Abstract
Virtually all cancer biological attributes are heterogeneous. Because of this, it is currently difficult to reconcile results of cancer transcriptome and proteome experiments. It is also established that cancer somatic mutations arise at rates higher than suspected, but yet are insufficient to explain all cancer cell heterogeneity. We have analyzed sequence variations of 17 abundantly expressed genes in a large set of human ESTs originating from either normal or cancer samples. We show that cancer ESTs have greater variations than normal ESTs for >70% of the tested genes. These variations cannot be explained by known and putative SNPs. Furthermore, cancer EST variations were not random, but were determined by the composition of the substituted base (b0) as well as that of the bases located upstream (up to b − 4) and downstream (up to b + 3) of the substitution event. The replacement base was also not randomly selected but corresponded in most cases (73%) to a repetition of b − 1 or of b + 1. Base substitutions follow a specific pattern of affected bases: A and T substitutions were preferentially observed in cancer ESTs. In contrast, cancer somatic mutations [Sjoblom T, et al. (2006) Science 314:268–274] and SNPs identified in the genes of the current study occurred preferentially with C and G. On the basis of these observations, we developed a working hypothesis that cancer EST heterogeneity results primarily from increased transcription infidelity.
Keywords: bioinformatics, transcription, oncology
Cancer is a genetic disease caused by sequential accumulation of mutations in oncogenes and tumor suppressor genes (1). Recent work reveals that cancer somatic mutations were more frequent than initially suspected. A large sequencing effort directed toward human colorectal and breast cancer DNA gene coding domains as well as exon–intron boundaries led to the identification of 1,307 novel confirmed somatic mutations (2). This study further showed that the subset of affected genes varies both with cancer type and within the same cancer type with individual tumors. Thus, the heterogeneity of cancer somatic mutations has clearly been established.
Virtually all biochemical, biological, and clinical attributes are heterogeneous within human cancer of the same histological subtype (3). Somatic mutations, although occurring at rates higher than suspected, remain relatively rare (3.1 per 106 bases), leading on average to 90 amino acid substitutions in a given tumor (2). Thus, somatic mutations alone cannot explain the large number of variants observed in systematic proteomic approaches.
The possibility that transcription [a process mediated by DNA-dependent RNA polymerases (RNAP)], as well as posttranscriptional enzymatic RNA base changes (4–6), might contribute to molecular heterogeneity of cancer has thus far not been considered. Recent work using in vitro transcription assays revealed that T7 RNAP, yeast RNAP II, and bacterial RNAP permit template–strand misalignment, leading to transcription infidelity (7, 8). This discovery led to the notion that in vivo transcription infidelity might increase when abasic sites or unrepaired DNA lesions are encountered (7, 8).
We sought to compare ESTs deriving from normal and cancer samples (9). It is known that a great deal of sequence variations are present in ESTs, specifically those available in noncurated databases (10, 11). EST sequence heterogeneity has thus far been considered as noise arising from (i) a high degree of sequencing errors, (ii) chimeric ESTs, (iii) intronic and interORF sequences, (iv) pseudogenes and paralogues, (v) SNPs, or (vi) somatic mutations.
Here we show a higher nonrandom heterogeneity in ESTs originating from cancer samples as compared with those from normal samples. Bioinformatic filtering of these sequences did not suppress increased cancer EST heterogeneity. Cancer EST sequence variation is determined by the sequence of the DNA template. The base substituted to the normal base (defined by Watson–Crick complementarities) occurs nonrandomly and corresponds in most cases to the base located immediately upstream or downstream of the substitution event.
Results
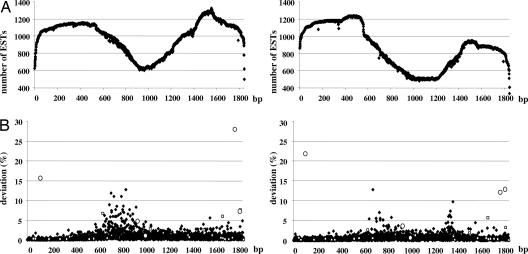
We accessed the noncurated EST database available on human dbEST from the National Center for Biotechnology Information (June 2005 version) (11) and classified ESTs according to sample sources. This led to the creation of three different sets of sequences containing ESTs from normal (N) tissue (≈2.8 × 106 sequences), cancer (C) tissue (≈2.6 × 106 sequences), and unknown origin (≈0.7 × 106 sequences). This last set was not considered in this analysis. We then selected 17 genes on the basis of their large representation in the database. Each EST sequence was then aligned against its mRNA Reference Sequence (RefSeq) by using MegaBLAST 2.2.13 software (12). We then measured the proportion of ESTs deviating from RefSeq at any given position. Fig. 1 provides a graphical representation of these variations occurring in the normal and cancer sets for a representative gene (VIM coding for vimentin: gene ID 7431). Supporting information (SI) Fig. 6 provides the same graphical representation for the remaining 16 genes. The data show that sequence variation occurred most frequently in the cancer set and further that the phenomenon appeared most predominant in specific mRNA sites. The very high number of variations could not be explained by SNPs. Putative SNPs (open squares in Fig. 1 and SI Fig. 6) and biologically validated SNPs (open circles in Fig. 1 and SI Fig. 6) are shown on the graph (dbSNP, build 126, September 2006) (13). Both putative and biologically validated SNPs leading to EST variations (n = 442) were excluded from further analysis.
