marker annotations, and STMN1 (represented by two independent reporters) and PTEN message levels are from Fig. 1 B and C. Stathmin IHC scores are centered to the median score, 6 (white boxes), with higher and lower scores colored in red and blue, respectively (key to right). (D) Scatterplot analyses show stathmin IHC levels to closely track STMN1 mRNA levels (average of the two reporters) and to be significantly inversely related to PTEN mRNA levels. Linear regression and the Pearson correlation r and P values are presented. (E) KM analysis indicates stathmin-high (score >10) tumors have a significantly higher rate of distant disease recurrence compared with stathmin-low (score 0–10) tumors among 191 BCs analyzed. Log-rank P values for complete follow up (top right corner) and the 2- and 5-year intervals are shown. Cox regression analysis is presented in SI Table 5. Representative stathmin IHC examples are presented in SI Fig. 10.
On the basis of these data, and that STMN1 message was also found to be one of the genes down-regulated upon PTEN induction or treatment with the PI3K-inhibitor LY294002 in U87MG cells (18), we considered stathmin to be a good candidate marker for PI3K pathway activation. We analyzed stathmin by semiquantitative IHC in 191 breast tumors (SI Fig. 10), of which 181 were also evaluated for PTEN protein levels (SI Table 8). Validating our microarray result, we found stathmin staining scores to be significantly higher in PTENIHC− tumors than in PTENIHC+ tumors (P = 0.005) (Fig. 3B). Although both PTEN loss and high stathmin have been shown independently to be associated to ER− BC (11, 16, 27, 28), stathmin immunoscores were also significantly higher in PTENIHC− tumors within the ER− subset (P = 0.042; Fig. 3B). Mapping the stathmin IHC data to 90 overlapping cases from the microarray analysis revealed stathmin protein levels to closely track STMN1 message levels (r = 0.648, P < 0.0001; Fig. 3 C and D) to be inversely correlated to PTEN message levels (r = −0.354, P = 0.0006; Fig. 3D) and to provide an accurate test for presence of the PTEN-loss signature (ROCarea = 0.809, P < 0.0001; Fig. 3C).
Clinical Implications of Stathmin Overexpression.
Although some reports have indicated that PTEN protein status can carry prognostic information in BC (11), others have not (12). In our material of 351 patients, PTEN protein status had no association to survival (data not shown), perhaps because of the relative homogeneity of this cohort (only stage II tumors) and confounded by other PI3K pathway-activating lesions. Because our data indicated stathmin to be a surrogate marker of the PTEN signature, we investigated whether stathmin protein levels were related to patient outcome. In the 191 patient set, the stathmin-high (score >10) group had a significantly worse DDFS at the 2- and 5-year follow-up intervals (Fig. 3E). Moreoever, the stathmin immunoscore was tested as a continuous variable (from 0 to 12) and found to be highly predictive of DDFS at 2 years (HR 1.16, P = 0.004; that is, for every one point increase in stathmin score, the hazard increases by 16%) and 5 years of follow-up (SI Table 5).
In multivariate Cox regression analyses, stathmin-high expression was independent of ER status and lymph node status at 2 and 5 years of follow-up (HR 2.45, P = 0.035 and HR 3.54, P = 0.019, respectively; SI Table 6). Among patients with lymph node negative disease, stathmin high was predictive for distant metastasis within 2 years of diagnosis, with a HR of 9.07 [95% confidence interval (CI), 1.76–46.84, P = 0.008], as well as among patients with lymph node-positive disease (HR 3.99, 95% CI 1.13–14.14, P = 0.032).
