Abstract
The Wnt/β-catenin signaling pathway regulates cell fate and behavior during embryogenesis, adult tissue homeostasis, and regeneration. When inappropriately activated, the pathway has been linked to colorectal cancer and melanoma, and when attenuated it may contribute to Alzheimer's disease and osteoporosis. Small molecules that modulate Wnt signaling will likely provide new insights into the regulation of this key developmental pathway and ultimately provide pharmacological agents to control Wnt signaling in vivo. To this end, we screened a library of 100,000 small molecules for activity in a cell-based assay of Wnt/β-catenin signaling and discovered a purine derivative, QS11, that synergizes with Wnt-3a ligand in the activation of Wnt/β-catenin signal transduction. Through affinity chromatography and subsequent functional assays, we showed that QS11 binds and inhibits the GTPase activating protein of ADP-ribosylation factor 1 (ARFGAP1), suggesting that QS11 modulates Wnt/β-catenin signaling through an effect on protein trafficking. Consistent with its function as an ARFGAP inhibitor, QS11 inhibits migration of ARFGAP overexpressing breast cancer cells.
Keywords: ARFGAP inhibitor, small-molecule screen, Wnt signaling, regenerative medicine
The canonical Wnt/β-catenin signaling pathway is evolutionarily conserved and plays key roles in development and disease (1–3). During development, Wnt signaling regulates cell morphology, motility, proliferation, and differentiation. Inappropriate regulation of the Wnt signaling pathway is implicated in tumorigenesis, osteoporosis, and neurodegenerative diseases. Since the discovery of the first Wnt family member, Wnt-1 (4), extensive studies have been carried out on the Wnt/β-catenin signaling pathway. Nonetheless, there remains a considerable need to further understand and better control the regulation and physiological effects of this pathway.
Cell-based screens of libraries of natural products and synthetic small molecules have provided useful tools for the study of complex cellular processes (5–8). Indeed, a number of small molecules have been identified that modulate Wnt/β-catenin signaling, including the agonists 6-bromoindirubin-3′-oxime (BIO) (9), LiCl (10), deoxycholic acid (11), and a pyrimidine derivative (12) and the antagonists quercetin (13), ICG-001 (14), and others (15, 16). To identify additional molecules that regulate the canonical Wnt/β-catenin signaling pathway, we carried out a cell-based screen for molecules that synergistically activate signaling in the presence of Wnt-3a. Because such small molecules activate Wnt signaling only in the presence of Wnt proteins, we anticipated that they would function through mechanisms that cross-talk with Wnt/β-catenin signaling and thus provide new insights into this pathway. One such molecule, QS11, has been identified, which modulates Wnt signaling through ARFGAPs and their roles in protein translocation.
Results and Discussion
Identification of a Wnt Synergist from a High-Throughput Chemical Screen.
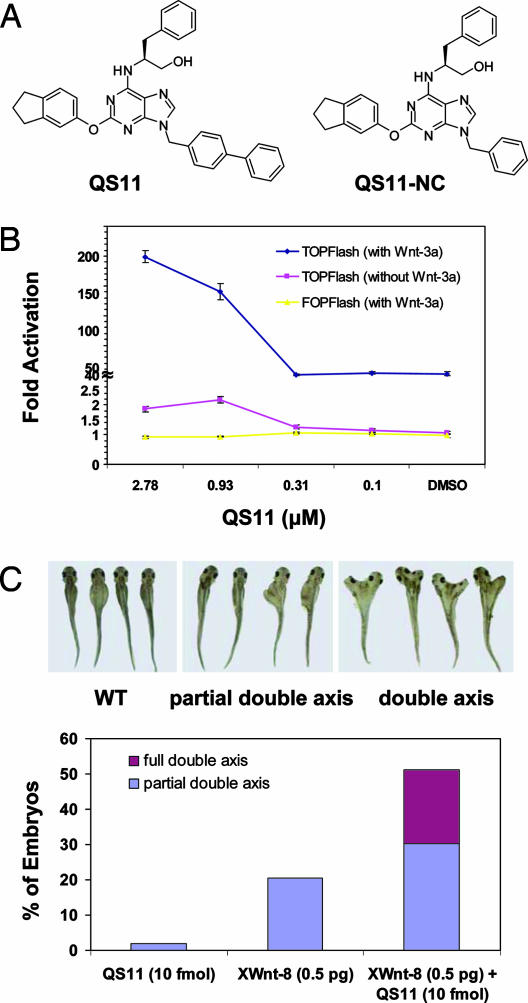
To carry out the screen [supporting information (SI) Methods], HEK293 cells stably harboring the Wnt/β-catenin reporter Super(8X)TOPFlash (17) were treated with a library of 100,000 heterocycles (final concentration of ≈7 μM) (18), stimulated with Wnt-3a conditioned medium (Wnt-3a CM), and assayed for luciferase activity after 36 h. Compounds that induced >3 SD from the mean luciferase activity for each plate were tested for their effect on the Super(8X)TOPFlash reporter in the absence of Wnt-3a stimulation, and those that did not appreciably activate the reporter in the absence of Wnt-3a CM were characterized further. From the compounds screened, a class of 2,6,9-trisubstituted purine derivatives was found to be the most potent Wnt synergists. A structure activity relationship study of a focused chemical library (SI Fig. 5 and SI Table 1) revealed that the biphenyl substitution at N9 is essential for activity, the aryloxy groups at C2 position generally are more active than their amino analogues, and the C6 position tolerates several different substituents. Purine derivative QS11 (Fig. 1A) showed potent activity (EC50 = 0.5 μM) with little cytotoxicity toward HEK293 and human primary fibroblast cells (≥10 μM, which is QS11's maximum solubility in culture media). A structurally similar but inactive analog, QS11-NC (Fig. 1A and SI Fig. 6A), was used as a negative control in subsequent experiments.
