Abstract
As a major agent of rapid speciation, interspecific hybridization has played an important role in plant evolution. When hybridization involves species that exhibit self-incompatibility (SI), this prezygotic barrier to self-fertilization must be overcome or lost to allow selfing. How SI, a normally dominant trait, is lost in nascent hybrids is not known, however. Here we demonstrate that hybrid self-fertility can result from epigenetic changes in expression of the S-locus genes that determine specificity in the SI response. We analyzed loss of SI in synthetic hybrids produced by crossing self-fertile and self-incompatible species in each of two crucifer genera. We show that SI is lost in the stigmas of A. thaliana–lyrata hybrids and their neo-allotetraploid derivatives and in the pollen of C. rubella–grandiflora hybrids and their homoploid progenies. Aberrant processing of S-locus receptor kinase gene transcripts as detected in Arabidopsis hybrids and suppression of the S-locus cysteine-rich protein gene as observed in Capsella hybrids are two reversible mechanisms by which SI might break down upon interspecific hybridization to generate self-fertile hybrids in nature.
THE origin of many plant species may be traced to sexual hybridization between more or less diverged species (Stebbins 1959; Rieseberg 1997, 2001). Stable self-fertile diploid (homoploid) hybrids are sometimes produced by hybridization between closely related species that have similar genomes and chromosome complements (Gross and Rieseberg 2005). More frequently, however, several barriers to gene flow between species must be overcome before fertile interspecific hybrids are generated. Most commonly discussed are postzygotic barriers that lead to sterility in F1 hybrids (Bushell et al. 2003), often resulting from aberrant meiotic pairing between highly divergent parental genomes. In these cases, chromosome doubling restores normal meiosis and generates fertile allopolyploids, a process that is thought to underlie at least 4% of speciation events in flowering plants (Rieseberg 2001). Neither homoploid nor allopolyploid hybrids can form, however, unless prezygotic barriers to hybridization are overcome, including pollination barriers that prevent pollen tubes from forming at the stigma surface, growing within the pistil, and reaching the ovules. A major prezygotic pollination barrier is genetic self-incompatibility (SI), which, although primarily known as an intraspecific barrier to self-fertilization in many obligate outcrossing plants, is also important in the context of interspecific hybridization. Indeed, when interspecific hybridization involves self-incompatible species, the generation of self-fertile hybrids, whether homoploid or allopolyploid, is dependent on the breakdown of SI.
The crucifer (Brassicaceae) family, which includes predominantly self-fertilizing species and self-incompatible species, provides several examples of self-fertile interspecific hybrids that occurred spontaneously or were produced artificially in breeding programs. Self-fertile allopolyploids are particularly common. Examples of allotetraploids include Brassica napus, derived by hybridization between B. oleracea and B. campestris (syn. B. rapa), and Arabidopsis suecica, derived by hybridization between self-fertile A. thaliana and self-incompatible A. arenosa (Mummenhoff and Hurka 1995; O'Kane et al. 1996). In addition, self-fertile homoploid F1 hybrids occur in this family. For example, crosses between self-incompatible Capsella grandiflora and self-fertile C. rubella produce self-fertile diploid F1 hybrids that can be selfed to establish F2 populations (Riley 1934; Acarkan et al. 2000; Koch and Kiefer 2005).
The crucifer family is particularly well suited for investigating the breakdown of SI in interspecific hybrid progenies of self-incompatible species, not only because of the prevalence of interspecific hybridization events in this family, but also because the crucifer SI system is well characterized. SI specificity has been shown to be determined by the highly polymorphic protein products of two genes that are tightly linked within the S-locus complex (Nasrallah 2005; Takayama and Isogai 2005). Allele-specific interaction between two proteins, the stigma-expressed S-locus receptor kinase (SRK) and its pollen ligand, the S-locus cysteine-rich protein (SCR, also known as SP11; Takayama and Isogai 2005), triggers a signaling cascade within the stigma epidermis that leads to pollen inhibition. Importantly, these two proteins have also been shown to be the primary determinants of the outcrossing mating system in the family as demonstrated by the successful transfer of the SI trait into self-fertile A. thaliana by transformation with an SRK–SCR gene pair from self-incompatible A. lyrata (Nasrallah et al. 2002, 2004).
To understand the molecular basis of breakdown of SI in interspecific hybrids, we focused on interspecific hybrids of Arabidopsis and Capsella. Previous studies had shown that crosses between A. thaliana and A. lyrata, on the one hand, and between C. rubella and C. grandiflora, on the other hand, produced interspecific hybrids in controlled pollinations (Acarkan et al. 2000; Nasrallah et al. 2000). Here, we document the behavior of pollen tubes in the hybrids upon self-pollinations and reciprocal crosses to the parental species. We also report on the isolation of S-locus genes from C. grandiflora and the use of these genes and of previously isolated A. lyrata SRK and SCR genes for a molecular analysis of interspecific hybrids. We demonstrate that both A. thaliana–lyrata and C. rubella–grandiflora hybrids exhibit a loss of SI, on the stigma side in the first case and on the pollen side in the second case. The expression patterns of S-locus genes in these hybrids suggest at least two different mechanisms by which SI might break down to generate fertile interspecific hybrids in nature.
MATERIALS AND METHODS
Plant materials and generation of interspecific hybrids:
For interspecific hybridization of Arabidopsis species, we used A. thaliana accession Col-0 and self-incompatible A. lyrata SaSb plants (described by Kusaba et al. 2001), which were descended from accessions collected in Michigan (kindly provided by Charles Langley, University of California at Davis). The generation of A. thaliana–lyrata hybrids by pollinating A. thaliana stigmas with pollen from A. lyrata, followed by ovule rescue, was described previously (Nasrallah et al. 2000). The C. rubella and C. grandiflora strains used in this analysis were described previously (Acarkan et al. 2000) and C. rubella–grandiflora hybrids were generated by manual cross-pollinations, which produced seed that could be germinated directly in soil.
