Abstract
Phytochromes confer the photoperiodic control of flowering in rice (Oryza sativa), a short-day plant. To better understand the molecular mechanisms of day-length recognition, we examined the interaction between phytochrome signals and circadian clocks in photoperiodic-flowering mutants of rice. Monitoring behaviors of circadian clocks revealed that phase setting of circadian clocks is not affected either under short-day (SD) or under long-day (LD) conditions in a phytochrome-deficient mutant that shows an early-flowering phenotype with no photoperiodic response. Non-24-hr-light/dark-cycle experiments revealed that a rice counterpart gene of Arabidopsis CONSTANS (CO), named PHOTOPERIOD SENSITIVITY 1 (Heading date 1) [SE1 (Hd1)], functions as an output of circadian clocks. In addition, the phytochrome deficiency does not affect the diurnal mRNA expression of SE1 upon floral transition. Downstream floral switch genes were further identified with rice orthologs of Arabidopsis FLOWERING LOCUS T (FT). Our RT-PCR data indicate that phytochrome signals repress mRNA expression of FT orthologs, whereas SE1 can function to promote and suppress mRNA expression of the FT orthologs under SD and LD, respectively. This SE1 transcriptional activity may be posttranscriptionally regulated and may depend on the coincidence with Pfr phytochromes. We propose a model to explain how a short-day plant recognizes the day length in photoperiodic flowering.
Keywords: Photoperiodism, flowering time, phytochrome, circadian clock, the external coincidence model, rice
Plant reproductive organs develop following the recognition of various environmental cues, in order for plants to survive and leave offspring efficiently. Light is one of the major environmental stimuli in the control of flowering. In addition to the intensity and quality of light, the duration per day, or photoperiod, is a major determinant of the timing of flowering. The molecular mechanisms of this photoperiodic response are largely unknown (Thomas and Vince-Prue 1997; Thomas 1998; Simpson et al. 1999; Samach and Coupland 2000).
In the long-day plant Arabidopsis, photoreceptor mutations that affect flowering time have been extensively analyzed (Levy and Dean 1998; Whitelam et al. 1998; Thomas 1999; Casal 2000; Lin 2000). The Arabidopsis blue-light receptor gene CRYPTOCHROME 2 (CRY2) promotes flowering under long-day (LD) conditions, indicating an important role of CRY2 in the photoperiodic control of flowering (Guo et al. 1998; Mockler et al. 1999). Under far-red-rich light conditions, PHYTOCROME A (PHYA) also promotes flowering of Arabidopsis in day-length extension experiments (Johnson et al. 1994). In contrast, analyses of phytochrome B (phyB) mutants have revealed that PHYB inhibits flowering under both LD and short-day (SD) conditions (Goto et al. 1991; Mazzella et al. 2001). Paradoxically, overexpression of PHYB also results in early flowering in Arabidopsis (Krall and Reed 2000). Redundancy of Arabidopsis phytochrome genes further makes the contribution of PHYB unclear: for example, PHYTOCHROME D and PHYTOCHROME E genes are known to have some overlapping functions with PHYB in floral induction (Devlin et al. 1998, 1999; Quail 1998). The CRY2 function in floral promotion requires red-light signaling mediated by PHYB under continuous light conditions, indicating that CRY2 antagonizes the PHYB inhibitory action of flowering (Guo et al. 1998; Mockler et al. 1999). Although the phytochrome signal transduction for flowering still remains to be revealed, nuclear localization (Sakamoto and Nagatani 1996; Kircher et al. 1999) and interaction with specific DNA-binding proteins (Martinez-Garcia et al. 2000) of the active Pfr form of phytochromes strongly suggest that phytochromes can function as transcription factors. Rapid changes of transcriptional profile mediated by PHYA may support this hypothesis in Arabidopsis (Tepperman et al. 2001).
Circadian clocks are also involved in the photoperiodic control of flowering. The Arabidopsis circadian clock component genes LATE ELONGATED HYPOCOTYL (LHY), CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), and TIMING OF CAB EXPRESSION 1 (TOC1) regulate their gene expression reciprocally to form a negative feedback loop as part of a central oscillator (Schaffer et al. 1998; Wang and Tobin 1998; Strayer et al. 2000; Alabadi et al. 2001). Ectopic expression of LHY and CCA1 results in late-flowering phenotypes with loss of various circadian rhythms (Schaffer et al. 1998; Wang and Tobin 1998).
The role of photoreceptors in the entrainment of circadian clocks has been well characterized in Arabidopsis (Devlin and Kay 2001). In the phyA mutant, periods of circadian rhythms in cab2::luc lengthen at low fluence rates of both red and blue lights (Somers et al. 1998). In phyB, cab2::luc periods lengthen at high fluence rates of red light (Somers et al. 1998). On the other hand, cab2::luc periods lengthen in cry1 under blue-light conditions, whereas cab2::luc periods are not much different in cry2 mutants from those in the wild-type plants (Somers et al. 1998). In contrast, the periods of cry1 and cry2 were longer in white-light conditions than those in the wild-type plants (Devlin and Kay 2000), indicating that red-light signaling modifies CRY function for the entrainment of circadian clocks. Regardless of the important roles of these photoreceptors in the entrainment of circadian clocks, these effects on circadian clocks are not enough to explain the flowering-time phenotypes in these photoreceptor mutants.
The CONSTANS (CO) gene encodes a zinc-finger-type transcription factor and is known to play a key role in the photoperiodic control of flowering in Arabidopsis (Putterill et al. 1995). Direct target genes of the CO transcription factor have been identified as FLOWERING LOCUS T (FT) and SUPRESSOR OF OVEREXPRESSION OF CO 1, both of which have roles in floral promotion in Arabidopsis (Onouchi et al. 2000; Samach et al. 2000). Recently, it has been shown that Arabidopsis CO mRNA expression is controlled by circadian clocks and is mainly detected at night (Suárez-López et al. 2001). In addition, CO mRNA expression levels are largely unaffected by photoperiod, although the daily duration of CO mRNA expression is longer under LD than SD conditions.
In addition to the molecular genetic studies in the past decade, many physiological studies on flowering have been performed since the first report of photoperiodism in 1920 (Garner and Allard 1920). The external coincidence model—a refined version of Bünning's hypothesis (Bünning 1960)—is one of the best-supported physiological models for explaining day-length measurement (Pittendrigh and Minis 1964; Thomas and Vince-Prue 1997; Carre 2001). In this model, light plays two distinct roles: one for the entrainment of circadian clocks to set a photoinducible phase in the day cycle, and the other as an external light signal to transduce the presence of sunlight to downstream pathways. The coincidence between the photoinducible phase and the external light signal produces promoting or inhibitory signals for floral differentiation in long-day and short-day plants, respectively. This model may bring to mind that CO is involved in the setting of the photoinducible phase, although it is not known whether the external coincidence model fits with molecular mechanisms of Arabidopsis floral transition.