Fig. 1.
QS11 is a small-molecule Wnt synergist in cell culture and Xenopus development. (A) Chemical structures of the purine derivative, QS11, and its negative control, QS11-NC. (B) Dose-dependent effects of QS11 on HEK293 cells transfected with Super(8X)TOPFlash reporter (with Wnt-3a CM, blue line; without Wnt-3a CM, red line) or SuperFOPFlash reporter (with Wnt-3a CM, yellow line). The cells were treated with QS11 at the indicated concentrations with or without Wnt-3a stimulation 24 h after transfection. Luciferase activity was measured 36 h after compound treatment. All data are normalized against renilla luciferase. Error bars are SD. (C) Synergistic effect of QS11 with XWnt-8 RNA on axis duplication in Xenopus. Xenopus embryos were treated with QS11 (10 μM, 10 nL), XWnt-8 RNA (0.5 pg), and QS11 (10 μM, 10 nL) plus XWnt-8 RNA (0.5 pg) for 24 h. (Upper) Representative pictures of Xenopus with different degrees of axis duplication. (Lower) Quantification of embryonic axis duplication on treatment with QS11 (10 μM, 10 nL) and/or XWnt-8 RNA (0.5 pg).
QS11 (2.5 μM) activated the Super(8X)TOPFlash reporter ≈200-fold in the presence of Wnt-3a CM, whereas Wnt-3a treatment alone increased reporter activity ≈40-fold; QS11 increased reporter activity only 2-fold in the absence of Wnt-3a (Fig. 1B). This synergistic effect was also observed when QS11 was combined with recombinant Wnt-3a protein (10–200 ng/ml) (data not shown), suggesting that the effective component in Wnt-3a CM is Wnt-3a protein. Moreover, when HEK293 cells were transfected with the negative control SuperFOPFlash (17) reporter, which has mutated β-catenin/T cell factor-binding elements, and treated with QS11 (0.011–25 μM) and Wnt-3a CM, no luciferase activity was observed (Fig. 1B). In addition, QS11 synergizes with the glycogen synthase kinase 3-β (GSK3β) inhibitor, BIO, in activating the Super(8X)TOPFlash reporter in HEK293 cells (SI Fig. 6B). Furthermore, quantitative RT-PCR indicates that QS11 synergizes with Wnt-3a to induce Axin2 and DKK1 expression (SI Fig. 7). To test whether QS11 also functions as a synergist of other signaling pathways, HEK293 cells were transfected with Gli-luc or NFκB-luc reporters and treated with the hedgehog protein or TNF-α, respectively. Synergistic activation by QS11 was not observed in these cells (data not shown). Taken together, these experiments indicate that QS11 specifically synergizes with Wnt-3a to activate the Wnt/β-catenin signaling pathway.