Pollination analyses and determination of SI phenotype:
Stigmas were examined for absence of contaminating pollen under a stereoscope, and appropriate pollen was manually applied to their surface. Two hours after pollination, flowers were fixed for 30 min in a 3:1 mixture of ethanol and acetic acid, softened for 30 min in 1 m NaOH at 65°, washed for 30 min in water, stained in decolorized aniline blue, and mounted on slides for examination by epifluorescence microscopy (Kho and Baer 1968). Under these conditions, an incompatible response is typically manifested by <10 pollen tubes per stigma while compatible pollinations exhibit numerous pollen tubes per stigma.
Construction and screening of genomic libraries:
For genomic library construction, C. grandiflora S7S8 DNA was partially digested with Sau3A1, and a fraction containing fragments of 9–20 kb was cloned into the BamHI site of the λDASHII vector (Stratagene, LaJolla, CA). The library was screened with a 32P-labeled probe containing a mixture of fragments derived from the first exons of the A. lyrata SRKa and SRKb genes (Kusaba et al. 2001).
DNA gel-blot analysis:
DNA was isolated from leaves according to Murray and Thompson (1980). Digested DNA (∼3 μg) was run on 0.8% (w/v) agarose gels, transferred to GeneScreen Plus membrane (DuPont-New England Nuclear, Boston) using an alkaline transfer method. The blots were prehybridized and hybridized at 65° in 10% (w/v) dextran sulfate, 330 mm sodium phosphate, pH 7.0, 10 mm EDTA, and 5% (w/v) SDS. Probes were labeled with 32P using the Random Priming kit (Roche, Indianapolis). After washing in a solution containing 0.2× SSC (1× SSC is 0.15 m NaCl and 0.015 m sodium citrate) and 0.1% (w/v) SDS at 65°, the blots were exposed to phosphor screens and developed using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager.
RNA analysis:
SRK transcripts were detected in poly(A)+ RNA isolated from stigmas as previously described (Kusaba et al. 2001), while SCR transcripts were detected in total RNA isolated from anthers using the TRIZOL reagent (Invitrogen, San Diego). RNA gel blot analysis was performed as previously described (Kusaba et al. 2001) by subjecting the RNA [∼1 μg of poly(A) RNA for SRK detection and ∼15 μg of total RNA for SCR detection] to denaturing electrophoresis on 1% (w/v) agarose, transfer to GeneScreen Plus membrane (DuPont-New England Nuclear, MA), and hybridization with 32P-labeled SRK or SCR probes as described above. Quantitation of SRK and SCR signal intensity was performed with a Molecular Dynamics PhosphorImager using the ImageQuant software package and normalization of hybridization signals was performed using an actin probe.
For reverse transcription–polymerase chain reaction (RT–PCR) analysis of SRKa transcripts, stigma RNA was treated with DNase I to eliminate contaminating genomic DNA, reverse transcribed, and amplified using the SuperScript one-step RT–PCR kit (Invitrogen) and SRKa-specific intron-flanking primers. The effectiveness of DNase digestion was verified by RT–PCR using actin intron-flanking primers, and only samples lacking contaminating DNA were used for RT–PCR of SRKa.
Sequence analysis and database searches:
DNA sequencing was performed at the Cornell University BioResource Center using an Applied Biosystems (Foster City, CA) automated sequencer. Sequences were manipulated and aligned using DNASTAR Lasergene software (DNASTAR, Madison, WI). BLAST searches were performed on the National Center for Biotechnology Information website (http://www.ncbi.nih.gov).
RESULTS
A.thaliana–lyrata interspecific hybrids:
Analysis of SI in A. thaliana–lyrata hybrid stigmas:
We used two hybrids produced from a cross between A. thaliana Col-0, which is homozygous for a defective S haplotype (Kusaba et al. 2001) designated S0, and an A. lyrata SaSb plant (Nasrallah et al. 2000). On the basis of gel-blot analysis of genomic DNA, the two hybrids were determined to have inherited the A. lyrata Sa haplotype (data not shown) and were therefore designated SaS0. These hybrids failed to produce pollen and were male sterile, consistent with the divergent chromosome number and genome organization of the two parental species (Kuittinen et al. 2004; Yogeeswaran et al. 2005). While male sterility precluded analysis of pollen from these SaS0 hybrids, hybrid stigmas were functional, allowing assays of cross-incompatibility responses by manual pollination with A. lyrata SaSa and SbSb pollen. Both pollinations resulted in equally prolific pollen tube growth (Table 1), demonstrating that the stigmas of A. thaliana–lyrata SaS0 hybrids failed to recognize and reject Sa pollen. Interestingly, backcrossing of these hybrids to A. lyrata restored the stigma SI response within one generation (Table 1).
TABLE 1.
Pollination analysis of Arabidopsis species hybrids and parental species
| ♂ A. thaliana–lyrata
|
||||
|---|---|---|---|---|
| ♂ A. lyrata SaSa | ♂ A. lyrata SbSb | F1b | Allotetraploid SaSaS0S0 | |
| ♀ A. lyrata SaSa | <10a | +++ | MS | <10 |
| ♀ A. lyrata SbSb | +++ | <10 | MS | +++ |
| ♀ A. thaliana–lyrata | ||||
| F1b | +++ | +++ | MS | ND |
| BCc | <10 | +++ | MS | ND |
| Allotetraploid SaSaS0S0 | +++ | +++ | MS | +++ |
The number of pollen tubes per pollinated stigma: <10, incompatible pollination; +++, compatible pollination; MS, male sterile; ND, not determined because previous generations were no longer available.