By analysis of a rice phytochrome-deficient mutant, termed photoperiod sensitivity 5 (se5), we previously showed that light-stable phytochromes confer photoperiodic control of flowering in rice (Oryza sativa), a short-day plant (Izawa et al. 2000). In addition, the Heading date 1 (Hd1) gene, a major flowering-time QTL (quantitative trait locus), was recently identified with a rice ortholog of the Arabidopsis CO gene and was shown to be allelic to a rice mutant termed photoperiod sensitivity 1 (se1; hereafter referred to as SE1 because the mutant line in this work was originally identified as an se1 mutant), although, unlike CO, SE1 may repress flowering under LD conditions and promote it under SD conditions (Yano et al. 2000). Therefore, in this study, we examined the roles of phytochrome signaling and circadian clocks in the photoperiodic flowering of rice. Our results show that phytochrome signals function as external signals to repress mRNA expression of rice floral-inducer genes homologous to Arabidopsis FT, whereas SE1 functions as an output of circadian clocks entrained by given photoperiods to control FT orthologs. These results led us to propose a model to explain the molecular mechanisms of day-length recognition in the photoperiodic flowering of rice.
Results
The se5 mutation does not affect the behavior of circadian clocks in LD and SD photoperiods
The null mutation in the SE5 gene, which is involved in phytochrome chromophore biosynthesis, results in complete loss of the photoperiodic response in rice (Izawa et al. 2000). Phytochromes could affect flowering by altering the phase of entrainment of the circadian clock (Somers et al. 1998), thus changing the coincidence between light and a critical circadian phase. Therefore, we examined the behavior of the circadian clock in the se5 mutant. First, rhythmic mRNA expression of circadian-clock-controlled genes (ccgs) of rice was analyzed by RNase protection assays. Two rice ccgs, OsLHY, which is homologous to the Arabidopsis LHY and CCA1 genes (Schaffer et al. 1998; Wang and Tobin 1998), and CAB1R (Luan and Bogorad 1992), were rhythmically expressed in se5 mutants as well as in the wild type in both constant light (LL) and constant darkness (DD; Fig. 1A). LHY and CCA1 genes are members of the Arabidopsis circadian clock's central components (Alabadi et al. 2001). Another ccg, CATA, of a rice catalase gene more robustly showed rhythmic mRNA expression in se5 mutants under LL than in the wild type (data not shown). These results indicate that a circadian clock functions in the se5 mutants.
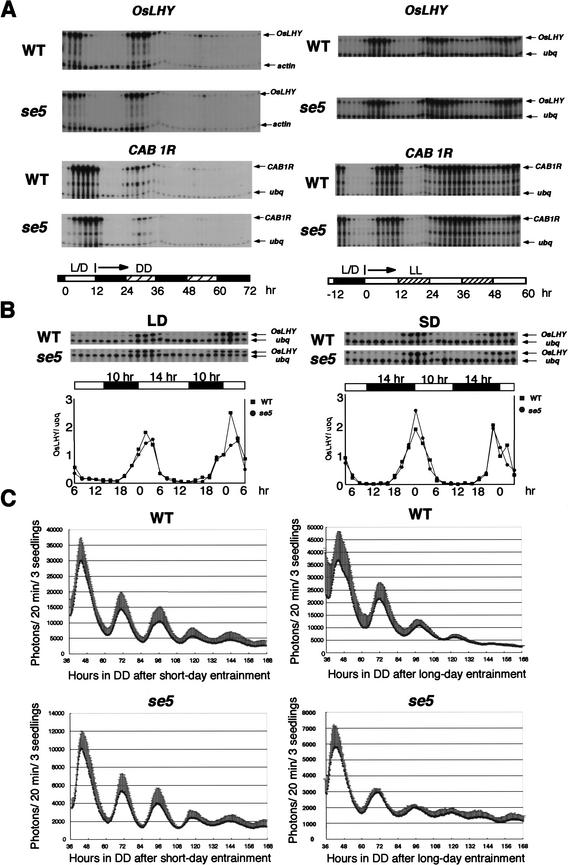
Figure 1.
Circadian clock behavior in the se5 mutant. (DD) 24 h darkness; (LL) 24 h light; (WT) wild type. (A) Rhythmic mRNA expression of two rice ccgs, OsLHY and CAB1R, under constant light conditions in RNase protection assays. RNA samples were harvested every 2 h from leaves of 1-month-old rice plants. Leaves from three to five plants were mixed. (Left, DD) After 5 d of entrainment (12L12D). (Right, LL) After the same entrainment. A rice actin or ubiquitin fragment was used as a control probe. (B) Diurnal mRNA expression of OsLHY under LD and SD conditions in RNase protection assays. RNA samples were harvested every 2 h. (Top) RNase protection assay. The ubiquitin is a control probe. (Bottom) Normalized data. (C) Rhythmic cab1r::luc expression in DD after SD and LD entrainments. Rhythmic cab1r::luc expression in 7-day-old rice seedlings was measured after 5 d of entrainment (10L14D for SD entrainment; 14L10D for LD entrainment). (Left, DD) After SD entrainment. (Right, LL) After LD entrainment. Averages of six or seven measurements were plotted with standard deviations. Representative of two or three independent experiments.
To determine the effect of the se5 mutation on the entrained phase of circadian clocks in different photoperiods, we next examined diurnal mRNA expression of OsLHY under LD and SD conditions (Fig. 1B). Diurnal mRNA expression of CAB1R and CATA was also examined (data not shown). The results clearly indicate that the phase of these ccgs was not affected in se5 mutants under these photoperiods. To evaluate the effects on circadian clocks more precisely, we further used a cab1r::luc bioluminescence reporter system of rice (Sugiyama et al. 2001) and examined rhythmic luciferase expression in DD and LL conditions after LD and SD entrainments (Fig. 1C; data not shown). The cab1r::luc expression patterns revealed no significant effect of the se5 mutation on the entrainment of circadian clocks under different photoperiods. We therefore conclude that the entrainment of circadian clocks is not affected by the se5 mutation under LD or SD conditions, and that the defect of photoperiodic flowering in se5 is not caused by defects in the phase of circadian clocks.