To further test this notion, we examined the effect of QS11 on Wnt/β-catenin signaling in vivo. Ectopic activation of the Wnt/β-catenin signaling pathway leads to duplication of the Xenopus embryonic axis (19). This developmental process has been established as an excellent experimental means to test modulation of Wnt signaling in vivo (20). Embryos were injected with QS11 (10 μM, 10 nL), XWnt-8 RNA (0.5 pg) [which possesses full axis-inducing activity in a manner similar to that of XWnt-3a (21)], or a combination of QS11 (10 μM, 10 nL) and XWnt-8 RNA (0.5 pg). Only 1.8% of embryos injected with QS11 alone and 20.5% of embryos treated with XWnt-8 RNA displayed partial axis formation (none formed a full double axis). In contrast, injection of a combination of XWnt-8 RNA and QS11 induced significantly more full (20.9%) and partial (30.2%) double axis formation (Fig. 1C). These experiments further support the notion that QS11 acts as a synergistic Wnt agonist.
Target Identification.
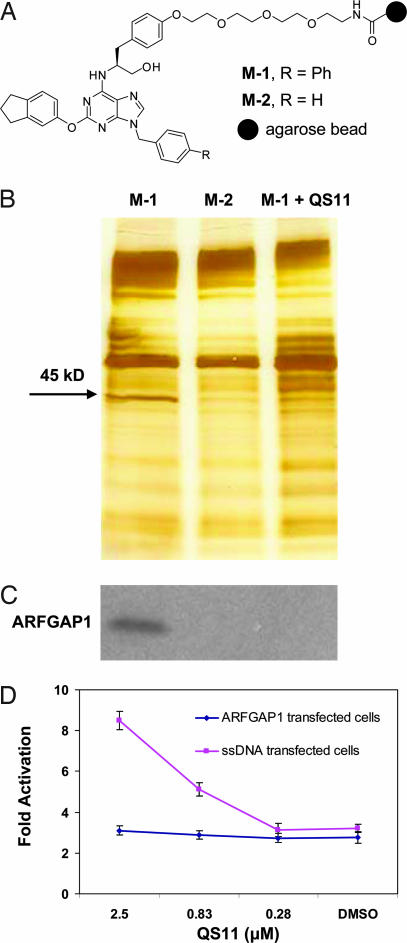
To identify the cellular target of QS11, a tetraethylene glycol linker was attached to the para- position of the phenyl group at C6 (which has little effect on activity) of QS11 and then coupled to Affi-Gel 10 resin under mildly basic conditions to afford the affinity matrix M-1 (Fig. 2A and SI Fig. 5B). The inactive analog QS11-NC was similarly coupled to resin and used as a negative control (affinity matrix M-2). Affinity matrices were incubated with HEK293 cell lysates and then washed, and bound proteins were eluted from resin by boiling in SDS-containing buffer. In addition, competition with soluble ligand was carried out by adding QS11 (50 μM) to the HEK293 cell lysates during binding to the M-1 matrix. The proteins retained by the affinity matrices were separated by SDS/PAGE and visualized by silver staining (Fig. 2B). A protein with an apparent molecular mass of 45 kDa was found to bind to M-1 and only weakly to M-2. Moreover, this 45-kDa protein could be competed off by the addition of 50 μM of QS11. Mass spectrometric analysis (SI Methods) identified this protein as a GTPase activating protein of ADP-ribosylation factor 1 (ARFGAP1) (SI Fig. 8). This result was independently confirmed by Western blotting with anti-ARFGAP1 antibody (Fig. 2C). To demonstrate that QS11 directly binds to ARFGAP1, purified ARFGAP1 protein was immobilized to a solid support by an amine coupling (SI Methods); surface plasmon resonance analysis afforded a dissociation constant (Kd) for QS11 of 620 nM (kon = 7.1 × 104 M–1s–1; koff = 4.4 × 10–3 s–1). In addition, overexpression of ARFGAP1 in HEK293 cells abolished synergistic activation of the Super(8X)TOPFlash reporter by QS11 with Wnt-3a CM (Fig. 2D), further supporting ARFGAP1 as a cellular target of QS11.
Fig. 2.
Affinity chromatography identified ARFGAP1 as the cellular target of QS11. (A) Chemical structures of affinity resins with QS11 (positive resin, M-1) or QS11-NC (negative resin, M-2) immobilized for target identification. (B) Pull-down experiments using the immobilized reagents M-1 [lane M-1, without soluble QS11; lane M-1 + QS11 (50 μM), with soluble QS11 at 50 μM] and M-2 (lane M-2). HEK293 cell lysates were incubated with the affinity matrices at 4°C for 1 h. Bound proteins were eluted, resolved on a 4–20% Tris-glycine gel, and visualized with silver staining. The band that contains ARFGAP1 is indicated by the arrow. (C) Western blot of ARFGAP1 resin-bound protein. Proteins that were resolved on a 4–20% Tris-glycine gel as in B were transferred to a nitrocellulose membrane and analyzed with an antibody against ARFGAP1. (D) Overexpression of ARFGAP1 cDNA blocks the synergistic effect of QS11 with Wnt-3a CM. HEK293 cells were transfected with Super(8X)TOPFlash reporter, pTK-RL plasmid, and ARFGAP1 cDNA (red line) or ssDNA (blue line) by using Fugene6. Cells were treated with QS11 at the indicated concentrations and Wnt-3a CM (1:1 vol/vol ratio to the growth medium) 24 h after transfection. Luciferase activities were measured 36 h after treatment with QS11 and Wnt-3a CM. The activation fold was normalized against renilla luciferase. Error bars are SD.