Fig. 1.
EST number and variations of Vimentin (VIM) gene. (A) The number of available ESTs at any given position of the mRNA RefSeq; ESTs deriving from cancer (Left) and normal samples (Right) are shown. (B) The percentage of ESTs differing from RefSeq at any given position (Left, cancer set; Right, normal set). Putative SNPs and biologically validated SNPs are shown as open squares and open circles, respectively.
We next (i) tested the statistical significance of the differences in sequence variation occurring between cancer and normal ESTs, and (ii) when the statistical test was significant we determined whether the difference arose from sequence variation occurring in the normal (N > C) or cancer (C > N) sample. The statistical method is provided in SI Fig. 7 (14).
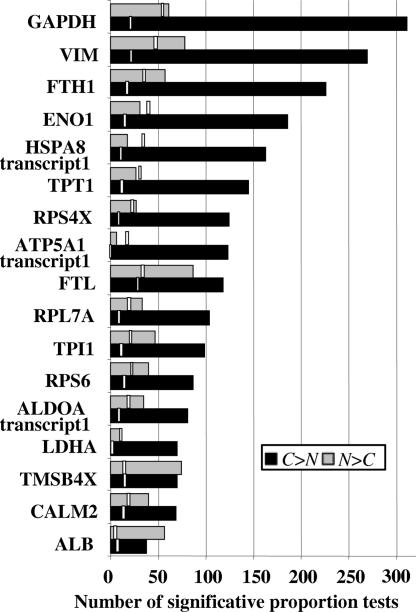
Data in Fig. 2 show that, for 15 of 17 abundantly expressed genes, statistically significant sequence variations arising from the cancer set, C > N, largely exceeded variations originating in the normal set, N > C. Furthermore, sequence variations C > N consistently and largely exceeded that of the estimated error resulting from multiple testing as defined by the location-based estimator (LBE) (shown as vertical bars). Variations N > C occurring at rates greater than that of LBE were found in 12 of 17 genes, but the ratio between statistically significant sequence variations and LBE was much lower in the normal set than in the cancer set. A more detailed analysis of these data is provided as SI Fig. 8.
Fig. 2.
Number of statistically significant deviations originating from normal and cancer ESTs. Statistically significant differences in the proportion of sequence variations originating from normal and cancer ESTs are shown by gray and black bars, respectively. The false positives estimator (LBE) is shown by vertical open bars.
Random sequencing errors cannot account for differences in sequence variation occurring between the normal and cancer sets. This interpretation is supported by the fact that libraries originating from cancer or from normal samples are processed essentially in the same manner. Mathematical analysis is consistent with this interpretation (SI Fig. 7).
Random sequencing errors being excluded, we sought to eliminate other sources of EST heterogeneity by filtering procedures. Our initial requirements were that EST aligned to RefSeq with 100% identity on at least 16 consecutive bases, and with ≥90% identity on at least 50 bases. As shown in Table 1, this yielded 2,281 and 725 statistically significant differences C > N and N > C, respectively, that were distinct from putative or biologically validated SNPs. The constraints of the statistical test were met at 7,644 positions of 23,930 cumulated positions of the 17 genes. The second filter required that each EST aligned to RefSeq continuously on >70% of its length. The third filter removed ESTs with sequence more closely related to paralogues and pseudogenes than to the bona fide RefSeq. The fourth filter deleted from analysis the first and last 50 bases of each EST alignment to remove mismatches at the 3′ and 5′ borders of EST created by the MegaBLAST program. As shown in Table 1, filters 2–4 decreased statistically significant sequence variation events C > N and N > C to 1,300 and 374, respectively. The C > N versus N > C ratio of sequence variations (C/N) increased from 3.15 to 3.48 (+10%), and the same ratio subtracted from LBE increased from 6.95 to 7.95 (+14%). The effect of filtering on C/N ratio subtracted from LBE was even more pronounced (25%) when one considered only the set of 13 genes with prefiltering C/N ratio >2 (SI Fig. 9).