Flowering responses in rice under non-24-h photoperiods
The absence of effects on the entrainment of circadian rhythms in se5 mutants led us to examine the effect of circadian clocks on the flowering response in rice under non-24-h photoperiods. The phase of circadian rhythms in non-24-h photoperiods was monitored in wild-type rice seedlings by using cab1r::luc reporter gene expression. Under SD (10 h light/14 h darkness, 10L14D) and LD (14 h light/10 h darkness, 14L10D) conditions, typical diurnal luc expression with a 24-h period was observed (data not shown). In a 36-h photoperiod comprising 24 h light/12 h darkness (24L12D), a clear rhythmic expression with a 36-h period was observed (Fig. 2A). In this photoperiod, an atypical waveform with two intervals of rising cab1r::luc expression in a cycle, once before light-off and once before light-on, was observed. In a 48-h photoperiod comprising 36 h light/12 h darkness (36L12D), a 48-h-period rhythmic luc expression was observed with no anticipation of light-on (Fig. 2B). Thus, there are apparent two peaks in the 48-h period, one after light-on and the other for subjective day. In constant light (24 h light, 24L), weak rhythmic cab1r::luc expressions with a 24-h period were observed (Fig. 2C). Under constant light, we used a temperature cycle comprising 12 h at 30°C/12 h at 25°C to entrain the circadian clock with a 24-h period. Therefore, the effects of these atypical photoperiods on the phase of circadian clocks may be explained by the following. In 24L12D, lights go out just after the cab1r::luc trough; thus, plants have a light night, then are reset and have a dark night on each cycle. In 36L12D, lights go out roughly at the cab1r::luc peak, only a few hours before the expected phase in the 24-h light/dark cycles such as SD and LD. In 24L, light does not function as a zeitgeber (or environmental cue to set the clock), but the 24-h temperature cycles may do so. If phases with low expression of cab1r::luc were considered as a subjective night for rice plants, tested plants under these 24L12D, 36L12D, and 24L conditions might be exposed to the external light longer than those under SD and LD, even in the subjective night.
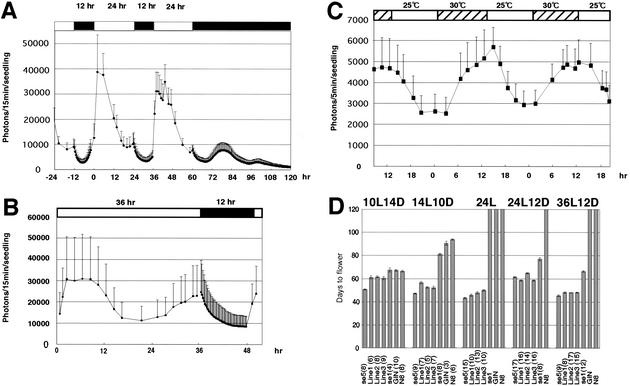
Figure 2.
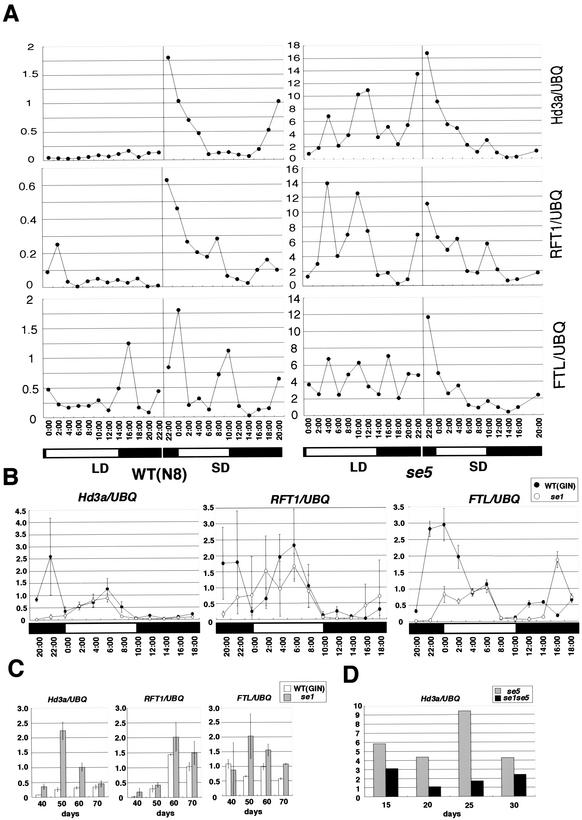
Circadian clock behavior and flowering responses in non-24-h photoperiods. (A) Rhythmic cab1r::luc expression in 24L12D and subsequent DD. Five-day-old seedlings were used for measurements. Averages of six seedlings are plotted with standard deviations. Representative of two experiments. (B) Rhythmic cab1r::luc expression in 36L12D. Five-day-old seedlings were used. Averages of six seedlings are plotted with standard deviations. (C) Rhythmic cab1r::luc expression in 24L. Five-day-old seedlings were used. Averages of six seedlings are plotted with standard deviations. Plants were grown under the 24-h temperature cycle (12 h at 25°C/12 h at 30°C) shown at the top. (D) Flowering time in non-24-h photoperiods. Heading dates after sowing were measured as flowering times. Averages were plotted with standard deviations. Plant numbers are shown in parentheses. Representative of two or three experiments. (se5) se5 mutant with Norin8 genetic background; (se1) se1 mutant with Ginbouzu genetic background; (N8) cv. Norin 8; (GIN) cv. Ginbouzu. Lines 1, 2, and 3 represent se1 se5 mutant lines derived from three F2 se1 se5 plants.
Flowering responses of rice mutants were next examined in these photoperiods (Fig. 2D). The wild-type rice plants (cv. Norin8, N8; cv. Ginbouzu, GIN) did not flower at 120 d after sowing under 24L, 24L12D, or 36L12D photoperiods. This indicates that these photoperiods caused extremely late flowering in the wild-type plants when compared with that under SD and LD conditions. On the other hand, se5 plants with the N8 genetic background flowered at ∼50 d in 24L and 36L12D photoperiods as well as in SD and LD conditions. In contrast, se5 plants took >60 d to flower in the 24L12D photoperiod. This result indicates that se5 can respond to the atypical phase setting of circadian clocks under 24L12D.
Flowering time of another rice mutant, se1, was also analyzed. Recently, se1 was shown to be allelic to the Hd1 gene of rice, a major flowering-time QTL that encodes a putative zinc-finger transcription factor orthologous to the Arabidopsis CO protein (Yano et al. 2000). The se1 mutant in this study contains a null allele of SE1 in the GIN background, because there is a 43-bp deletion in the coding region (data not shown). The se1 plants did not flower at 120 d in the 24L photoperiod condition, showing a remaining photoperiodic response in se1. In addition, se1 plants flowered at ∼60–80 d in light conditions of SD, LD, 24L12D, and 36L12D, indicating that the atypical phase settings of circadian clocks did not cause any drastic delay of flowering in se1 plants, unlike in the wild-type plants.
To analyze se1 se5 double mutants, we tested three lines derived from different F2 individuals of a cross between se1 and se5, to be sure about the phenotypes of the se1 se5 double mutant. Because an F2 population between N8 and GIN did not show segregation at flowering time, there is no major QTL for flowering between them (Okumoto et al. 1991). Double-mutant se1 se5 plants showed dramatically early-flowering phenotypes under these photoperiods, as did se5 plants, but flowered slightly later than se5 plants in SD, LD, 24L, and 36L12D conditions (Fig. 2D), indicating that SE1 can promote flowering slightly in the se5 background. No significant difference in flowering time was observed between se1 se5 and se5 plants under the 24L12D condition. These results indicate that se5 is completely epistatic to se1 in the 24L12D-photoperiod condition; in all other conditions, the se1 se5 double mutant is intermediate but closer to the se5 single mutant. Therefore, SE5 inhibits flowering in all tested conditions, whereas SE1 inhibits flowering except in SD and in 24L (where the se1 mutation has no effect) or in the se5 background (where its promoting effect is revealed). Phytochrome's inhibition of flowering is apparently strong in rice and can exist independently of SE1. It is, however, possible that SE1 may require phytochrome signal transduction to inhibit flowering, because SE1 promotes flowering in the se5 mutant background.