Mechanism of Action.
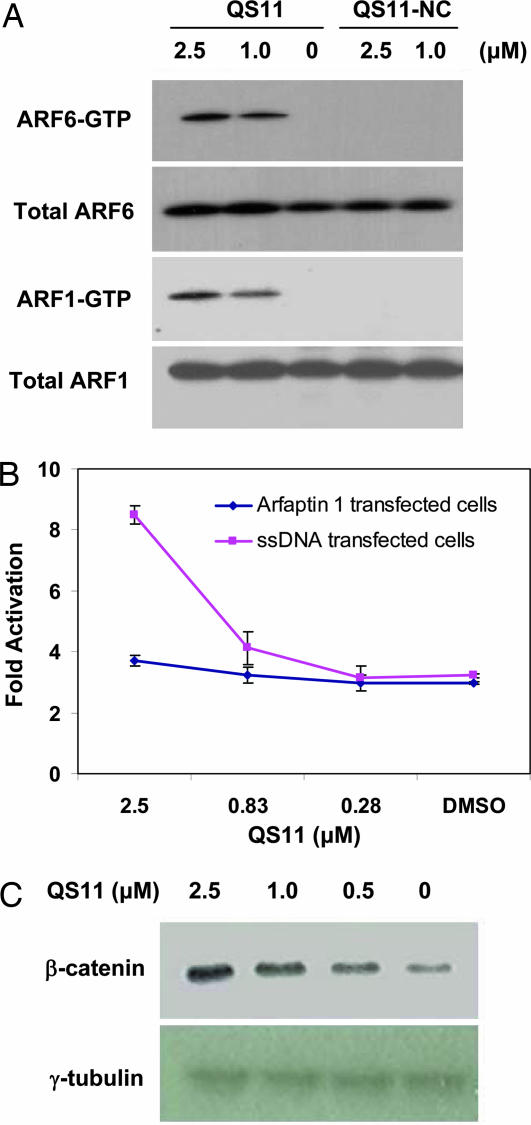
ARFGAPs form a family of GTPase activating proteins that regulate the small GTPase ADP-ribosylation factors (ARFs) (22). There are six different ARFs in mammals that mediate a number of biological processes, including vesicle trafficking and cytoskeleton reorganization through their ability to cycle between an inactive GDP-bound form and an active GTP-bound form (23). ARFGAPs promote ARF inactivation by stimulating GTP hydrolysis, whereas guanine nucleotide exchange factors of ARF (ARFGEFs) catalyze the formation of active GTP-bound ARFs. To test whether QS11 leads to increased ARF activity, the levels of endogenous ARF1-GTP and ARF6-GTP in cells treated with QS11 and QS11-NC were measured by a biochemical ARF-GTP pull-down assay (24), in which ARF-GTP is recovered from cell lysates based on its affinity for the ARF binding domain of the effector protein golgi-associated γ-adaptin ear containing ARF binding protein 3 (GGA3). Endogenous levels of ARF1-GTP and ARF6-GTP were significantly increased in NIH 3T3 cells treated with QS11 (1.0 or 2.5 μM) relative to those treated with inactive QS11-NC (1.0 or 2.5 μM) or DMSO control, consistent with inhibition of ARFGAP1 by QS11 (Fig. 3A).
Fig. 3.