These results were validated in the independent Dutch BC data set of 295 tumors: STMN1 mRNA levels were higher in the SP-classified poor-prognosis class (P < 0.0001) and directly related to the signature score (r = 0.690, P < 0.0001; SI Table 7). STMN1 mRNA itself was prognostic, with STMN1-high (top 50th percentile) cases having a significantly worse DDFS and OS at 5 years and complete follow-up (P = 0.0002 and P = 0.0003, respectively, for both time intervals; SI Fig. 11). Moreover, supporting our hypothesis that the signature and the signature marker stathmin are pathway-integrative, we note that classification by PTEN mRNA levels by itself was not as prognostic (DDFS: 5 year, P = 0.038 and complete follow-up, P = 0.111; OS: 5 year, P = 0.005 and complete follow-up, P = 0.012; SI Fig. 11), whereas the best prognostic separation was achieved with the full signature (Figs. 2 A and B and SI Table 5).
Stathmin Is PTEN/PI3K Pathway-Regulated in Vitro and in Vivo.
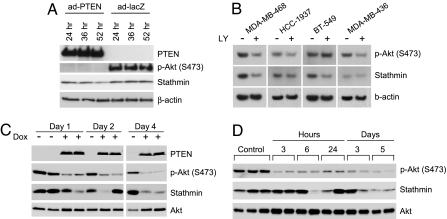
A marker of PTEN/PI3K pathway activity could have clinical utility, particularly one that demonstrates pharmacodynamic properties. Therefore, we determined whether stathmin protein expression was regulated by the pathway. To test this, we expressed PTEN in PTEN-null MDA-MB-468 cells and monitored stathmin levels over time. Compatible with its reported half-life of ≈28 h (29) stathmin levels were 23–45% down-regulated from 24 to 52 h after infection (Fig. 4A). Moreover, despite variable down-regulation of p-Akt (S473) by PI3K pathway inhibition using LY294002, stathmin levels were reduced robustly in the three PTEN-mutant BC cell lines (Fig. 4B), MDA-MB-468 (56% diminished), HCC-1937 (29%), and BT-549 (32%), and moderately in MDA-MB-436 cells (15%), which are PTEN wild-type but have undetectable PTEN and low levels of stathmin at steady state (data not shown). These results were further validated in vivo by using two mouse xenograft models: stathmin levels were appreciably down-regulated upon doxycyclin-induced PTEN expression (30) in MDA-468TR-PTEN tumors (Fig. 4C) and were similarly down-regulated in MDA-MB-468 tumors in mice following administration of the wortmannin-analog PI3K-inhibitor PWT-458 (Fig. 4D).
Fig. 4.
The PTEN/PI3K pathway regulates stathmin protein levels. (A) Immunoblotting analysis of stathmin down-regulation after adenovirus-mediated PTEN expression (ad-PTEN) in MDA-MB-468 PTEN-null cells compared with control (ad-lacZ). (B) Stathmin is down-regulated in BC lines by treatment with the PI3K-inhibitor LY294002 (LY; 20 μM) for 4 days as detected by immunoblotting. (C) Immunoblotting analyses show that, in vivo, stathmin is down-regulated upon PTEN induction with doxycyclin (Dox) in MDA-468TR-PTEN xenograft tumors and (D) after treatment of mice with MDA-MB-468 tumors with the PI3K-inhibitor PWT-458 (100 mg/kg). For C and D, independent xenografts from separate animals were analyzed in each sample lane.
Thus, evaluation of stathmin may be an effective way to quantitatively measure PI3K pathway activity, stratify patients, and monitor treatment response. Moreover, stathmin's role in regulating microtubule dynamics, promoting cell motility and proliferation (31), and conferring resistance to antimicrotubule drugs (32) are consistent with it being downstream of the PI3K pathway and may indicate that it is also a potential therapeutic target. Whether stathmin is dose-responsive or predicts sensitivity to PI3K-targeting agents are important questions which require further study.
In conclusion, we describe an in vivo gene expression signature for PTEN and find PTEN protein levels to be primarily dictated by its message level in BC. Our data suggest that quantitative measurements of PTEN mRNA may identify BCs with pathway activation more reliably than by PTEN IHC. PTEN mRNA levels may also be correlated to clinical outcome, which warrants further study. We elucidated differential activation of the signature by different types of PIK3CA mutations and by amplified HER2, which appears to depend on the dose of PTEN expression. Interestingly, our data suggest that most ER− tumors have pathway activation; however, it may be possible to further stratify BC subtypes according to degree of pathway activity quantitatively.