F1, interspecific hybrids from the A. thaliana × A. lyrata cross.
BC, plants from a backcross of A. thaliana–lyrata hybrids to A. lyrata.
Loss of SI was also exhibited by a neo-allotetraploid that arose spontaneously on one A. thaliana–lyrata SaS0 hybrid (Nasrallah et al. 2000). This allotetraploid, which was shown by cytological analysis to have 26 chromosomes (or double the chromosome number of the A. thaliana–lyrata F1 hybrids) (Nasrallah et al. 2000), produced functional pollen due to restoration of normal meiosis upon chromosome doubling. Selfing of this plant generated self-fertile allotetraploid progeny (Nasrallah et al. 2000). Reciprocal pollinations to A. lyrata SaSa plants demonstrated that the pollen of the allotetraploid and its progeny retained Sa specificity while its stigmas allowed confluent Sa pollen tube growth (Table 1). This pollination phenotype was stable and persisted over four allotetraploid generations analyzed. Thus, loss of SI in allotetraploids—and, by inference, in the original A. thaliana–lyrata hybrid—was stigma specific.
Molecular basis of SI breakdown in A. thaliana–lyrata hybrids:
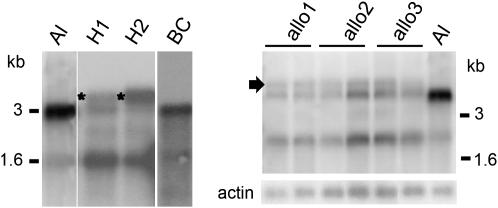
To investigate the molecular basis of breakdown in hybrid stigmas, we compared expression of the SRKa gene in each of the two A. thaliana–lyrata SaS0 hybrids and in A. lyrata by gel-blot analysis of stigma poly(A)+ RNA using an SRKa-specific probe derived from the SRKa first exon. Self-incompatible A. lyrata Sa stigmas exhibit an SRK transcript profile (Kusaba et al. 2001) consisting of a fully spliced 3-kb SRKa transcript that encodes the full-length SRKa receptor and a 10-fold less abundant 1.6-kb alternative transcript derived from the first exon of the gene, which results from the use of an alternative poly(A) addition site within the first intron of SRK and encodes a soluble form of the SRK ectodomain. The stigmas of A. thaliana–lyrata SaS0 hybrids and allotetraploids differed from those of A. lyrata Sa stigmas in two respects (Figure 1). They exhibited an additional 4-kb transcript species (Figure 1), at least some of which correspond to unspliced SRKa transcripts (expected size: 3.920 kb) on the basis of RT–PCR using intron-flanking primers (data not shown). They also exhibited dramatically reduced levels of the fully spliced 3-kb transcripts, which at best equaled those of the alternative 1.6-kb transcripts (Figure 1). Importantly, these 3-kb SRKa transcripts accumulated to levels that were <10% (6.4% ± 0.5) of those detected in A. lyrata Sa stigmas.
Figure 1.—
Association of aberrant SRK transcript processing with self-fertility in A. thaliana–lyrata hybrids and allotetraploid derivatives. Gel blots of stigma poly(A)+ RNA were hybridized with a probe derived from exon 1 of A. lyrata SRKa as previously described (Kusaba et al. 2001). Al, A. lyrata; H1 and H2, first-generation hybrids; BC, a first-generation plant from a backcross of H2 to A. lyrata; allo1, allo2, and allo3, three consecutive allotetraploid generations. The 3- and 1.6-kb transcripts typically produced by SRK genes are indicated. Note the 4-kb aberrant SRKa transcripts produced by stigmas of F1 hybrids (asterisks) and allotetraploid derivatives (arrow).
To examine further the association of aberrant SRK transcript profiles with loss of SI, we analyzed SRKa expression in the stigmas of self-incompatible plants produced by backcrossing each of the A. thaliana–lyrata SaS0 hybrids to the A. lyrata parent. Interestingly, the correct SRK transcript profile (Figure 1) was restored along with SI (Table 1) in first-generation backcross plants. These observations suggest that aberrant splicing of SRKa transcripts, and in particular the dramatic reduction in correctly spliced full-length transcripts, is the cause of the breakdown of SI in A. thaliana–lyrata SaS0 hybrids.
C. rubella–grandiflora interspecific hybrids:
A cross between a C. rubella plant and a C. grandiflora plant produced fertile F1 hybrids that set seed spontaneously, as previously described (Riley 1934; Acarkan et al. 2000; Koch and Kiefer 2005). The fertility of C. rubella–grandiflora hybrids is not surprising, given that these plants have the same basic chromosome number (n = 8) and are very similar at the molecular level (Hurka and Neuffer 1997). Despite these similarities, however, C. rubella and C. grandiflora are recognized as separate species (Hurka and Neuffer 1997). In this context, it should be noted that the concept of a strict biological species, which is defined by the ability of its members to produce viable and fertile progeny upon cross-hybridization, has not been recognized by plant biologists as a useful species concept (Grant 1981), and hybridization between plants belonging to well-recognized species is common (Arnold 1997).