QS11 synergizes with Wnt-3a in activating Wnt signaling through ARF activation. (A) QS11 increased cellular ARF-GTP levels. NIH 3T3 cells were treated with QS11, DMSO, or QS11-NC at the indicated concentrations for 36 h. The cell lysates were analyzed with an antibody against ARF1 or ARF6 before (total ARF1 or ARF6) and after (ARF1-GTP or ARF6-GTP) incubating with a GST-fusion protein GGA3VHS-GAT. (B) HEK293 cells were transfected with the Super(8X)TOPFlash reporter, pTK-RL plasmid, and cDNA of arfaptin 1 (red line) or ssDNA (blue line) using Fugene6. The cells were treated with QS11 at the indicated concentrations and Wnt-3a CM (1:1 vol/vol ratio to the growth medium) 24 h after transfection. Luciferase activities were measured 36 h after QS11 treatment. The activation fold was normalized against renilla luciferase. Error bars are SD. (C) The effect of QS11 on nuclear β-catenin. HEK293 cells were treated with QS11 at the indicated concentrations and Wnt-3a CM (1:1 vol/vol ratio to the growth medium) for 24 h. Nuclear β-catenin was analyzed with an antibody against β-catenin. γ-tubulin was used as the loading control.
To further show that the synergistic effect of QS11 results from ARF activation, Arfaptin 1 (25, 26), which interacts with and inhibits ARFs specifically in their active GTP-bound form, was overexpressed in HEK293 cells. The synergistic effect of QS11 with Wnt-3a CM was abolished in these cells (Fig. 3B) in agreement with the notion that QS11 functions by modulating ARF-GTP levels. Treatment of HEK293 cells with 0.25 μM of brefeldin A, an ARFGEF inhibitor (27), reduces the activity of the Super(8X)TOPFlash reporter 50% in Wnt-3a CM, compared with that observed with the DMSO control. This antagonistic effect is rescued by adding 1 μM of QS11 (SI Fig. 9A), further supporting its ability to inhibit ARFGAP activity. In addition, overexpression of the constitutively active mutant ARF1(Q71L) (24) in HEK293 cells activated the Super(8X)TOPFlash reporter ≈2-fold, whereas overexpression of the dominant-negative mutant ARF1(T34N) (24) reduced the activity of the Super(8X)TOPFlash reporter 44%, compared with the control in the presence of Wnt-3a CM (Fig. 9B).
It has been reported that the activation of ARFs promotes the dissociation of membrane-bound β-catenin (28). Thus, one possible mechanism by which QS11 acts as a Wnt synergist is to inhibit ARFGAP1, thereby leading to an increase in activated ARF and subsequent β-catenin translocation. The released β-catenin is degraded in the absence of Wnt signaling, but accumulates and translocates to the nucleus when cells are stimulated with Wnt-3a CM. To test this model, we overexpressed the cDNAs for the β-catenin interacting cadherins 1, 5, or 16 in HEK293 cells and observed that the synergy of QS11 with Wnt-3a is blocked (SI Fig. 10). Furthermore, QS11 does not synergize with overexpressed β-catenin in HEK293 cells (data not shown), indicating that QS11 works upstream of β-catenin in the signaling pathway. Finally, increased levels of nuclear β-catenin were observed following QS11 (0.5–2.5 μM) treatment in the presence of Wnt-3a CM (Fig. 3C). Although these results support a β-catenin translocation model, ARFs may also affect Wnt signaling through other mechanisms (e.g., endocytosis of frizzled and/or low-density lipoprotein receptor-related proteins).
Inhibition of Migration of MDA-MB-231 Cells.
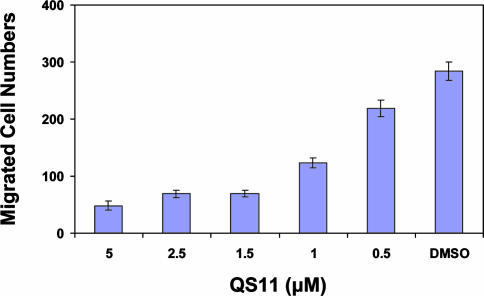
If QS11 functions by inhibiting ARFGAP, it may modulate other biological processes mediated by ARFGAPs. For example, AMAP1 is an ARFGAP that plays an essential role in the invasive activities of MDA-MB-231 breast cancer cells (29). Specifically, siRNA-mediated silencing of AMAP1 in MDA-MB-231 cells inhibits cell migration (29). The ARFGAP domain in AMAP1 shares 62% homology with that in ARFGAP1, raising the possibility that QS11 may also inhibit AMAP1. Indeed, when tested in a transwell cell migration assay, QS11 inhibited the migration of MDA-MB-231 cells in a dose-dependent manner, with 80% of the migration inhibited at a concentration of 2.5 μM, compared with the DMSO control (Fig. 4). Purified His-tagged [325–724]AMAP1 protein that contains the ARFGAP domain was then immobilized to a solid support by an amine coupling; surface plasmon resonance analysis provided a dissociation constant (Kd) of 364 nM. Finally, we carried out an initial survey of the selectivity of QS11 by assaying the effects of overexpression of ARFGAP cDNAs on the activity of QS11 in the Super(8X)TOPFlash reporter assay. Overexpression of 13 ARFGAPs encoded by the human and mouse genomes blocks the synergistic effect of QS11 with Wnt-3a by ≥50% (SI Fig. 11), indicating that QS11 might be a relatively broad specificity ARFGAP inhibitor.