Importantly, we find that activated PTEN/PI3K pathway signaling is a biological correlate of poor prognosis in breast, prostate, and bladder carcinoma. In BC, prognostic prediction by the signature and the signature surrogate stathmin is independent of node status, indicating that activation of this pathway may promote cancer spread by hematogenous routes. Because loss of PTEN protein expression has been associated with resistance to trastuzumab in HER2+ patients, we suggest that the signature may improve prediction of trastuzumab response, because it captures other mechanisms of pathway activation, e.g., PIK3CA KD mutations. There appears to be a continuum of PI3K pathway contribution in several cancer types, in which tumors with the highest correlation to the activated pathway profile are most likely to cause patient mortality. Intriguingly, these same tumors are perhaps also the most “addicted” to the pathway and thus potentially the most sensitive to therapies that specifically attack this pathway. Exploitation of this fortuitous situation could lead to effective therapy for the worst-prognosis tumors and could have a major impact on patient survival. Our results do not exclude the contribution of other oncogenic pathways that may work in parallel, cooperate with PI3K signaling, or be associated to PTEN loss.
The PTEN signature identifies the majority of the same good and poor prognosis breast tumors as the Dutch 70-gene classifier (3), a highly predictive prognostic signature that was built without any regard to the underlying biological mechanisms driving the metastastic process (2). Recently, the Dutch 70-gene test and three other independent signatures were shown to essentially have similar prognostic power when tested against the same Dutch microarray data set of 295 BC patients (23). Our results shed some light into the prognostic “black box” and suggest that activation of oncogenic PI3K signaling is present in as many as 74% of the poor-prognosis patients identified by the 70-gene signature. Conversely, our data indicate that the less aggressive tumors identified by the 70-gene classifier may have a more favorable outcome precisely because they do not exhibit significant activation of the PI3K pathway. Moreover, our results appear consistent with other prognostic signatures such as the fibroblast response to serum “wound healing” signature (33), the chromosome instability signature (34), and the CD44+/CD24−/low signature (35), because serum potently activates the PI3K pathway, PTEN is integral to maintaining genomic stability (36, 37), and the PTEN/PI3K pathway has been shown to promote maintenance of cancer stem cell compartments (9).
The signature and signature genes could have significant impact on the development and clinical testing of PI3K pathway-specific therapies. For example, stathmin and other signature genes may be useful in identifying patients who have PTEN/PI3K pathway involvement to aid in the design and execution of clinical trials. Moreover, we demonstrate the signature of PTEN loss to be common among carcinomas and strongly associated to poor outcome, thereby providing the in vivo rationale for prioritization of anti-PI3K pathway drug development.
Methods
Patient Material.
Formalin-fixed paraffin-embedded tumor specimens were retrieved for 361 Swedish patients with stage II primary BC treated uniformly with 2 years of adjuvant tamoxifen (SI Methods), of which 351 were analyzed for PTEN protein expression, 191 were analyzed for stathmin protein expression, and snap-frozen samples corresponding to 105 patients were selected for microarray analysis. The study was approved by the Lund University Hospital Ethics Board.
IHC and Genetic Analyses.
Evaluations were performed blinded to all clinical and biological variables. Methods for PTEN IHC, PIK3CA mutational screening, and HER2 chromogenic in situ hybridization have been described (16, 38). Stathmin IHC is described in SI Methods.
Microarrays and Data Analyses.