Genetic analysis of SI in C. grandiflora, C. rubella, and interspecific hybrid populations:
Prior to analysis of C. rubella–grandiflora interspecific hybrids, it was necessary to perform a genetic analysis of the parental strains used in our interspecific cross. First, we generated a C. grandiflora population of plants that segregated for SI specificity by forced self-pollination in young floral buds prior to the developmental onset of SI in stigmas. These plants were analyzed by microscopic examination of pollen tube growth in self-pollinations and reciprocal cross-pollinations among all siblings and with the parental plant. On the basis of these pollinations, we confirmed early reports (Bateman 1955) that the SI system of Capsella, like that of other crucifers, is under single-locus sporophytic control, whereby the pollen SI phenotype is determined by the diploid genotype of its parent plant. We also determined that the C. grandiflora plant used in our interspecific cross carried two SI specificities, arbitrarily designated S7 and S8, with S7 and S8 exhibiting codominance in the stigma and S7 being dominant to S8 in pollen (Table 2). Finally, we analyzed several progeny derived from the spontaneous selfing of the C. rubella parent and confirmed that all of these plants were self-compatible, as expected (Table 2).
TABLE 2.
Pollination analysis of Capsella species hybrids and parental species
| ♂ F2
|
|||||
|---|---|---|---|---|---|
| ♂ C. rubella S0S0 | ♂ C. grandiflora S7S7b | S0S0 | S7S0 | S7S7 | |
| ♀ C. rubella S0S0 | +++a | +++ | +++ | +++ | +++ |
| ♀ C. grandiflora S7S7b | +++ | <10 | +++ | +++ | <10 |
| ♀ F2 | |||||
| S0S0 | +++ | +++ | +++ | +++ | +++ |
| S7S0 | +++ | <10 | +++ | +++ | <10 |
| S7S7 | +++ | <10 | +++ | +++ | <10 |
The number of pollen tubes per pollinated stigma: <10, incompatible pollination; +++, compatible pollination; MS, male sterile; ND, not determined because previous generations were no longer available.
C. grandiflora S7S7 homozygotes were used because the C. rubella–grandiflora F2 population segregated for the S7 allele and only pollination assays with S7S7 plants are relevant.
Genetic analysis was then carried out on the C. rubella × C. grandiflora cross. Starting with a C. rubella–grandiflora F1 hybrid, we produced an F2 population of 74 plants that segregated for SI and self-compatibility (SC). Analysis of the F1 and F2 plants by self-pollinations, autonomous seed set, and reciprocal pollinations with C. grandiflora S7S7 and S8S8 tester plants showed that this population inherited the S7 allele and that SC segregated as a simple dominant trait (53 SC:21 SI, approximating a 3:1 ratio; χ2 = 0.45; P = 0.5) linked to the S locus (Table 2). Reciprocal pollinations of the self-incompatible F2 plants with tester C. grandiflora S7S7 and S8S8 homozygotes showed that pollinations with S8S8 plants were compatible while those with S7S7 plants were incompatible, indicating that the plants expressed S7 specificity in stigma and pollen (Table 2). The stigmas of 36 (or 2/3) of the self-compatible F2 plants inhibited pollen from the S7S7 tester, but the pollen of these plants germinated and produced pollen tubes on S7S7 stigmas (Table 2). The remaining 17 (or 1/3) of the self-compatible F2 plants were reciprocally cross-compatible with both S8S8 and S7S7 plants (Table 2).
The data, confirmed by seed counts and self-pollinations in 106 additional F2 plants, are consistent with the following interpretation. The self-fertile C. rubella parent is homozygous for a nonfunctional S haplotype, designated S0, and crossing it to the C. grandiflora S7S8 plant produces an S7S0 F1 hybrid. In the F2 generation, S7S7 plants are self-incompatible, S0S0 plants are self-compatible, and S7S0 plants express S7 specificity in the stigma (their stigmas inhibit pollen from S7S7 plants) but are self-compatible due to the breakdown of SI in pollen (their pollen germinates and produces tubes on S7S7 stigmas).
Molecular cloning of S-locus genes from C. grandiflora:
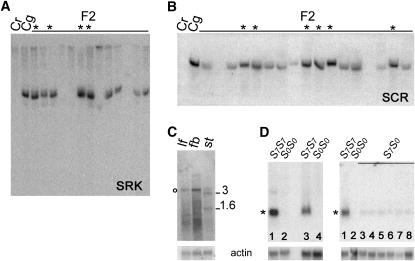
The availability of SRK and SCR genes is essential for a molecular analysis of SI and its breakdown. To isolate SRK and SCR genes from the C. grandiflora S7 haplotype, a C. grandiflora genomic library constructed from an S7S8 plant was screened with a probe derived from the first exons of the A. lyrata SRKa and SRKb genes (Kusaba et al. 2001). Sequence analysis of a subset of positive clones revealed them to contain an SRK-like gene in close proximity to an SCR-like gene. DNA gel-blot analysis with probes from these genes confirmed that they were derived from the S7 haplotype. A probe corresponding to exon 1 of the C. grandiflora SRK-like gene hybridized most strongly with plants previously classified by pollination analysis as carrying the S7 haplotype, and it detected a restriction fragment length polymorphism that cosegregated with the S7 haplotype in C. grandiflora F2 plants segregating for the S8 and S7 haplotypes (data not shown). In the C. rubella × C. grandiflora F2 population, the SRK and SCR probes hybridized only to plants carrying the S7 haplotype (Figure 2B) but not to plants predicted to be homozygous for the C. rubella-derived S0 haplotype. This lack of hybridization, even with the SRK probe, suggests that the nonfunctional S0 haplotype carries either deleted or highly diverged SI genes.