Table 1.
Variations in ESTs before and after sequential application of electronic filters
| Filtering procedure | C > N | LBE | N > C | LBE | (C > N)/(N > C) | [(C > N) − LBE]/[(N > C) − LBE] |
|---|---|---|---|---|---|---|
| EST alignment > 50 bp | 2,281 | 259 | 725 | 488 | 3.15 | 6.95 |
| >70% EST aligned | 2,065 | 230 | 722 | 429 | 2.86 | 5.59 |
| Removal of paralogues and pseudogenes | 2,065 | 223 | 694 | 428 | 2.98 | 6.08 |
| Fifty base pairs deleted at each extremity | 1,300 | 132 | 374 | 266 | 3.48 | 7.95 |
The C > N column provides the number of positions where statistically significant sequence variations are in excess in the cancer set. N > C provides the number of positions where sequence variations originate from the normal set. (C > N)/(N > C) ratio was calculated from pooled data. [(C > N) − LBE] and [(N > C) − LBE] were calculated for each gene. Negative results were treated as 0. Results of all genes were pooled to yield [(C > N) − LBE]/[(N > C) − LBE] ratio. The data show the results of each filtering procedure applied sequentially on the 17 studied genes.
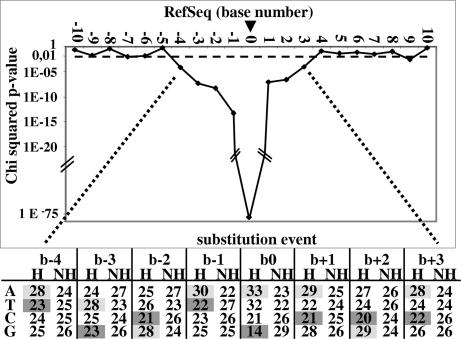
We next hypothesized that cancer EST variations occurred because of differences in the base composition on genomic DNA template. This analysis was performed exclusively by using EST variations where C > N was statistically significant. To avoid bias that might be introduced by the filtering procedures, we used all available nonfiltered data. Results of Fig. 3 show the difference in base composition observed when comparing sequences upstream and downstream of substitution events (heterogeneous, n = 2,281) with those where no substitution event was detected (nonheterogeneous, n = 12,273). The criteria for nonheterogeneous sites were cancer set variations <0.5% and not statistically different from normal set variations. It must be emphasized that the 2,281 C > N heterogeneous positions used at this stage of analysis are defined on a statistical basis and hence contain a limited number of false positives as well as false negatives. Furthermore, the populations are necessarily incomplete because statistical tests cover only 32% of gene lengths. In this analysis we refer to the base undergoing substitution as b0, bases located on pre-mRNA (pmRNA) sequence 5′ end are referred to as b − n, and bases located on 3′ end are referred to as b + n. The data show first that not all four bases were equally susceptible to variation: b0 = A (33%) ≈ T (32%) ≫ C (21%) ≫ G (14%). Furthermore, the compositions of the four bases upstream and three bases downstream of the site of event were statistically different from those of the sites without significant EST variation (Fig. 3). Specifically, sites where variations occur were more frequently preceded and followed by A ≥ G > T ≈ C. Thus, the occurrence of cancer EST heterogeneity is not random, but is determined first by the nature of the base undergoing substitution and second by the nature of the bases that immediately precede and follow the event.
Fig. 3.
Effect of pmRNA sequence base composition on base b0 heterogeneity. (Upper) The results of χ2 analysis of the differences in pmRNA base composition at any given position, as well as upstream and downstream of C > N statistically significant sequence variations (n = 2,281) versus that where no EST heterogeneity was observed (n = 12,273). The dotted line is the P value threshold (P = 0.01). (Lower) The base composition for sites on pmRNA corresponding to significant C > N EST heterogeneity (H) and nonheterogeneous sites (NH). Gray shading shows enriched bases; darker gray shows paucity bases.