The se5 mutation does not affect the diurnal mRNA expression of SE1 upon floral transition
The flowering-time experiments showed that flowering under atypical entrainment conditions is not drastically delayed in se1 mutant plants. In addition, it has recently been shown that the CO gene functions as an output of circadian clocks to control flowering in Arabidopsis (Suárez-López et al. 2001). Therefore, we examined SE1 mRNA expression in se5 mutants by RT-PCR analysis. First, SE1 mRNA expression was examined in 20-day-old rice plants (Fig. 3A), because inflorescence meristems become microscopically visible in se5 plants under LD at 20 d after sowing (data not shown). SE1 mRNA expression occurred mainly at night under LD conditions, although some SE1 mRNA expression was detected in the daytime. SE1 mRNA levels under LD conditions were similar to those under SD conditions, although the duration of SE1 mRNA expression was slightly longer under LD than under SD conditions (data not shown). In se5 mutant plants, no apparent difference in SE1 mRNA expression was detected under LD conditions compared with expression in the wild type (Fig. 3A).
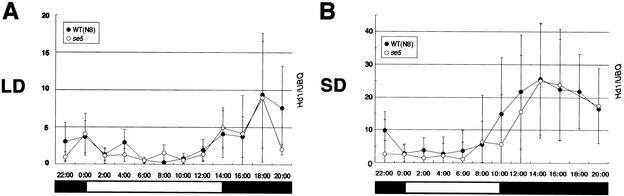
Figure 3.
SE1 mRNA expression in se5 mutant. (A) Diurnal SE1 mRNA expression under long-day (LD) conditions by RT-PCR analysis. RNA samples were harvested from 20-day-old rice plants every 2 h from leaves of plants grown under LD conditions after sowing. Leaves from three to five plants were mixed into one sample. The DNA blot was hybridized with specific radioactive probes to perform a quantitative analysis. Radioactive signals were quantified with a BAS 2000 image analyzer. The normalized data for ubiquitin expression are plotted. Average values and standard deviations from three RT-PCR data are shown. Representative of two independent experiments. (B) Diurnal SE1 mRNA expression under short-day (SD) conditions by RT-PCR analysis. RNA samples were harvested from 28-day-old rice plants in the same way as in A. One RNA sample of se5 in SD conditions is missing, so we do not show one data at 18:00. The DNA blot was hybridized with specific radioactive probes to perform a quantitative analysis. Radioactive signals were quantified with a BAS 2000 image analyzer. The normalized data for ubiquitin expression are plotted. Average values and standard deviations from four RT-PCR data are shown. Representative of two independent experiments.
We next examined SE1 mRNA expression in 28-day-old rice plants (Fig. 3B), because se5 shoot apical meristems may change to inflorescence meristems under SD conditions at ∼28 d after sowing. Note that se5 plants flowered 1 wk later under SD than under LD conditions (Fig. 2D). In 28-day-old rice plants, SE1 mRNA expression was also detected mainly at night and was similar to expression in the 20-day-old plants, but the diurnal expression patterns were more robust under both LD and SD conditions compared with those in 20-day-old rice plants (data not shown). In se5 plants, SE1 mRNA expression was very similar under SD conditions to that in the wild type (Fig. 3B). This indicates that SE1 expression was not affected upon the floral transition of se5 plants under SD conditions. These results of SE1 expression indicate that the se5 mutation does not greatly affect the diurnal expression of SE1 mRNA upon floral transition. In addition, clear reduction of SE1 mRNA expression was observed under LD conditions in 28-day-old se5 plants compared with that in the wild type (data not shown). But, we believe that this reduction is not related to floral transition of se5 plants, because RNA samples were obtained 1 wk after the floral transition.
FTL, a member of the FT-like small gene family, is involved in floral promotion in rice
Because SE1 mRNA expression was not greatly affected upon floral transition by the se5 mutation, we searched for rice genes homologous to the Arabidopsis flowering-time genes in databases and examined the mRNA gene expression of the homologous genes in se5. This search led us to find several rice genes homologous to the Arabidopsis FT gene (hereafter termed FT-like genes; Fig. 4A). A phylogenic analysis revealed that the FT-like genes comprise a small gene family in the rice genome (Fig. 4B). One of the FT-like genes was identical to the Hd3a gene, a flowering-time QTL that was recently isolated in rice (Yano et al. 2001). Hd3a has been genetically shown to have a floral promotion effect under SD conditions (Monna et al. 2002).
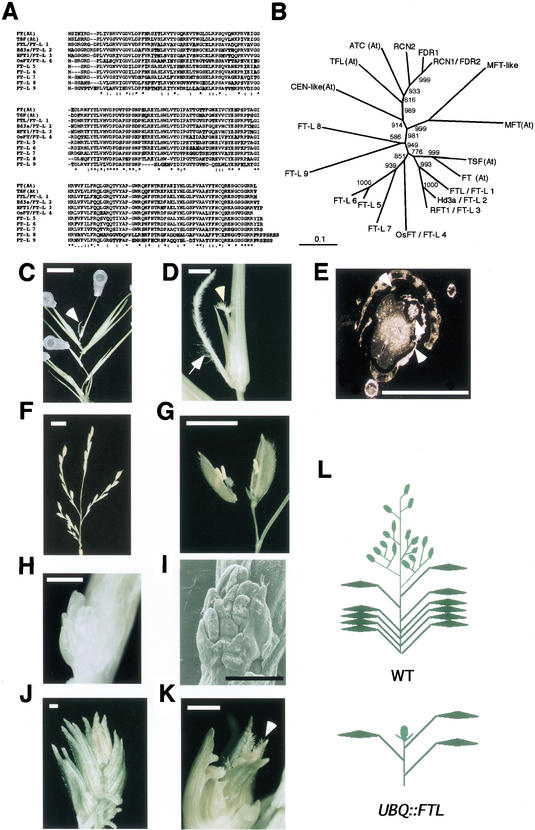
Figure 4.