Fig. 4.
QS11 inhibits migration of AMAP1 overexpressing MDA-MB-231 cells. Dose-dependent inhibition of the migration of AMAP1 overexpressing MDA-MB-231 cells by QS11 treatment in a transwell assay. Migration was measured with a modified Boyden chamber containing Transwell filters (Coastar) coated on the underside with 5 μg/cm2 Matrigel. Migrated cells were counted under a microscope.
Conclusion
In summary, a small-molecule Wnt synergist, QS11, was identified from an unbiased, cell-based screen. We identified ARFGAP1 as the molecular target of QS11, and further demonstrated that QS11 inhibits ARFGAP1 function and, as a consequence, modulates ARF activity and β-catenin localization in the cells. Because this molecule is the only ARFGAP inhibitor reported to date, it may not only be useful in studies of the Wnt/β-catenin signaling pathway, but it may also provide a useful tool to explore novel functions of ARFGAPs in cell culture and whole organisms. Along these lines, we note that QS11 effectively reduces in vitro migration of metastatic human breast cancer cells. Experiments are ongoing to determine the molecular basis for inhibition of ARFGAP1 by QS11 and explore its activity in zebrafish development.
Materials and Methods
Cell Culture and Plasmids.
HEK293, NIH 3T3, MDA-MB-231, and L-Wnt3a cells were grown according to instructions from ATCC (www.atcc.org). Wnt-3a CM was prepared according to the protocol from ATCC. Human ARFGAP1 and ARFAPTIN1 cDNAs were obtained from Origene (Rockville, MD). ARF1(Q71L), ARF1(T34N), and His-[325–724]AMAP1 were generous gifts from P. Randazzo (National Cancer Institute, Bethesda, MD), V. M. Hsu (Harvard University, Cambridge, MA), and J. Donaldson (National Heart, Lung, and Blood Institute, Bethesda, MD).
Western Blot.
The following primary antibodies were used for Western blot: β-catenin (1:1,000; BD Biosciences Transduction Laboratories, Lexington, KY), γ-tubulin (1:1,000; Sigma–Aldrich, St. Louis, MO), ARFGAP1 (1:500; Abgent, San Diego, CA), ARF1 (1:500; Chemicon International, Temecula, CA), and ARF6 (1:200; Santa Cruz Technology, Santa Cruz, CA). HRP-conjugated anti-mouse or anti-rabbit IgG was used as a secondary antibody (1:2,000; Amersham Biosciences, Piscataway, NJ), and the ECL plus Western Blotting Detection Kit (Amersham Biosciences) was used for detection.
ARFGAP1 cDNA Complementation.
A mixture of human ARFGAP1 cDNA (180 ng), Super(8X)TOPFlash (60 ng), and pTK-RL (6 ng) was incubated with Fugene6 (0.79 μl; Roche Diagnostics, Indianapolis, IN) in serum-free DMEM (50 μl; Invitrogen) at 25°C for 45 min in one well of a 96-well plate. HEK293 cells (20,000 cells per well) were then added and incubated with 5% CO2 at 37°C for 24 h. The transfection efficiency was 50–60% as estimated by GFP transfection in a control well. QS11 (2.5 μM) and Wnt-3a CM (1:1 vol/vol ratio to the growth medium) were then added. Luciferase activities were measured 36 h later by first removing the media and then adding Dual-Glo assay solutions (Promega, Madison, WI) according to the manufacturer's instructions. The fold activation was normalized against renilla luciferase. This same protocol was also applied to a cDNA panel (SI Fig. 11). Three independent data points were averaged for each treatment.
Nuclear Protein Extraction.
HEK293 cells were grown in a six-well plate for 24 h and then treated with QS11 or DMSO for 24 h. The cells were washed with cold PBS, transferred to an Eppendorf tube (1.5 ml; Boulder, CO), and centrifuged (3,000 × g at 4°C for 5 min). The cell pellet was resuspended in a buffer containing 10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, protease inhibitors (Sigma–Aldrich) and phosphatase inhibitors (Sigma–Aldrich), and the resulting suspension was incubated at 4°C for 30 min. The cells were then lysed by pipetting up and down six to eight times. After centrifugation (16,000 × g for 5 min at 4°C), the supernatant was removed, and the cell pellet was washed twice with the above buffer; resuspended in a buffer containing 20 mM Hepes (pH 7.9), 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 0.5 mM DTT, protease, and phosphatase inhibitors; and incubated with shaking at 4°C for 30 min. The mixture was then centrifuged (16,000 × g at 4°C for 5 min), and the supernatant was isolated as a nuclear fraction of proteins.