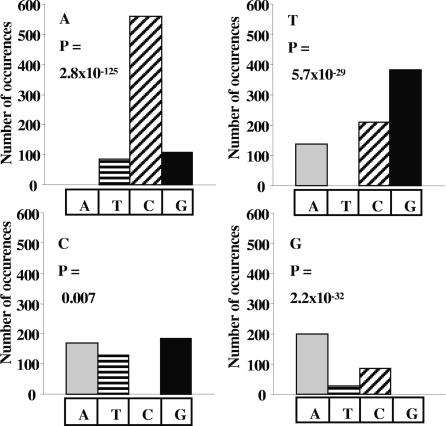
It is clear from Fig. 4 that the replacement base was also not selected randomly. A was preferentially replaced by C (P = 2.8 × 10−125), T by G (P = 5.7 × 10−29), and G by A (P = 2.2 × 10−32). Substitution of C showed a more even distribution, with a slight paucity of T (P = 0.007).
Fig. 4.
Composition of replacement base for each substituted base. The figure represents, for each substituted base C > N, the composition of the most common replacement base. Statistical significance of difference in proportions was calculated with the null hypothesis that the replacement base is selected randomly.
To identify the underlying causes of such preferential base replacement, we distinguished two sets of informative and noninformative events.
Informative events were situations where the substituted base was different from either the preceding base (b − 1) or the following base (b + 1) (n = 1,676) (SI Fig. 10). Noninformative events were situations not matching these criteria. When informative events were analyzed, two cases were encountered: substituted base was replaced by b − 1 or b + 1 (79%) or by another base, different from b − 1 and b + 1 (21%). (i) In the first subset, replacement base was identical to b − 1 (n = 799) or b + 1 (n = 530). When replacement base was b − 1, then b0 = A (36%) > C (30%) ≫ T (21%) ≫ G (13%). A was preferentially replaced by C (71% of the cases). When replacement base was b + 1, then b0 = T (47%) ≫ A (21%) > C (19%) ≫ G (13%) (SI Fig. 11). T was preferentially replaced by G (71%). For b − 1 substitutions, the pattern of relative influence of base composition was b0 > b − 1 > b − 2 > b + 1 > b − 3 > b + 2. For b + 1 substitutions, the relative influence of the surrounding base followed a pattern of b0 > b + 1 > b − 1 > b + 2 > b − 3 > b − 2. (ii) In the second subset of informative events, the replacement base did not correspond to either b − 1 or b + 1 (n = 347). Affected bases were in the following order: A (47%) > T (29%) > C (14%) > G (10%). A was most commonly replaced by C (91% of the cases), T by C (50%) and A (42%), C by G (46%), and G by C (73%). Thus, when replacement base does not correspond to b − 1 or b + 1, the replacement base is not randomly selected, but C is in large excess.
We next considered the set of noninformative events, i.e., situations where (i) b − 1 = b + 1 and where (ii) b − 1 = b0 = b + 1 (SI Fig. 10). When b − 1 and b + 1 were identical but different from b0 (n = 339), substituted bases were in the order T (34.8%) > G (23.6%) > C (21.2%) > A (20.4%) and followed the same pattern of preference as in Fig. 4: T→G, G→A, and A→C. Substitutions occurring on the central base of repeat of three identical bases (n = 266) were observed in the following order: A (46.2%) > T (36.9%) > G (10.5%) > C (6.4%). In this case, the most common substitution events were A→C and T→C and A. Rare GGG substitutions were most commonly replaced by GCG and CCC by CAC.
Thus, when substitutions occur within three consecutive identical bases and when substitutions did not correspond to either b − 1 or b + 1, then C was the most common replacement base. When the replacement base corresponds to b − 1, the most common substitution was A→C; when the replacement base corresponds to b + 1, the most common substitution was T→G. It can therefore be concluded that neither the base undergoing substitution nor the replacement base was selected randomly. Both phenomena followed predictable patterns defined by the composition of the base undergoing substitution and that of bases located upstream and downstream of this event.