FT-like gene family in rice and phenotypes of UBQ::FTL plants. (A) Amino acid comparison among rice FT-like genes. Two genes, FT and TSF, belong to the FT family in Arabidopsis. Only amino acids that are not conserved are highlighted. (B) A phylogenic analysis of the FT-TFL family in Arabidopsis and rice. All six members in Arabidopsis are shown with (At) in this tree. Clustal W and NJ methods were used for the alignment and phylogenic tree, respectively. Bootstrap values are shown at branches. (C) A regenerated rice plant transformed with UBQ::FTL. A terminal flower is visible at the tip of the plant (arrowhead). All axial buds produced several leaves with terminal tissue. Bar, 1 cm. (D) Magnification of the terminal flower in C. There is a pistil-like structure at the center (arrowhead). A root was developed at the base of this floret (arrow). Bar, 1 mm. (E) Cross-section of the terminal flower in C. Stamen-like filamentous structures were observed around the pistil-like organ (arrowheads). Bar, 1 mm. (F) A developing panicle in the wild-type rice plant. Bar, 1 cm. (G) A mature floret of the wild-type rice. The floret was dissected to reveal internal organs. Bar, 5 mm. (H) Another rice terminal flower of a UBQ::FTL plant. Glumes were removed. Bar, 5 mm. (I) SEM image of the terminal flower in H. Bar, 5 mm. (J) A terminal tissue with many glumes that developed alternately. Several roots developed at the bases of this terminal tissue. Bar, 1 mm. (K) A terminal flower in glumes. An anther-like structure protruded next to the pistil-like organ (arrowhead). Bar, 1 mm. (L) Schematic presentation of UBQ::FTL phenotypes. Rice leaves are represented as thin lines (sheaths) with attached diamond shapes (blades). Florets are shown as oval shapes. The wild-type (WT) rice panicle is composed of primary and secondary branches with several florets. The terminal floret of UBQ::FTL plants is shown as a floret with additional glumes (UBQ::FTL). For simplification, axillary buds with terminal tissues are not shown in UBQ::FTL plants.
To determine whether these FT-like genes are involved in rice floral induction, we transformed cDNA of one FT-like gene, termed FTL, driven by a maize ubiquitin promoter (UBQ::FTL), into rice. All of the regenerated UBQ::FTL plants produced a terminal tissue at the tips in culture boxes after three to five leaves had developed (Fig. 4C,L). In the control group of regenerated plants, no panicles formed when they grew in culture boxes. Then the plants were transferred into growth chambers, and flowered after more than 10 leaves had developed (Fig. 4F). All UBQ::FTL plants showed elongation of internodes, which is a morphological marker of floral transition in rice. In addition, most axial meristems also produced similar terminal tissues in UBQ::FTL plants (Fig. 4C), whereas the axial meristems became dormant in the control plants (data not shown). Many of the terminal tissues were composed of many glumes that alternately developed (Fig. 4J). In some cases, the terminal tissues contained a terminal floret at the tip, instead of a panicle (Fig. 4D,H,I,K). These terminal florets were composed of several stamen-like organs and a pistil-like organ (Fig. 4D,E,H,I,K). This terminal-bud phenotype was similar to Arabidopsis terminal flower (tfl) mutants and LEAFY and FT co-overexpressers of Arabidopsis (Bradley et al. 1997; Kobayashi et al. 1999). This indicates that common molecular mechanisms are involved in floral architecture formation between rice and Arabidopsis and that antagonistic action between TFL and FT may also be conserved in rice. Our data therefore indicate that the ectopic expression of FTL resulted in floral induction, elongation of internodes, loss of apical dominance, and production of terminal flowers at the tip (Fig. 4L). Thus, like Arabidopsis FT, rice FTL expression is related to floral promotion.
FT-like gene expression is regulated by interaction between SE5 and SE1 genes in rice
Preliminary RT-PCR analyses showed that eight of the FT-like genes were expressed in rice leaves and six of them were up-regulated in leaves of se5 plants grown under 24L, compared with the wild type (data not shown). Therefore, the mRNA expression patterns of three FT-like genes, Hd3a, RFT1, and FTL, were examined in detail by a quantitative RT-PCR method. We first used the same RNA samples as those used for assessing SE1 expression to determine the effect of the se5 mutation on FT-like mRNA expression. Ectopic expression of rice Hd3a and RFT1 genes resulted in early-flowering phenotypes (Yano et al. 2001; M. Yano, pers. comm.) as in UBQ::FTL plants (Fig. 4), indicating that all three FT-like genes are involved in floral promotion in rice. In the wild-type plants, Hd3a and RFT1 mRNA expression was up-regulated under inductive SD conditions, but not under LD conditions (Fig. 5A). Under SD conditions, Hd3a and RFT1 mRNA was diurnally expressed with a peak before dawn. FTL mRNA expression was detected under both LD and SD conditions, although mRNA levels were higher under SD than under LD conditions (Fig. 5A). These results indicate that these FT-like gene expressions are responding to different photoperiods, although the responses to a certain photoperiod may vary among them. In se5 plants, all three genes were more highly expressed under both LD and SD conditions than in wild-type plants (Fig. 5A). Under SD conditions, the mRNA expression patterns of FT-like genes were similar in se5 plants to those in wild-type plants. Under LD conditions, mRNA expression of FT-like genes in se5 plants was strong but not clearly diurnal. These results indicate that phytochrome signals repress FT-like gene expression under both LD and SD conditions, but in different ways.
Figure 5.
FT-like gene expression is controlled by SE5 and SE1. (A) Diurnal mRNA expression of three FT-like genes in wild-type (WT) and se5 plants under LD and SD conditions. RNA samples were obtained from 28-day-old plants (partly, the same RNA samples were used with those in Fig. 3B). Quantitative RT-PCR was performed twice to get average values. The normalized data are plotted. One time-point sample, for 18:00, is missing in se5 under SD conditions. Similar results were obtained with RNA samples harvested from 20-day-old plants. Different scales are used between the left and right graphs because of the differences in expression levels. (B) Diurnal mRNA expression of three FT-like genes under SD conditions in se1. RNA samples were obtained from 30-day-old plants. Quantitative RT-PCR was performed two to four times to get average values and standard deviations. The normalized data are plotted. (C) mRNA expression of three FT-like genes under LD conditions in se1 at different developmental stages by quantitative RT-PCR. Leaves were harvested at 09:30 in the morning every 10 d. RT-PCRs were performed several times to get averages and standard deviations. Representative data are shown from four, two, and two independent experiments on Hd3a, RFT1, and FTL expression, respectively. (D) mRNA expression of Hd3a in se1 and se1se5 plants under LD conditions at different developmental stages. Leaves were harvested at 10:30 every 10 d. Representative data from two independent experiments.
In se1 mutants under SD conditions, all tested FT-like mRNA expression levels were reduced before dawn, whereas mRNA expression levels in the daytime were not greatly affected (Fig. 5B). These results indicate that the SE1 gene functions as an activator of FT-like genes before dawn under SD conditions. Interestingly, although the second peak of Hd3a expression in wild-type GIN plants was apparent in daytime under SD, there were no such apparent daytime peak of Hd3a expression in the wild-type N8 plants (Fig. 5A,B). This may be owing to genetic variations between those varieties. Under LD conditions, in contrast, significant effects of the se1 mutation on FT-like gene expression were not detected because of the very low levels of FT-like gene expression of 30-day-old plants in both se1 and wild type plants (data not shown). Therefore, we next examined mRNA expression of FT-like genes under LD conditions at different developmental stages. In the wild type, RFT1 mRNA expression gradually increased under LD conditions from 40 to 60 d and then slightly decreased at 70 d (Fig. 5C). In contrast, Hd3a and FTL mRNA expression was not greatly increased under LD conditions until 70 d after sowing. The results showed that mRNA expression levels of the RFT1 gene expression were developmentally regulated under LD conditions, suggesting distinct roles of FT-like genes in rice development. In the se1 mutant, mRNA expression levels of all three FT-like genes were higher than those in the wild type plants (Fig. 5C). These results indicate that the SE1 gene functions as a repressor in FT-like gene expression under noninductive LD conditions.