Figure 2.—
Molecular analysis of self-incompatibility and self-fertility in Capsella species hybrids. (A and B) DNA gel blots of C. rubella (Cr), C. grandiflora (Cg), and two populations of C. rubella–grandiflora F2 plants probed with the CgSRK7 exon 1 probe (A) and CgSCR7 (B). Only F2 plants indicated with asterisks were self-incompatible and genotypically S7S7 on the basis of pollination assays. All other F2 plants were self-compatible and could be grouped into two classes on the basis of pollination analysis: self-fertile F2 plants that hybridized with the probes were S7S0 while those that did not hybridize were S0S0, as with C. rubella. The small differences in electrophoretic mobility of the hybridizing fragments observed between some of the lanes are due to irregular migration of DNA fragments, which is often associated with the use of relatively impure DNA obtained by mini-preparation methods. (C) Stigma-specific expression of the 3.0- and 1.6-kb CgSRK7 transcripts detected by gel-blot analysis of poly(A)+ RNA from leaf tissue (lf), floral buds with pistils removed (fb), and stigmas (st) of C. grandiflora S7S7. The cross-hybridizing band (circle) common to the three samples represents transcripts from an SRK-related gene. (D) Gel-blot analysis of CgSCR7 in total RNA from C. grandiflora (S7S7) and C. rubella (S0S0) whole floral buds (left, lanes 1 and 2) and anthers (left, lanes 3 and 4) and in the anthers of a representative sample of C. rubella–grandiflora F2 plants (right). Note the lack of hybridization signal in S0S0 plants and the drastic reduction in the steady-state levels of CgSCR7 in the anthers of S7S0 F2 plants (right, lanes 3–8). Hybridization with actin was used as a loading control.
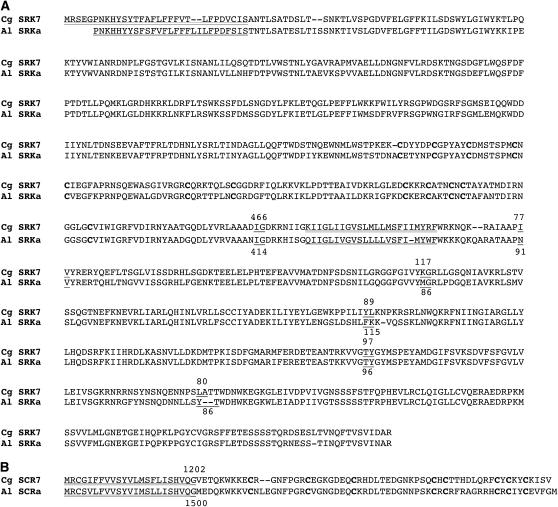
Our molecular cloning of S-locus genes from C. grandiflora confirmed the expectation that SI in this genus has the same molecular basis as other crucifers. The C. grandiflora SRK7 and SCR7 genes (hereafter designated CgSRK7 and CgSCR7), which represent the first SRK–SCR gene pair identified in the Capsella species, contain all sequence motifs characteristic of SRK and SCR genes (Figure 3). The CgSRK7 sequence represents a new allele distinct from the partial C. grandiflora SRK sequences that have been reported recently (Paetsch et al. 2006). BLAST searches show CgSRK7 and CgSCR7 to be most similar to the SRKa and SCRa genes of A. lyrata (Kusaba et al. 2001). The exons of CgSRK7, on average, share ∼84% nucleotide and ∼86% amino-acid sequence identity with the corresponding exons of AlSRKa, while CgSCR7 exons 1 and 2 are ∼85% and ∼74% identical to AlSCRa exons 1 and 2 in nucleotide and amino-acid sequence, respectively. Interestingly, the C. grandiflora S7 haplotype is also similar to the A. lyrata Sa haplotype in the position and spacing of the SRK and SCR genes relative to each other (Kusaba et al. 2001). In both S haplotypes, the two genes are arranged tail to tail and are separated by a relatively small intergenic region (1006 bp in C. grandiflora S7 and 1500 bp in A. lyrata Sa). However, neither the region between CgSRK7 and CgSCR7 nor the introns of the two genes, which are similar in size and placement (Figure 3), share appreciable sequence identity according to pairwise BLAST alignments (data not shown).
Figure 3.—
Alignment of the predicted SRK (A) and SCR (B) amino-acid sequences from the C. grandiflora S7 haplotype (CgSRK7 and CgSCR7; accession nos. 892031 and 894476) and the A. lyrata Sa haplotype (Kusaba et al. 2001) [AlSRKa (accession no. AB052755) and AlSCRa (accession no. AB052753)]. Cysteine residues that are conserved in SRKs and SCRs are shown in boldface type. Signal sequences and transmembrane domains are double underlined. The location of introns in the genomic sequences is shown by underlined residues that flank each intron with numbers indicating intron size in base pairs.
Molecular basis of SI breakdown in C. rubella–grandiflora hybrids:
The molecular cloning of CgSRK7 and CgSCR7 allowed us to compare their expression in self-incompatible and self-compatible plants. In self-incompatible plants, these genes exhibited hybridization patterns characteristic of other SRK and SCR genes. CgSRK7 transcripts were detected in the pistils, but not in the leaves or anthers, of plants carrying the S7 haplotype (Figure 2C), irrespective of whether the plants were self-incompatible (i.e., S7S7) or self-compatible (i.e., S7S0). CgSCR7 transcripts were detected in anthers or young floral buds of C. grandiflora S7S7 plants and self-incompatible S7S7 C. rubella–grandiflora F2 plants (Figure 2D). In contrast, all 4 S0S0 F2 plants analyzed failed to hybridize with the CgSCR7 probe and all 10 S7S0 F2 plants analyzed produced only a weak hybridization signal that was 10- to 12-fold lower in self-compatible S7S0 plants relative to their self-incompatible S7S7 sibs (Figure 2D).