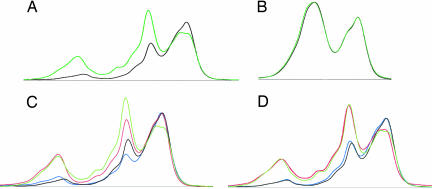
Because EST sequences used in this analysis originated from many patients and several laboratories, we next sought to obtain biological validation of the differences in mRNA heterogeneity between normal and cancer cells isolated from the same patient. Preliminary analysis of denaturing HPLC (DHPLC) profiles revealed the ability to detect three bases cDNA heterogeneity occurring at a rate of 2.5–5% (data not shown). We selected the ENO1 gene because (i) it appears clearly affected by the increased EST heterogeneity in the cancer set (Fig. 2) and (ii) its sequence allowed identification of primers that do not match on a genome-wide basis with paralogues or pseudogenes. The difference in cumulated percentage of base substitutions between cancer and normal ESTs for the ENO1 gene-amplified fragment that was calculated from dbEST data is 7.25%, i.e., within DHPLC sensitivity range. We also selected TMSB4X as the negative control because we found no statistical difference in EST heterogeneity between normal and cancer sets and were also able to identify genome-specific primers. The results show a difference in heterogeneity of RT-PCR-amplified products of ENO1 gene isolated from kidney normal and cancer cells of the same patient (Fig. 5A). This contrasted with the absence of difference observed with the TMSB4X gene using the same samples (Fig. 5B). The lengths of amplified fragments were similar for both genes, and the same polymerases were used in both experiments. PCR infidelity did not create variations for the TMSB4X gene, so one can therefore reasonably assume that it is also the case for ENO1 and thus that the observed difference between normal and cancer profiles reflects the heterogeneity of mRNA sequence. To further establish this conclusion, we repeated the analysis of ENO1 using two different PCR preparations and again observed significant differences between cancer and normal tissue of the same patient (Fig. 5C). Finally, we verified the reproducibility of the loading procedure to DHPLC (Fig. 5D). These data establish that mRNA heterogeneity is different when one compares RNA from cancer to normal tissues isolated from the same patient. We have repeated the procedure using different genes and different cancer types and obtained results indicating that cancer mRNA heterogeneity is both cancer- and gene-specific (data not shown). Because DHPLC does not allow probing the extent and position of sequence heterogeneity, a detailed analysis of cancer versus normal mRNA sequences using highly accurate sequencing procedures is needed.
Fig. 5.
DHPLC elution profiles of double-stranded DNA amplified from cDNA obtained from kidney cancer and adjacent normal tissues. mRNA isolated from cancerous and adjacent normal kidney tissues from the same patient were reverse-transcribed, and the double-stranded cDNA PCR-amplified by using primers specific to the ENO1 gene (A) and the TMSB4X gene (B) was loaded on a DHPLC system. DNA elution was followed by absorbance (mV) in function of retention time (minutes). Normal and cancerous elution profiles are shown in green and black, respectively. Variations due to repeated PCR applied to ENO1 cDNA are shown in C (green and red, normal; black and blue, cancer). Variations due to DHPLC loading of the ENO1 PCR products are shown in D with the same color coding.
Discussion
The primary conclusions of this analysis are that cancer EST heterogeneity is greatly increased compared with that of normal ESTs and is not random noise, but follows a specific pattern defined by the genomic DNA template and determined first by the composition of the base undergoing substitution and second by the composition of the bases that immediately precede or follow this event. Applying stringent filters did not erase the differences between cancer and normal groups. Therefore, cancer EST variations reflect the fact that a small but significant proportion of cancer cell mRNA are heterogeneous and do not carry the information predicted by simple base-pairing to the human genome.
Direct comparison of mRNA heterogeneity between cancer and normal cells from the same patient indicate that differences exist for some but not all mRNA. It is also most probable that genes affected are cancer-specific. We speculate that a large sequencing effort of normal and cancer cDNA libraries from the same patient using accurate pyrosequencing method (15, 16) will lead to the definition of cancer-specific molecular signature of mRNA base substitution.
There are five limitations of our current study. First, the method of analysis does not take into account variations of EST sequences other than single base substitutions. Testing the possibility that deletion(s) and/or insertion(s) are also present requires a different analytical strategy. Second, the analysis is restricted to substitution events considered independent of one another. A longitudinal analysis of the relationship between substitution events is needed. Third, the statistical method chosen for this first study was conservative. The less stringent Fisher exact test will be needed to achieve complete coverage of gene lengths and allow testing of low-abundance genes. A large proportion (76%) of tested genes showed increased cancer EST heterogeneity and C/N > 2. However, selection criteria are biased toward genes highly transcribed and with relatively short transcripts. Hence, the conclusion may not extend to low-abundance genes and mRNA. Fourth, the analysis did not discriminate between different cancer types or cancer cell lines. We verified that a greater statistically significant EST heterogeneity persisted in the cancer set after removing ESTs produced from cultured cancer cells (data not shown). Fifth, we are currently unable to determine whether EST heterogeneity occurs in normal cells following a pattern similar to or different from that of cancer cells. Indeed, the number of N > C events is not in large excess of the estimated rate of false positives. Thus, N > C events contain a large proportion of false positives that prevent meaningful data interpretation. To address this important issue we need larger sets of data comparing the mRNA sequence of cancer and normal cells from the same individual.