Affinity Chromatography.
HEK293 cells stably transfected with the Super(8X)TOPFlash reporter were lysed with homogenization buffer [60 mM β-glycerophosphate, 15 mM p-nitrophenyl phosphate, 25 mM Mops (pH 7.2), 15 mM EGTA, 15 mM MgCl2, 1 mM DTT, protease inhibitors (Sigma–Aldrich), and 0.5% Nonidet P-40]. Cell lysates were centrifuged at 16,000 × g for 20 min at 4°C, and the supernatant was collected. The total protein concentration in the supernatant was determined by using a BCA protein assay kit (Pierce Chemical, Rockford, IL). The lysate (1 mg) was then added to the packed affinity matrix (30 μl), and bead buffer [50 mM Tris·HCl (pH 7.4), 5 mM NaF, 250 mM NaCl, 5 mM EDTA, 5 mM EGTA, protease inhibitors (Sigma–Aldrich), and 0.1% Nonidet P-40] was added up to a final volume of 1 ml (for the competition experiment, QS11 was added to a final concentration of 50 μM). After rotating at 4°C for 1 h, the mixture was centrifuged at 16,000 × g for 1 min at 4°C, and the supernatant was removed. The affinity matrix was then washed (six times) with cold bead buffer and eluted by boiling with Laemmli sample buffer at 95°C for 3 min. Samples were loaded and separated on a 4–20% Tris-glycine gel (Invitrogen). The gel was then sequentially treated with solutions of 50% MeOH/10% HAc/40% H2O (40 min), 20% EtOH/80% H2O (10 min), H2O (10 min), and aqueous Na2S2O3 (200 mg/L, 1 min). After washing twice with H2O, the gel was soaked in aqueous AgNO3 (2 g/L) for 30 min and then washed twice with H2O. To visualize the proteins, the gel was developed in a solution of Na2CO3 (30 g/L), Na2S2O3 (10 mg/L), and formaldehyde (52 mg/L) in H2O. Acetic acid (1%) was then added to stop the staining process.
Assay for ARF-GTP.
ARF-GTP levels were measured by using the GGA binding assay as previously described (30). Briefly, NIH 3T3 cells were treated with QS11 or QS11-NC at the indicated concentrations for 36 h. Cells were lysed in ARF assay lysis buffer [50 mM Tris·HCl (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 10% glycerol, and protease inhibitors (Roche Diagnostics)]. GTP-bound ARF was assayed by its binding to a GST fusion protein, which contains the VHS domain to the GAT region of an ARF effector GGA3 as described previously (30). Total and GTP-bound ARFs were analyzed by Western blot with anti-ARF1 (1:500; Chemicon International) or anti-ARF6 (1:200; Santa Cruz Biotechnology) antibodies.
Transwell Cell Migration Assay.
Migration was measured by using modified Boyden chambers containing Transwell filters (Coastar, Cambridge, MA) coated on the underside with 5 μg/cm2 matrigel. MDA-MB-231 cells were plated at the density of 5,000 cells per well in 0.5 ml of RPMI1640 (Invitrogen) supplemented with 0.5% BSA and QS11 at the indicated concentrations to the upper chamber of the Transwell filter in a 24-well plate. The lower chamber of the filter contained 0.5 ml of medium containing DMEM (Invitrogen) supplemented with 10% FBS. After 12 h, cells remaining on the upper surface of the filter were removed, whereas cells on the lower side were quantified by counting under a microscope.
Xenopus Injection.
In total, 0.5 pg of XWnt-8 RNA and 10 nl of water or 10 nl of QS11 (10 μM) was injected into a ventral blastomere at the 16- to 32-cell stage. Axis duplication was scored at 2 days.