DISCUSSION
We investigated two cases of breakdown of SI upon interspecific hybridization in Arabidopsis and Capsella. In both cases, loss of SI was due to reversible changes in expression of the S-locus recognition genes, SRK or SCR. In A. thaliana–lyrata SaS0 hybrids and their allotetraploid derivatives, stigmas exhibited aberrant SRKa transcript profiles. In particular, the levels of the fully spliced 3.0-kb SRKa transcripts were reduced to ∼6% of the levels detected in A. lyrata Sa stigmas, while the levels of the 1.6-kb transcripts were slightly enhanced. The role of the soluble forms of the extracellular domain of SRK (designated eSRK) that are encoded by the 1.6-kb transcripts is not understood. However, there is no evidence that they have a negative effect on SRK function; rather, they might contribute to the stabilization of the full-length SRK receptor, as previously suggested for the Brassica S-locus glycoprotein SLG, which shares a high degree of sequence similarity with the extracellular domain of SRK (Dixit et al. 2000). Thus, any increased accumulation of eSRKa that might result from the slight increase in 1.6-kb transcript levels observed in A. thaliana–lyrata SaS0 hybrid stigmas is not expected to cause loss of SI through dominant-negative effects. In contrast, the ∼94% reduction in the levels of fully spliced 3.0-kb SRKa transcripts, which is much greater than the 75% reduction that was previously shown to cause loss of SI in Brassica (Conner et al. 1997), is expected to result in absence or suboptimal accumulation of SRKa protein and, consequently, in the breakdown of SI in the stigmas of A. thaliana–lyrata hybrids and their neo-allotetraploid derivatives. Strong support for this conclusion is provided by the observation that reestablishment of SI in the first-generation backcross of A. thaliana–lyrata SaS0 hybrids to A. lyrata was accompanied by restoration of normal SRKa transcript profiles.
Species-specific differences in pre-mRNA splicing have been reported (Laverdiere et al. 2000; Pan et al. 2005), but their consequences for gene expression in interspecific hybrids had not been previously explored. The production of some 3-kb SRKa transcripts in A. thaliana–lyrata hybrids indicates that the correct SRKa initiation, termination, and splice sites are utilized, albeit inefficiently, by the transcription and processing machinery of hybrid stigmas. Similarly, the production of 1.6-kb SRKa transcripts indicates that this machinery recognizes and utilizes the alternative poly(A) addition site within the SRKa first intron that generates this transcript species. At present, the cause of aberrant SRKa transcript processing is unclear. It cannot be due to global defects in transcript synthesis and processing in all tissues of A. thaliana–lyrata hybrids, because these hybrids do not exhibit major developmental or physiological abnormalities. Nor can it be ascribed to dilution of the A. lyrata-derived machinery by A. thaliana factors that cannot process A. lyrata SRK transcripts, because the SRKb transgene is processed correctly in A. thaliana stigmas (Nasrallah et al. 2002). Although analysis of additional A. lyrata SRK alleles and their processing in A. thaliana–lyrata hybrids is required, it is possible that this phenomenon reflects incompatibilities between the A. thaliana and A. lyrata RNA processing machineries resulting from independent changes that accumulated in the two species since their divergence from a common ancestor ∼5 million years ago (Koch et al. 2000).
While breakdown of SI occurred in the stigmas of A. thaliana–lyrata hybrids, it occurred in pollen of C. rubella–grandiflora hybrids in correlation with a 10- to 15-fold reduction in CgSCR7 gene expression in S7S0 anthers. This suppression of CgSCR7 is similar to, albeit not as complete as, the silencing of SCR alleles from pollen-recessive S haplotypes observed in intraspecific heterozygotes (Kusaba et al. 2002; Shiba et al. 2002, 2006; Fujimoto et al. 2006). Notably, a functional SCR allele has been reported in Brassica, which is silenced in the presence of a nonfunctional SCR allele (Fujimoto et al. 2006), similar to the situation described here. Why particular combinations of SCR alleles cause silencing while others do not is not understood. In any case, it is evident that pollen recessiveness based on SCR suppression, which allows an S haplotype to evade SRK surveillance in intraspecific pollinations, can also cause loss of SI upon interspecific hybridization.
Interestingly, the sequences of CgSRK7 and CgSCR7 are most similar to the SRK and SCR genes of the A. lyrata Sa haplotype, which also exhibits pollen recessiveness resulting from silencing of its SCRa gene, at least in SaSb heterozygotes (Kusaba et al. 2001). In previous studies, comparison of S haplotypes from Brassica and Raphanus identified several cases of trans-genus polymorphisms, whereby pairs of S haplotypes were found to determine the same recognition specificity (Sato et al. 2004). In one of these intergeneric pairs, the SRKs and SCRs exhibited ∼88% and ∼70% amino-acid sequence identity, respectively (Sato et al. 2004). CgSRK7 and CgSCR7 exhibit similarly high 86 and 74% amino-acid sequence identity to the A. lyrata SRKa and SCRa genes, respectively. On the basis of this extensive sequence identity and on the basis of a similar genomic organization of SRK and SCR genes, it is possible that the C. grandiflora S7 and A. lyrata Sa haplotypes may be descended from one ancestral haplotype that existed before the divergence of Capsella and Arabidopsis species ∼10 million years ago (Koch et al. 2000).