The next issue is to define the origin of cancer EST heterogeneity. The first source is mutations affecting genomic DNA. We have excluded from our analysis all putative and biologically validated SNPs. Thus, except if a large number of SNPs are currently unsuspected, it is unlikely that SNPs are responsible for cancer mRNA heterogeneity. Furthermore, within the SNP pool of the 17 studied genes, C and G are preferentially affected (68% of 442 SNPs), whereas A and T are preferentially affected in cancer ESTs (65% of 2,281 substitution events).
Alternatively, one can speculate that somatic mutations of cancer cell DNA account for mRNA heterogeneity. Similar to SNPs of the current study, colon and breast cancer somatic mutations were shown to affect preferentially C and G bases (80.6% of cases). However, in-depth efforts of breast and colon cancer DNA sequencing that included 14 of 17 genes used in our study led to an estimated somatic mutation rate of 3.1 mutations per 106 bases (2). Sites of cancer EST variations are 3 to 4 orders of magnitude more commonly encountered than those of somatic mutations. This is not to say that all cancer cell mRNA carries 10 mutations every 100 bases but rather that up to 10 bases per 100 bases can be substituted on any given cancer mRNA. Thus, it is likely that EST heterogeneity occurs not at the genomic level, but rather at the pmRNA level.
Two mechanisms are envisioned. First, RNA editing is able to change C to U, U to C, and A to I read as G (4–6). In the cancer set we observed at the cDNA level 5.7% C→T, 9.2% T→C, and 4.7% A→G changes. Thus, mRNA editing cannot account for >20% of single base substitutions described here. Indeed, the most common base substitutions, A→C (24.6%) and T→G (16.8%), represent base family changes that are not explained by known human enzymatic RNA editing processes.
The alternate hypothesis is therefore to consider that cancer mRNA heterogeneity occurs as a result of transcription infidelity, leading most frequently (73%) to repeat of b − 1 or b + 1.
Cases of transcription infidelity have been reported. In the Brattleboro rat with diabetes insipidus resulting from a + 2 frameshift in the vasopressin gene, GA deletion of the GAGAG sequence of the mRNA reverts part of the transcript to normal and improves the phenotype (17, 18). Transcriptional frameshift events affecting repetitive A sequence of β-amyloid and ubiquitin B yield proteins with alternate reading frames that are detected by immunological staining of Alzheimer's disease plaques (19, 20). Transcriptional infidelity of dog AP3B1 gene yields to the addition or removal of a single A within a poly(A) stretch (21). In the rat p53 gene A6 transcription leads to the insertion of an extra A in 9% of cloned transcripts (22).
In vitro assays of transcription infidelity using multisubunit yeast RNAP II show misalignment and b + 1 base replacement (7, 8). Forward misalignment results from extrahelical flipping out of the substituted base on DNA template. Crystal structure data show that space is available within the RNAP active site to transiently accommodate the flipped-out base and hence allow misalignment. Thus, a molecular mechanism explaining cases where replacement base is b + 1 is documented. However, no molecular model currently explains our most common observation, i.e., repetition of b − 1. Consistent with the hypothesis that cancer EST heterogeneity is due to transcription infidelity is the finding that bases most influential of the substitution events are not only the substituted base itself (b0), but also the four bases located upstream and the three downstream of this event. This pattern corresponds in the elongation complex with the first four RNA–DNA base-pairing and to the three transcription-driven melted bases (23). In vitro studies of RNAP infidelity established that DNA grip three bases downstream of the misalignment event is critical at controlling transcription fidelity (24, 25).
Although this is not the first report of transcription infidelity, our analysis introduces the notion that the phenomenon might occur on a previously unsuspected scale in cancer samples where it could affect a large proportion of genes. Furthermore, we show that transcription infidelity represents a nonrandom phenomenon driven in part by genomic DNA sequence. The underlying causes of the increase of transcription infidelity in cancer samples are currently unknown.
The relatively low selectivity of RNAP makes misincorporation unavoidable (26). Transcript-assisted transcriptional proofreading with strong Mg2+ dependence corrects base misincorporation events (27). Most of the RNA in erroneous complexes are cleaved, but a fraction is extended past the misincorporation site (28). We compared in vitro efficacy of transcription-assisted transcriptional proofreading by measuring the kcat/Kd ratio from data of Zenkin et al. (27) obtained with an in vitro model where b − 1 = G and b + 1 = C. Interestingly, the highest efficacy of repair was for substitution with A. Among 138 substitutions that in our pool correspond to the same sequence context as that of Zenkin et al. (27), only seven substitutions with A were observed. Furthermore, Zenkin et al. (27) show that C→G substitutions led to equal ratio of cleavage versus extended RNA; we observed 67 C→G substitutions of 138. We thus speculate that increased transcription infidelity in the cancer set is due in part to defective transcription-assisted transcriptional proofreading. Alternatively, heterogeneous mRNA might accumulate in cancer cells because of a decrease in their degradation rate.