To evaluate the interaction between phytochrome signaling and SE1 in FT-like gene expression, we examined the effect of SE1 on Hd3a gene expression in the se5 background. In the se1 se5 double mutant, Hd3a gene expression levels were reduced compared with those in se5 single mutants (Fig. 5D). These results indicate that the SE1 gene functions as an activator of Hd3a in the se5 mutant, correlated to the flowering data (Fig. 2D).
Discussion
Distinct regulation of the entrainment of circadian clocks and photoperiodic flowering in rice
Phytochromes and cryptochromes are involved in both light entrainment of plant circadian clocks and the photoperiodic control of flowering (Lin 2000; Samach and Coupland 2000; Devlin and Kay 2001). Using se5, a phytochrome-deficient mutant of rice, we have shown that phytochromes mediate the external light signal to repress mRNA expression of major floral-inducer genes (Fig. 5A) but may not be necessary for the entrainment of circadian clocks under LD and SD conditions (Fig. 1). In addition to circadian clock outputs such as CAB1R, mRNA expression of OsLHY, which is homologous to members of the Arabidopsis central circadian clock components (Schaffer et al. 1998; Wang and Tobin 1998), may provide us with strong evidence regarding the behavior of circadian clocks in se5 plants.
In recent years, the entrainment of circadian clocks has been extensively analyzed by using Arabidopsis photoreceptor mutants to show redundant and distinct roles of photoreceptor genes in phytochrome and cryptochrome families (Somers et al. 1998; Devlin and Kay 2000). Only in continuous red light do phytochrome mutants show longer periods in cab2::luc Arabidopsis seedlings. In hy1, an Arabidopsis counterpart mutant of se5, no effect on circadian clock behavior was observed under white-light conditions as in other phytochrome mutants of Arabidopsis (Millar et al. 1995; Somers and Kay 1998). In contrast, cry1 cryptochrome mutants show longer periods under red-, blue-, and white-light conditions, whereas cry2 mutants significantly lengthen periods only under white-light conditions (Somers et al. 1998; Devlin and Kay 2000). Furthermore, other photoreceptors may be involved in the entrainment of circadian clocks, because light entrainment has been observed in a quadruple photoreceptor mutant phyA phyB cry1 cry2 (Yanovsky et al. 2000). These data indicate that phytochromes redundantly play a role for the entrainment of circadian clocks in Arabidopsis. Thus, not only in rice, as shown in this study, but also in Arabidopsis, phytochrome deficiencies do not affect the entrainment of circadian clocks in white-light photoperiods enough to explain the effects on floral induction. Considering the completely photoperiod-insensitive flowering in se5 plants (Fig. 2D; Izawa et al. 2000), an alternative phototransduction pathway that is not mediated by circadian clocks—the external light pathway—must exist in the photoperiodic control of flowering in rice.
SE1 functions as an output of the circadian clock
Recent studies based on analyses of circadian-clock-related mutants such as early flowering 3, gigantea, and lhy have revealed that the Arabidopsis CO gene mediates the circadian clock to control flowering time (Suarez-Lopez et al. 2001). Here, we have shown that the wild-type's complete inhibition of flowering under atypical entrainment conditions such as 24L12D and 36L12D is lost in the se1 mutant, a rice counterpart mutant of Arabidopsis co (Fig. 2D). We used the non-24-h light/dark cycles to affect the entrainment of the circadian clock and introduce light at unconventional phases. Then we tested the phase of circadian rhythms (Fig. 2A–C) and the flowering response (Fig. 2D) under the non-24-h light/dark cycles to determine whether flowering can be explained in terms of interaction between the circadian phase and light. Therefore, lack of flowering response in se1 mutants to two atypical entrainment conditions, 24L12D and 36L12D, strongly suggests that SE1 function is normally phase-specific because of output from the circadian clock. In addition, diurnal SE1 mRNA expression patterns also suggest that SE1 gene expression is regulated by circadian clocks (Fig. 3A,B). Because the acute light response was also affected in the non-24-h cycles, the effects of acute light responses should be considered in the evaluation of flowering phenotypes in non-24-h photoperiods. In Arabidopsis, phyA mediates far-red light to control mRNA levels of CO in a microarray analysis (Tepperman et al. 2001). Thus, the effects of acute light responses on SE1 mRNA expression remain to be examined.
The se5 deficiency in phytochrome signaling resulted in no response to photoperiods, but in an ∼2-wk delay of flowering under 24L12D conditions (Fig. 2D), suggesting that a signal transduction pathway other than the phytochrome signaling pathway was modulated under 24L12D. Together with the flowering response in se1 plants, the flowering response in se5 plants under the 24L12D condition indicates that the other pathway is likely to be regulated by the circadian clock (Fig. 2D), although it is not known whether SE1 is the sole output of circadian clocks in the photoperiodic flowering of rice. The flowering data indicating that se5 is completely epistatic to se1 under 24L12D (Fig. 2D) is consistent with this idea. In addition, upon floral transition, SE1 mRNA expression was not much affected by the se5 mutation (Fig. 3A,B). These results indicate that both a circadian-clock-associated pathway and the phytochrome-phototransduction pathway must exist to control photoperiodic flowering in rice.
FT-like genes downstream of phytochrome and SE1
The Arabidopsis FT gene is expressed in inductive LD conditions, but not under SD conditions, at the early developmental stage, and is directly regulated by CO (Kardailsky et al. 1999; Kobayashi et al. 1999; Samach et al. 2000). Here, we have shown that several rice FT-like genes, Hd3a, FTL, and RFT1, are preferentially expressed in inductive SD conditions but not under LD conditions, indicating a clear difference between short-day and long-day plants at the transcriptional level. Recent identification of a rice flowering-time QTL, termed Hd3a, with an FT homolog and early-flowering phenotypes in ectopic expression of FT-like genes indicate a conserved function of FT orthologs for promotion of flowering between rice and Arabidopsis (Fig. 4; Yano et al. 2001). RT-PCR analyses revealed that, in wild-type plants, Hd3a, FTL, and RFT1 all showed diurnal mRNA expression patterns and had a peak at about dawn under SD conditions (Fig. 5A). Because these mRNAs were detected even in the daytime, unlike FT, the phase settings of rice FT-like gene expression are different from that of FT in LD conditions (Harmer et al. 2000; Suárez-López et al. 2001).