The 3:1 ratio of self-compatible to self-incompatible progeny observed in the C. rubella–grandiflora F2 population is noteworthy for two reasons. First, the dominance of SC over SI inferred from this ratio is unusual for a loss-of-function trait and it also violates the general, albeit not absolute, rule of dominance of SI over SC inferred from intraspecific crosses (Nasrallah 1974; Nasrallah et al. 1992, 2004). Second, the 3:1 ratio, although clearly consistent with the segregation of self-fertility as a single S-locus-linked trait, is not a simple case of dominance of one allele over another. Rather, this ratio has a more complex basis, with two different phenomena contributing to the majority self-fertile class: homozygosity for the nonfunctional C. rubella S0 locus, on the one hand, and suppression of the SCR7 gene in S7S0 heterozygotes, on the other hand. In any case, the segregation of self-fertility as a single S-locus-linked trait in the C. rubella–grandiflora F2 population indicates that the C. rubella strain used in our study has not suffered major mutations at other loci required for SI. The strain is therefore similar to the A. thaliana C24 accession, which becomes self-incompatible when transformed with functional SRK and SCR genes from A. lyrata (Nasrallah et al. 2004). Whether other strains of C. rubella, like other accessions of A. thaliana (Nasrallah et al. 2002, 2004), have accumulated additional mutations at SI modifier loci remains to be determined.
In summary, our analysis has revealed two mechanisms for loss of SI upon interspecific hybridization in crucifers. While the well-documented changes in genome structure and gene expression (Song et al.1995; Soltis and Soltis 1999; Wendel 2000; Ranz et al. 2004; Lai et al. 2006; Wang et al. 2006) that occur upon merging of divergent genomes have been reasonably proposed to underlie phenotypic variability in interspecific hybrids and their allopolyploid derivatives, our results provide a concrete case in which de novo changes in expression of specific genes are correlated directly with a specific and adaptive change in phenotype. Although analysis of additional interspecific hybrids generated by crossing plants carrying different S haplotypes is required, it is unlikely that the aberrant SRK RNA processing in hybrid stigmas and downregulation of SCR in hybrid anthers that we observed are restricted to the AlSRKa and CgSCR7 alleles analyzed in this study. Indeed, C. rubella–grandiflora S8S0 heterozygotes also exhibit breakdown of SI in pollen, suggesting that the CgSCR8 allele might also be silenced in heterozygous anthers. However, confirming this hypothesis will require the isolation of the CgSCR8 allele, which has not been accomplished as yet. Because of the extensive polymorphisms of SRK and SCR, the behavior of individual S haplotypes in interspecific hybrids will have to be determined empirically on a case-by-case basis.
The two mechanisms underlying breakdown of SI described here do not result from changes in DNA sequence, and they are reversible and therefore epigenetic in nature. These mechanisms can allow establishment of stable self-fertile hybrid genotypes, promote their reproductive isolation from parental species, and thus facilitate hybrid speciation in nature. Unlike mutation or deletion of S-locus recognition genes, which cause irreversible evolutionary switches from SI to self-fertility, these epigenetic processes can cause reversible loss of SI. In addition to allowing self-fertile interspecific hybrids to act as bridges for transfer of functional S haplotypes between species, this reversibility would provide nascent interspecific hybrids with flexibility in their selection of mating system, possibly contributing to reproductive success in the face of uncertain ecological conditions.
Acknowledgments
This material is based upon work supported by the National Science Foundation under grant no. 0414521 to M.E.N. and J.B.N.
References
- Acarkan, A., M. Rossberg, M. Koch and R. Schmidt, 2000. Comparative genome analysis reveals extensive conservation of genome organization for Arabidopsis thaliana and Capsella rubella. Plant J. 23: 55–62. [DOI] [PubMed] [Google Scholar]
- Arnold, M. L., 1997. Natural Hybridization and Evolution. Oxford University Press, London/New York/Oxford.
- Bateman, A.J., 1955. Self-incompatibility systems in angiosperms. III. Cruciferae. Heredity 9: 53–68. [Google Scholar]
- Bushell, C., M. Spielman and R. J. Scott, 2003. The basis of natural and artificial postzygotic hybridization barriers in Arabidopsis species. Plant Cell 15: 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner, J. A., T. Tantikanjana, J. C. Stein, M. K. Kandasamy, J. B. Nasrallah et al., 1997. Transgene-induced silencing of S-locus genes and related genes in Brassica. Plant J. 11: 809–823. [Google Scholar]
- Dixit, R., M. E. Nasrallah and J. B. Nasrallah, 2000. Post-transcriptional maturation of the S receptor kinase of Brassica correlates with co-expression of the S-locus glycoprotein in the stigmas of two Brassica strains and in transgenic tobacco plants. Plant Physiol. 124: 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto, R., T. Sugimura, E. Fukai and T. Nishio, 2006. Suppression of gene expression of a recessive SP11/SCR allele by an untranscribed SP11/SCR allele in Brassica self-incompatibility. Plant Mol. Biol. 61: 577–587. [DOI] [PubMed] [Google Scholar]
- Grant, V., 1981. Plant Speciation. Columbia University Press, New York.