Cancer mRNA heterogeneity might explain the relative lack of reproducibility of cancer microarray transcriptome experiments (29). If heterogeneous cancer mRNA are translated, this would cause the occurrence of a myriad of protein variants, possibly explaining the complexity of current cancer proteomic results (30).
If the hypothesis of increased transcription infidelity during carcinogenesis is confirmed, we will have to recognize that EST sequencing efforts contributed by many laboratories and often considered of limited value have allowed the emergence of a new paradigm, perhaps opening new avenues toward a better understanding of cancer biology.
Materials and Methods
Gene Selection.
Genes were selected solely on the basis of their high abundance in Unigene clusters and without consideration for their putative or established function, as well as their association with diseases. Selected genes were VIM (NM_003380.2), GAPDH (NM_002046.3), FTH1 (NM_002032.2), ENO1 (NM_001428.2), HSPA8 (NM_006597.3), TPT1 (NM_003295.1), RPS4X (NM_001007.3), RPL7A (NM_000972.2), RPS6 (NM_001010.2), TMSB4X (NM_021109.2), ALB (NM_000477.3), FTL (NM_000146.3), ALDOA (NM_000034.2), ATP5A1 (NM_001001937.1), CALM2 (NM_001743.3), LDHA (NM_005566.1), and TPI1 (NM_000365.4).
Filtering Procedure.
ESTs that did not align continuously with RefSeq on 70% of its length were removed.
For pseudogenes and paralogues filter, pseudogene sequences were downloaded from the http://pseudogene.org database by Ensembl identification number. Paralogue sequences were generated by sequence alignment of RefSeq against human RefSeq mRNA by using BLASTN, MegaBLAST, or Discontiguous MegaBLAST with default parameters (except W = 16 for MegaBLAST). Homologous sequences were defined by BLASTN results completed by MegaBLAST and Discontiguous MegaBLAST results. ESTs deriving from pseudogene or paralogue sequences (Ps/Pa) were removed based on the following calculation: Cost 1 corresponds to the number of mismatches between RefSeq and EST. Cost 2 is calculated as the number of mismatches between EST and RefSeq + the number of mismatches between Ps/Pa and RefSeq − [2 × the number of common mismatches (with the same replacement base) between Ps/Pa and EST] − the number of common mismatches (with different replacement base) between Ps/Pa and RefSeq. When Cost 2 is lower than Cost 1, it is assumed that the EST sequence corresponds more closely to Ps/Pa, and the EST is removed.
Fifty bases were deleted on both 5′ and 3′ extremities of each aligned EST.
DHPLC Profiles of Kidney Cancer Tissue Versus Normal Adjacent Tissue.
cDNA from cancerous and adjacent normal kidney tissues obtained from the same individual (BioChain; CliniSciences, Montrouge, France) were amplified by PCR with oligonucleotides complementary to the ENO1 gene (positions 1505–1524 and 1788–1812), the TMSB4X gene (positions 311–335 and 590–614), and the Pfx polymerase (Invitrogen, Cergy-Pontoise, France). These cDNA of 300 bp were then purified on Nucleospin Extract II columns (Macherey-Nagel, Hoerdt, France), visualized on agarose gel, quantified, and normalized. Another PCR cycle was performed by using 50 ng of each cDNA with a Pfx DNA polymerase. Samples were then denatured for 5 min at 95°C and slowly renatured by decreasing the temperature by 1°C for 38 seconds and 72 cycles. The double-stranded DNA samples were then injected on a DHPLC system (Transgenomic, Elancourt, France). The temperature of the oven for each gene was selected by using Navigator software. For the ENO1 gene the temperature was 61.5°C, and for TMSB4X the temperature was 55°C. DNA elution was followed by absorbance (millivolts) in function of retention time (minutes). Curves were visualized and normalized with Navigator software.
Supplementary Material
Acknowledgments
We thank Prof. L. Méjean for stimulating scientific discussions, Prof. P. Jonveaux for suggesting use of the DHPLC method and his team for technical assistance, and L. Bonnard for technical assistance. This work was supported by Genclis; and by grants from the Région Lorraine, Communauté Urbaine du Grand Nancy, Cancéropôle du Grand Est, Institut National du Cancer; and by a Ministry of Research and Higher Education fellowship (to M.B.).