Supplementary Material
Acknowledgments
We thank Drs. V. W. Hsu (Harvard University, Cambridge, MA), P. Randazzo (National Cancer Institute, Bethesda, MD), and J. Donaldson (National Heart, Lung, and Blood Institute, Bethesda, MD) for expression plasmids and proteins; and Drs. M. Mukherji, W. Xiong, Y. Zhao, and C. Cho for helpful discussions. This work was supported by the Novartis Research Foundation and the Skaggs Institute for Chemical Biology, and in part by National Institute of General Medical Sciences Grant PHS NRSA T32 GM 07270 (to N.D.C.). This is manuscript no. 18513 of The Scripps Research Institute. R.T.M. is an investigator, and M.B.M. is an associate of the Howard Hughes Medical Institute.
Abbreviation
- CM
conditioned medium.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702136104/DC1.
References
- 1.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. Nat Rev Genet. 2004;5:689–699. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 2.Bejsovec A. Cell. 2005;120:11–14. doi: 10.1016/j.cell.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 4.Nusse R, Varmus HE. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 5.Dove A. Nat Biotechnol. 2003;21:859–864. doi: 10.1038/nbt0803-859. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, et al. PLoS Biol. 2005;3:764–766. doi: 10.1371/journal.pbio.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- 8.Kwok TCY, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, McCourt P, Cutler SR, Roy PJ. Nature. 2006;44:91–95. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- 9.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 10.Klein RS, Melton DA. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pai R, Tarnawski AS, Tran T. Mol Biol Cell. 2004;15:2156–2163. doi: 10.1091/mbc.E03-12-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Wu X, Mitchel B, Kintner C, Ding S, Schultz PG. Angew Chem Int Ed. 2005;44:1987–1990. doi: 10.1002/anie.200462552. [DOI] [PubMed] [Google Scholar]
- 13.Park CH, Chang JY, Hahm ER, Park S, Kim HK, Yang CH. Biochem Biophys Res Commun. 2005;328:227–234. doi: 10.1016/j.bbrc.2004.12.151. [DOI] [PubMed] [Google Scholar]
- 14.Emami KH, Nguyen C, Ma H, Kim DH, Jeong KW, Eguchi M, Moon RT, Teo JL, Oh SW, Kim HY, et al. Proc Natl Acad Sci USA. 2004;101:12682–12687. doi: 10.1073/pnas.0404875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lepourcelet M, Chen YNP, France DS, Wang H, Crews P, Petersen F, Bruseo C, Wood AW, Shivdasani RA. Cancer Cell. 2004;5:91–102. doi: 10.1016/s1535-6108(03)00334-9. [DOI] [PubMed] [Google Scholar]
- 16.Shan J, Shi D, Wang J, Zheng J. Biochemistry. 2005;44:15495–15503. doi: 10.1021/bi0512602. [DOI] [PubMed] [Google Scholar]
- 17.DasGupta R, Kaykas A, Moon RT, Perrimon N. Science. 2005;308:826–833. doi: 10.1126/science.1109374. [DOI] [PubMed] [Google Scholar]
- 18.Ding S, Gray NS, Wu X, Ding Q, Schultz PG. J Am Chem Soc. 2002;124:1594–1596. doi: 10.1021/ja0170302. [DOI] [PubMed] [Google Scholar]
- 19.McMahon A, Moon RT. Cell. 1989;58:1075–1084. doi: 10.1016/0092-8674(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 20.Moon RT, Kimelman D. BioEssays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 21.Christian JL, McMahon JA, McMahon AP, Moon RT. Development (Cambridge, UK) 1991;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- 22.Makler V, Cukierman E, Rotman M, Admon A, Cassel D. J Biol Chem. 1995;270:5232–5237. doi: 10.1074/jbc.270.10.5232. [DOI] [PubMed] [Google Scholar]
- 23.Bernards A, Settleman J. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanoh H, Williger BT, Exton JE. J Biol Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- 26.Tsai SC, Adamik R, Hong JX, Moss J, Vaughan M, Kanoh H, Exton JH. J Biol Chem. 1998;273:20697–20701. doi: 10.1074/jbc.273.33.20697. [DOI] [PubMed] [Google Scholar]
- 27.Klausner RD, Donaldson JG, Lippincott-Schwartz J. J Cell Biol. 1992;116:1071–1080. doi: 10.1083/jcb.116.5.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palacios F, Schweitzer JK, Boshans RL, D'Souza-Schorey C. Nat Cell Biol. 2002;4:929–936. doi: 10.1038/ncb881. [DOI] [PubMed] [Google Scholar]
- 29.Onodera Y, Hashimoto S, Hashimoto A, Morishige M, Mazaki Y, Yamada A, Ogawa E, Adachi M, Sakurai T, Manabe T, et al. EMBO J. 2005;24:963–973. doi: 10.1038/sj.emboj.7600588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santy LC, Casanova JE. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.