In the se5 mutant, FT-like genes were expressed under both LD and SD conditions (Fig. 5A), correlating with the early flowering of the mutants (Fig. 2D). Even under SD conditions, mRNA levels of FT-like genes in the se5 mutant were much higher than those in the wild-type plant. In contrast, mRNA levels of SE1 diurnal expression were not affected much by the se5 mutation upon floral transition (Fig. 3A,B). Thus, phytochrome signals strongly repress FT-like gene expression, but this repression does not reflect SE1 mRNA levels. Instead, SE1 mRNA expression is likely to be controlled by the circadian clock (Fig. 3A,B), as CO is in Arabidopsis. It is noteworthy that phytochromes can inhibit flowering independently of SE1, because se1 plants did not flower at 120 d under 24L conditions, whereas se5 and se1 se5 plants flowered at ∼50 d (Fig. 2D). Therefore, a genetic factor other than SE1 may be involved in the photoperiodic control of flowering in rice.
The RT-PCR analyses using the se1 mutant revealed that the SE1 gene promotes mRNA expression of FT-like genes in the inductive SD conditions and represses FT-like mRNA expression in the noninductive LD conditions (Fig. 5B,C). This antagonistic action of SE1 activity may be regulated posttranscriptionally, because the diurnal SE1 mRNA expression pattern was not greatly changed by different photoperiods (data not shown). Furthermore, this SE1 activity may be regulated by phytochrome signaling because SE1 promotes Hd3a gene expression under noninductive LD treatment in the se5 background (Fig. 5D). Comparison of flowering times between se5 and se1 se5 mutants supports this idea (Fig. 2D). Therefore, phytochrome plays an important role in day-length recognition, partly through the control of SE1 activity in rice. The possible interaction between SE1 and the phytochromes may include formation of a nuclear transcriptional complex and/or phosphorylation of SE1 by phytochromes (Neff et al. 2000).
Flowering time of se1 plants was not different under SD conditions from that of wild-type plants (Fig. 2D). Because the se1 mutation affected FT-like mRNA expression mainly around dawn (Fig. 5B), FT-like expression in the daytime under SD conditions might be enough to induce floral transition in se1 and wild-type plants. Given that SE1 can promote flowering under SD conditions in a cv. Nipponbare background (Yano et al. 2000), the effect of SE1 on FT-like mRNA expression under SD conditions may be related to flowering response in the cv. Nipponbare. In addition, it is noteworthy that FT-like gene expression pattern may depend on genetic background, because there was no apparent daytime expression of Hd3a detected in wild-type N8 plants under SD conditions, for example (Fig. 5A).
The external coincidence model in rice
Theoretically, at least two diurnal patterns that are differently shaped by a certain photoperiod (likely caused by light-on and light-off signals) are required for day-length recognition. At a molecular level, the diurnal pattern may include gene expression and/or protein modification. With the support of many physiological experimental data, the external coincidence model was proposed in the 1960s (Pittendrigh and Minis 1964). In this model, the above two diurnal patterns are composed of the following: one is a photoinducible phase in a day cycle set by the circadian clock, and the other is an external light signal reflecting the presence or absence of sunlight. The duration of overlap (or coincidence) between the photoinducible phase and the external signal is thought to be a determinant of day-length measurement in the model. It is noteworthy that light functions as a zeitgeber to set the photoinducible phase, in addition to having a role as the external signal. When different fluence rates of light pulses were used in a night-break experiment, two distinct roles of light were suggested in Japanese morning glory, although the molecular nature of these roles of light is completely unknown (Lumsden and Furuya 1986). Recently, it has been shown that transcriptional activity of CO protein may have a light effect in Arabidopsis, although responsible photoreceptors were not identified (Suárez-López et al. 2001).
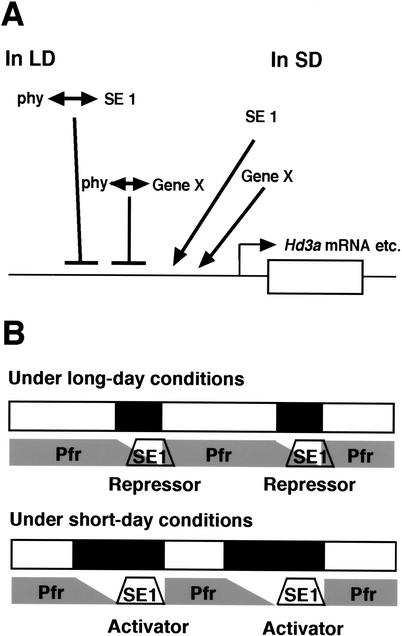
Our results have led us to propose a molecular mechanism, reminiscent of the external coincidence model, to explain day-length recognition in the photoperiodic control of flowering in rice (Fig. 6). In our model, the SE1 protein level corresponds to the photoinducible phase, whereas the active Pfr form of phytochromes corresponds to the external signal. Because the Arabidopsis CO protein is unstable and its expression is likely to reflect CO mRNA diurnal expression (Suárez-López et al. 2001), the rice counterpart SE1 protein may exist mainly at nighttime, according to SE1 mRNA levels (Fig. 3A,B). Because the phase settings of plant circadian clocks are regulated by both light-on and light-off (Millar and Kay 1996; Sugiyama et al. 2001), SE1 protein levels may be affected by both light-on and light-off signals of given photoperiods. On the other hand, in Arabidopsis hypocotyl elongation, phyB can transmit the light signal for several hours in darkness after light-off, reflecting the amount of Pfr in darkness (McCormac et al. 1993; Shinomura et al. 2000). In addition, it has been shown in vitro that the Pfr form of phytochromes gradually reverts to the Pr form in darkness (Elich and Chory 1997; Eichenberg et al. 2000). Thus, the amounts of the Pfr form of light-stable phytochromes in rice may be stable during the light photoperiod and decrease gradually in darkness (Fig. 6B). Consequently, the diurnal pattern of Pfr phytochromes becomes distinct from circadian-clock-controlled SE1 expression. Therefore, the duration of overlap between SE1 and Pfr phytochrome depends heavily on the photoperiod (Fig. 6B). Because we have shown a genetic interaction between phytochrome signals and SE1 activity (Figs. 2D, 5D), there might be an interaction between them during the overlap. As shown in Figure 6, the interaction levels should be higher under noninductive LD conditions than under SD conditions. Our results suggest that, in interacting with Pfr, SE1 may function as a repressor of FT-like gene expression (Fig. 5C). In contrast, SE1 may function as its activator without the interaction (Fig. 5A,B,D). The dual SE1 activity that is switched by phytochrome signals may be involved in the setting of critical night length as the acute threshold often observed in short-day plants (Thomas and Vince-Prue 1997).
Figure 6.