Abbreviations
- RNAP
DNA-dependent RNA polymerase
- pmRNA
pre-mRNA
- LBE
location-based estimator
- DHPLC
denaturing HPLC.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0611076104/DC1.
References
- 1.Vogelstein B, Kinzler KW. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 2.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 3.Nelkin B, Pardell D, Robinson S, Small D, Vogelstein B. In: Tumor Cell Heterogeneity: Origins and Implications. Owens A, Coffey DS, Baylin SB, editors. New York: Academic; 1982. pp. 441–460. [Google Scholar]
- 4.Gott JM, Emeson RB. Annu Rev Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 5.Maas S, Rich A. BioEssays. 2000;22:790–802. doi: 10.1002/1521-1878(200009)22:9<790::AID-BIES4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 6.Niswender CM. Cell Mol Life Sci. 1998;54:946–964. doi: 10.1007/s000180050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomerantz RT, Temiakov D, Anikin M, Vassylyev DG, McAllister WT. Mol Cell. 2006;24:245–255. doi: 10.1016/j.molcel.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashkina E, Anikin M, Brueckner F, Pomerantz RT, McAllister WT, Cramer P, Temiakov D. Mol Cell. 2006;24:257–266. doi: 10.1016/j.molcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Lodish H, Berk A, Zipursky L, Matsudaira P, Baltimore D, Darnell J. Molecular Cell Biology. New York: Freeman; 2000. pp. 216–223. [Google Scholar]
- 10.Hillier LD, Lennon G, Becker M, Bonaldo MF, Chiapelli B, Chissoe S, Dietrich N, DuBuque T, Favello A, Gish W, et al. Genome Res. 1996;6:807–828. doi: 10.1101/gr.6.9.807. [DOI] [PubMed] [Google Scholar]
- 11.Boguski MS, Lowe TM, Tolstoshev CM. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Schwartz S, Wagner L, Miller W. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 13.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmasso C, Broet P. J Soc Fr Stat. 2005;146:63–75. [Google Scholar]
- 15.Ng P, Tan JJ, Ooi HS, Lee YL, Chiu KP, Fullwood MJ, Srinivasan KG, Perbost C, Du L, Sung WK, et al. Nucleic Acids Res. 2006;34:e84. doi: 10.1093/nar/gkl444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, Garraway LA, LaFramboise T, Lee JC, Shah K, et al. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 17.Evans DA, van der Kleij AA, Sonnemans MA, Burbach JP, van Leeuwen FW. Proc Natl Acad Sci USA. 1994;91:6059–6063. doi: 10.1073/pnas.91.13.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Leeuwen FW, Fischer DF, Kamel D, Sluijs JA, Sonnemans MA, Benne R, Swaab DF, Salehi A, Hol EM. Neurobiol Aging. 2000;21:879–891. doi: 10.1016/s0197-4580(00)00151-2. [DOI] [PubMed] [Google Scholar]
- 19.Vogel G. Science. 1998;279:174. doi: 10.1126/science.279.5348.174. [DOI] [PubMed] [Google Scholar]
- 20.van Leeuwen FW, van Tijn P, Sonnemans MA, Hobo B, Mann DM, Van Broeckhoven C, Kumar-Singh S, Cras P, Leuba G, Savioz A, et al. Neurology. 2006;66:S86–S92. doi: 10.1212/01.wnl.0000193882.46003.6d. [DOI] [PubMed] [Google Scholar]
- 21.Benson KF, Person RE, Li FQ, Williams K, Horwitz M. Nucleic Acids Res. 2004;32:6327–6333. doi: 10.1093/nar/gkh974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ba Y, Tonoki H, Tada M, Nakata D, Hamada J, Moriuchi T. Mutat Res. 2000;447:209–220. doi: 10.1016/s0027-5107(99)00209-2. [DOI] [PubMed] [Google Scholar]
- 23.Armache KJ, Kettenberger H, Cramer P. Curr Opin Struct Biol. 2005;15:197–203. doi: 10.1016/j.sbi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Cramer P. Curr Opin Genet Dev. 2004;14:218–226. doi: 10.1016/j.gde.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Westover KD, Bushnell DA, Kornberg RD. Cell. 2004;119:481–489. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 27.Zenkin N, Yuzenkova Y, Severinov K. Science. 2006;313:518–520. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 28.Cramer P. Science. 2006;313:447–448. doi: 10.1126/science.1131205. [DOI] [PubMed] [Google Scholar]
- 29.Marshall E. Science. 2004;306:630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 30.Master SR. Clin Chem. 2005;51:1333–1334. doi: 10.1373/clinchem.2005.053686. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.