- Gross, B. L., and L. H. Rieseberg, 2005. The ecological genetics of homoploid hybrid speciation. J. Hered. 96: 241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurka, H., and B. Neuffer, 1997. Evolutionary processes in the genus Capsella (Brassicaceae). Plant Syst. Evol. 206: 295–316. [Google Scholar]
- Kho, Y. O., and J. Baer, 1968. Observing pollen tubes by means of fluorescence. Euphytica 17: 298–302. [Google Scholar]
- Koch, M., and M. Kiefer, 2005. Genome evolution among cruciferous plants—a lecture from the comparison of the genetic maps of three diploid species: Capsella rubella, Arabidopsis lyrata ssp. petraea and Arabidopsis thaliana. Am. J. Bot. 92: 761–767. [DOI] [PubMed] [Google Scholar]
- Koch, M., B. Haubold and T. Mitchell-Olds, 2000. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17: 1483–1498. [DOI] [PubMed] [Google Scholar]
- Kuittinen, H., A. A. de Haan, C. Vogl, S. Oikarinen, J. Leppala et al., 2004. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics 168: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M., K. D. Dwyer, J. Hendershot, J. Vrebalov, J. B. Nasrallah et al., 2001. Self-incompatibility in the genus Arabidopsis: characterization of the S locus in the outcrossing A. lyrata and its autogamous relative A. thaliana. Plant Cell 13: 627–643. [PMC free article] [PubMed] [Google Scholar]
- Kusaba, M., C.-W. Tung, M. E. Nasrallah and J. B. Nasrallah, 2002. Monoallelic expression and dominance interactions in anthers of self-incompatible Arabidopsis lyrata. Plant Physiol. 128: 17–20. [PMC free article] [PubMed] [Google Scholar]
- Lai, Z., B. L. Gross, Y. Zou, J. Andrews and L. H. Rieseberg, 2006. Microarray analysis reveals differential gene expression in hybrid sunflower species. Mol. Ecol. 15: 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverdiere, M., J. Beaudoin and A. Lavigneur, 2000. Species-specific regulation of alternative splicing in the C-terminal region of the p53 tumor suppressor gene. Nucleic Acids Res. 28: 1489–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenhoff, K., and H. Hurka, 1995. Allopolyploid origin of Arabidopsis suecica (Fries) Norrlin: evidence from chloroplast and nuclear genome markers. Bot. Acta 108: 449–456. [Google Scholar]
- Murray, M. G., and W. F. Thompson, 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, J. B., 2005. Recognition and rejection of self in plant self-incompatibility: comparisons to animal histocompatibility. Trends Immunol. 26: 412–418. [DOI] [PubMed] [Google Scholar]
- Nasrallah, M. E., 1974. Genetic control of quantitative variation in self-incompatibility proteins detected by immunodiffusion. Genetics 76: 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, M. E., M. K. Kandasamy and J. B. Nasrallah, 1992. A genetically defined trans-acting locus regulates S-locus function in Brassica. Plant J. 2: 497–506. [Google Scholar]
- Nasrallah, M. E., K. Yogeeswaran, S. Snyder and J. B. Nasrallah, 2000. Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol. 124: 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, M. E., P. Liu and J. B. Nasrallah, 2002. Generation of self-incompatible Arabidopsis thaliana by transfer of two S-locus genes from A. lyrata. Science 297: 247–249. [DOI] [PubMed] [Google Scholar]
- Nasrallah, M. E., P. Liu, S. Sherman-Brolyes, N. A. Boggs and J. B. Nasrallah, 2004. Natural variation in expression of self-incompatibility in Arabidopsis thaliana: implications for the evolution of selfing. Proc. Natl. Acad. Sci. USA 101: 16070–16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane, S. L., Jr., B. A. Schaal and I. Al-Shehbaz, 1996. The origins of Arabidopsis suecica (Brassicaceae), as indicated by nuclear rDNA. Syst. Bot. 21: 559–566. [Google Scholar]
- Paetsch, M., S. Mayland-Quellhorst and B. Neuffer, 2006. Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97: 283–290. [DOI] [PubMed] [Google Scholar]
- Pan, Q., M. A. Bakowski, Q. Morris, W. Zhang, B. J. Frey et al., 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 21: 73–77. [DOI] [PubMed] [Google Scholar]
- Ranz, J. M., K. Namgyal, G. Gibson and D. L. Hartl, 2004. Anomalies in the expression profile of interspecific hybrids of Drosophila melanogaster and Drosophila simulans. Genome Res. 14: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg, L. H., 1997. Hybrid origins of plant species. Annu. Rev. Ecol. Syst. 28: 359–389. [Google Scholar]
- Rieseberg, L. H., 2001. Polyploid evolution: keeping the peace at genomic reunions. Curr. Biol. 11: R925–R928. [DOI] [PubMed] [Google Scholar]
- Riley, H. P., 1934. A further test showing the dominance of self-fertility to self-sterility in shepherd's purse. Am. Nat. 68: 60–64. [Google Scholar]
- Sato, Y., S. Okamoto and T. Nishio, 2004. Diversification and alteration of recognition specificity of the pollen ligand SP11/SCR in self-incompatibility of Brassica and Raphanus. Plant Cell 16: 3230–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba, H., M. Iwano, T. Entani, K. Ishimoto, H. Shimosato et al., 2002. The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14: 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba, H., T. Kakizaki, M. Iwano, Y. Tarutani, M. Watanabe et al., 2006. Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat. Genet. 38: 297–299. [DOI] [PubMed] [Google Scholar]
- Soltis, D. E., and P. S. Soltis, 1999. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Song, K., P. Lu, K. Tang and T. C. Osborn, 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploidy evolution. Proc. Natl. Acad. Sci. USA 92: 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins, G.L., 1959. The role of hybridization in evolution. Proc. Am. Philos. Soc. 103: 231–251. [Google Scholar]
- Takayama, S., and A. Isogai, 2005. Self-incompatibility in plants. Annu. Rev. Plant Biol. 56: 467–489. [DOI] [PubMed] [Google Scholar]
- Wang, J., L. Tian, H. S. Lee, N. E. Wei, H. Jiang et al., 2006. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J. F., 2000. Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Yogeeswaran, K., A. Frary, T. L. York, A. Amenta, A. H. Lesser et al., 2005. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 15: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]