Microarrays with 27,648 reporters (cDNA clones mapping to >14,000 unique clusters based on UniGene build 188) were fabricated by the SWEGENE Microarray Facility, Lund University. Experimental protocols are described in SI Methods. Microarray data are available through the National Center for Biotechnology Information Gene Expression Omnibus database (accession GSE5325) and from http://icg.cpmc.columbia.edu/faculty_parsons.htm. Data processing was performed within BASE (39) and resulted in 16,174 reporters that were used for all subsequent analyses. The 3-fold cross-validation design (40) using the Mann–Whitney test and support vector machines, gene set enrichment analysis (17), Gene Ontology analysis, and hierarchical clustering are described in SI Methods.
Prediction of Signature Activation in Independent Data Sets.
Five independent carcinoma data sets were tested for presence of the signature and its association to available clinical outcome (1, 3, 4, 25, 26). The NCC training and classification procedure are described in SI Methods. In brief, within each data set, matched genes were centralized, and for each classification, the SP and SA centroids were generated by using our data as defined in Fig. 1B. A test sample was classified based on which centroid it is most correlated to by using Pearson correlation. A continuous signature score was calculated by subtracting the SA correlation from the SP correlation.
Statistical Analysis.
The Mann–Whitney test was used to assess the difference between two observed continuous variables, the Pearson χ2 test was used to assess association between categorical variables in contingency tables unless the expected count in one or more cells was less than five, in which case Fisher's exact test was used, and the Pearson correlation was used for testing association between two continuous variables. For survival analysis, the KM method was used to estimate relevant event variables, and the log-rank test was used to compare survival between two strata. The Cox proportional hazards model was used for univariate and multivariate survival analyses. Results from all outcome event variables tested are presented. Survival analyses were carried out by using Stata 9.2 (Stata Corporation, College Station, TX). All tests were two-sided, and P ≤ 0.05 was considered significant.
Additional methods are described in SI Methods.
Supplementary Material
Acknowledgments
We acknowledge the South Sweden Breast Cancer Group for providing samples and thank Kristina Lövgren, Cecilia Hegardt, Mervi Jumppanen, Bruce Stillman, Michael Whitfield, and Vivek Mittal for assistance. Funding was provided in part from the National Institutes of Health (Medical Scientist Training Grant 5T32 GM07367-29 to L.H.S. and Grant CA097403 to R.P.), the Avon Foundation (H.H. and R.P.), American Association for Cancer Research -Amgen (M.M.), the Swedish Cancer Society (M.R. and Å.B.), and the Mrs. Berta Kamprad Foundation, Gunnar Nilsson Cancer Foundation, Lund University Hospital Foundations, King Gustav V Jubilee Foundation, and the Ingabritt and Arne Lundberg Foundation (Å.B.).
Abbreviations
- APV
average P value
- BC
breast cancer
- BCS
BC-specific survival
- CD
C2 domain
- DDFS
distant disease-free survival
- HD
helical domain
- IHC
immunohistochemistry
- KD
kinase domain
- KM
Kaplan–Meier
- NCC
nearest centroid classifier
- OS
overall survival
- PI3K
phosphatidylinositol 3-kinase
- SA
Signature Absent
- SP
Signature Present
- ER
estrogen receptor.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE5325).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702507104/DC1.
References
- 1.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, et al. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 2.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 3.van de Vijver MJ, He Y. D., van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, et al. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 4.Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, et al. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quackenbush J. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 6.Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, et al. Cancer Cell. 2004;6:117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Shaw RJ, Cantley LC. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 8.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, et al. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 9.Janzen V, Scadden DT. Nature. 2006;441:418–419. doi: 10.1038/441418b. [DOI] [PubMed] [Google Scholar]
- 10.Oda K, Stokoe D, Taketani Y, McCormick F. Cancer Res. 2005;65:10669–10673. doi: 10.1158/0008-5472.CAN-05-2620. [DOI] [PubMed] [Google Scholar]
- 11.Depowski PL, Rosenthal SI, Ross JS. Mod Pathol. 2001;14:672–676. doi: 10.1038/modpathol.3880371. [DOI] [PubMed] [Google Scholar]
- 12.Panigrahi AR, Pinder SE, Chan SY, Paish EC, Robertson JF, Ellis IO. J Pathol. 2004;204:93–100. doi: 10.1002/path.1611. [DOI] [PubMed] [Google Scholar]
- 13.Shah A, Swain WA, Richardson D, Edwards J, Stewart DJ, Richardson CM, Swinson DE, Patel D, Jones JL, et al. Clin Cancer Res. 2005;11:2930–2936. doi: 10.1158/1078-0432.CCR-04-1385. [DOI] [PubMed] [Google Scholar]
- 14.Tang JM, He QY, Guo RX, Chang XJ. Lung Cancer. 2006;51:181–191. doi: 10.1016/j.lungcan.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, et al. Am J Pathol. 1999;155:1253–1260. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, et al. Cancer Res. 2005;65:2554–2559. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, et al. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stolarov J, Chang K, Reiner A, Rodgers L, Hannon GJ, Wigler MH, Mittal V. Proc Natl Acad Sci USA. 2001;98:13043–13048. doi: 10.1073/pnas.221450598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minaguchi T, Kuramoto H, Nishida M, Satoh T, Tanaka T, et al. Cancer Res. 2001;61:3741–3749. [PubMed] [Google Scholar]
- 20.Majumder PK, Febbo PG, Bikoff R, Berger R, Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, et al. Nat Med. 2004;10:594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy S, Nakamura N, Sansal I, Bergeron L, Sellers WR. Cancer Cell. 2002;2:81–91. doi: 10.1016/s1535-6108(02)00086-7. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, et al. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DS, Nobel A. B., van't Veer LJ, Perou CM. N Engl J Med. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 24.Ringner M, Veerla S, Andersson S, Staaf J, Hakkinen J. Bioinformatics. 2004;20:2305–2306. doi: 10.1093/bioinformatics/bth089. [DOI] [PubMed] [Google Scholar]
- 25.Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blaveri E, Simko JP, Korkola JE, Brewer JL, Baehner F, Mehta K, Devries S, Koppie T, Pejavar S, et al. Clin Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 27.Brattsand G. Br J Cancer. 2000;83:311–318. doi: 10.1054/bjoc.2000.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curmi PA, Nogues C, Lachkar S, Carelle N, Gonthier MP, Sobel A, Lidereau R, Bieche I. Br J Cancer. 2000;82:142–150. doi: 10.1054/bjoc.1999.0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerner C, Vejda S, Gelbmann D, Bayer E, Gotzmann J, Schulte-Hermann R, Mikulits W. Mol Cell Proteomics. 2002;1:528–537. doi: 10.1074/mcp.m200026-mcp200. [DOI] [PubMed] [Google Scholar]
- 30.She QB, Solit DB, Ye Q, O'Reilly KE, Lobo J, Rosen N. Cancer Cell. 2005;8:287–297. doi: 10.1016/j.ccr.2005.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iancu-Rubin C, Atweh GF. Trends Cell Biol. 2005;15:346–348. doi: 10.1016/j.tcb.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Alli E, Bash-Babula J, Yang JM, Hait WN. Cancer Res. 2002;62:6864–6869. [PubMed] [Google Scholar]
- 33.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, et al. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 35.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, et al. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 36.Puc J, Keniry M, Li HS, Pandita TK, Choudhury AD, Memeo L, Mansukhani M, Murty VV, Gaciong Z, et al. Cancer Cell. 2005;7:193–204. doi: 10.1016/j.ccr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 37.Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 38.Tanner M, Gancberg D, Di Leo A, Larsimont D, Rouas G, Piccart MJ, Isola J. Am J Pathol. 2000;157:1467–1472. doi: 10.1016/S0002-9440(10)64785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-software0003. SOFTWARE0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavey S, Johansson P, Packer L, Taylor J, Stark M, Pollock PM, Walker GJ, Boyle GM, Harper U, et al. Oncogene. 2004;23:4060–4067. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.