Model for day-length measurement in the photoperiodic control of flowering in rice. (A) In our model, the interaction between phytochrome signaling and the SE1 transcription factor makes SE1 a repressor of FT-like gene expression. In contrast, SE1 protein alone functions as an activator of FT-like gene expression. The interaction levels are higher under LD conditions than under SD conditions, as shown in B. The molecular basis of the interaction could be the formation of a transcriptional complex, because phytochrome is a nuclear factor. Phosphorylation of SE1 protein by phytochromes is an alternative. Gene X is postulated as a transcription factor that has a similar function to SE1, because mRNA expression of FT-like genes was detected under SD conditions even in the se1 mutant. (B) The amount of the active Pfr form of phytochromes is stable in the daytime and may gradually decrease with dark reversion. On the other hand, SE1 mRNA is nocturnally expressed by circadian clocks, and SE1 protein may exist mainly at a certain phase of subjective night. Phase setting of circadian clocks is determined by both light-on and light-off signals, whereas the decrease in levels of the Pfr form starts with the light-off signal. Thus, the timing of the coincidence of the Pfr form and the SE1 protein may differ among photoperiods. When the coincidence occurs, the interaction between Pfr and SE1 may make a repressor of FT-like genes. Without the coincidence, SE1 protein may function as an activator of FT-like genes to promote flowering in rice. Possible interactions between phytochrome and other factors than SE1 are omitted from this panel to focus on the relationship between SE1 and phytochrome.
Another coincidence model, termed the internal coincidence model, has been proposed (Thomas 1998). In this model, two distinct circadian rhythms produce a coincidence to recognize day length. Given that PHYB and PHYA mRNA expression is affected by circadian clocks (Bognar et al. 1999; Hall et al. 2001; Toth et al. 2001) and that the acute response of cab2::luc expression is gated by circadian clocks in Arabidopsis (Millar and Kay 1996) and (weakly) in rice (Sugiyama et al. 2001), the diurnal Pfr expression pattern may be partly regulated by circadian clocks. Therefore, the essence of the internal coincidence model is also included in our model.
Molecular mechanisms of flowering conserved between short-day and long-day plants
Here we also have shown that one of the key differences in floral induction between short-day and long-day plants is the different transcriptional regulation of FT orthologs. In rice, a short-day plant, FT orthologs are expressed mainly under SD conditions (Fig. 5). In contrast, FT is expressed preferentially under LD conditions in Arabidopsis, a long-day plant (Suárez-López et al. 2001). The FT orthologs as floral inducers are evolutionarily conserved between these species, which diverged about 200 million years ago (Fig. 4; Kardailsky et al. 1999; Kobayashi et al. 1999). Furthermore, SE1 and CO mRNA diurnal expression patterns under SD and LD conditions are also conserved evolutionarily (Fig. 3A,B; Samach and Gover 2001; Suárez-López et al. 2001). In addition, both SE1 and CO function as transcription factors for FT orthologs, although direct binding of CO and SE1 to cis elements in target promoters has not yet been examined. Because CO functions as a transcriptional activator (Simon et al. 1996) and the Arabidopsis PHYB gene may not be involved in FT mRNA expression (although diurnal patterns were not examined; Blazquez and Weigel 1999), SE1 repressor activity through the interaction with phytochromes may be specific to floral induction in rice.
In Arabidopsis, CRY2 plays an important role in floral induction (Guo et al. 1998; Mockler et al. 1999). Recently, a QTL analysis between Ler and Cvi ecotypes of Arabidopsis revealed a novel allele of CRY2 that has a mutation at the chromophore-binding site and functions as a constitutively active form of cryptochrome to promote floral induction (El-Din El-Assal et al. 2001). In contrast, phytochromes are likely to have more critical roles in determining flowering time in rice (Izawa et al. 2000), although Arabidopsis PHYB is required for floral promotion by CRY2 (Guo et al. 1998; Mockler et al. 1999). Therefore, the roles of rice cryptochromes in floral induction remain to be analyzed.
Materials and methods
Plant materials
The se5 mutant plant line was obtained as previously described (Izawa et al. 2000). The se1 mutant line (Line 4H313A) was originally isolated from a γ-ray-treated collection and backcrossed four times with the GIN wild type.
RNase protection assays
Total RNA was prepared by an SDS-phenol method as previously described (Sugiyama et al. 2001). Then 2–5 μg of total RNA was subjected to an RNase protection reaction by using an RPA III kit (Ambion). After electrophoresis, radioactive signals were quantified by the BAS 2000 system (Fuji Photo Film). Calibration was performed with MacBAS Version 2.52 software. Protected mRNA sizes were 395 bp for CAB1R (Luan and Bogorad 1992), 321 bp for OsLHY (accession no. D48887), 317 bp for CATA (accession no. D29966), and 245 bp for UBQ (accession no. X13908).
Light entrainment of circadian clocks
For RNase protection assays, rice plants were grown in a greenhouse for 30 d after sowing and then transferred to a chamber with 12 h light/12 h darkness (12L12D) for 5 d. White light from a fluorescent lamp at a photon fluence rate of 50 μmole/m2 sec was used for this light entrainment. For cab1r::luc bioluminescence assays, sterile rice seeds were imbibed and germinated on MS medium containing 3% sucrose at 30°C under conditions of long day, 14L10D, or short day, 10L14D, for 5 d. On the third day after sowing, luciferin was added to the germinating seeds, and seeds expressing luc activity were selected. Selected seeds were transferred to 6-well plates with an MS medium containing 3% sucrose and 5 mM luciferin for bioluminescence measurement.
cab1r::luc bioluminescence measurements
The same cab1r::luc lines used in a previous study (Sugiyama et al. 2001) were used. After entrainment, 6-well plates were placed in a camera obscura set with a VIM camera in an ARGUS 50 system (Hamamatsu Photonic Systems) for photon-counting measurements in real-time sampling mode, as described previously (Sugiyama et al. 2001).
Flowering-time measurement
For flowering-time measurements, sterile seeds were germinated in the chamber with given photoperiod conditions until flowering. Heading dates after sowing were recorded as the flowering times of the plants. For short-day, 10L14D, and long-day, 14L10D, light conditions, white light from a mixed-light source of halogen and Na lamps at a photon fluence rate of 300 μmole/m2 sec was used (Izawa et al. 2000). For 24L, 24L12D, and 36L12D light conditions, white fluorescent lamps were used as light sources at a photon fluence rate of 150 μmole/m2 sec (Izawa et al. 2000). Light quality was verified with a spectroradiometer LI1800 (Li-Cor).
Database search
Public databases, including the Monsanto rice database, were searched with various Arabidopsis flowering-time genes. By using the FT amino acid sequence as a query, we identified nine rice FT homologs. The FTL (FT-L 1) ORF was found in a PAC clone sequence, P0665D10 (accession no. AP002861), on chromosome 1. Upon this database search, eight FT homolog ORFs were further identified on Monsanto contigs: FT-L 2 on OSM123786; FT-L 3 on OSM15346; FT-L 4 on OSM12638; FT-L 5 on OSM15106; FT-L 6 on OSM12071; FT-L 7 on OSM139582; FT-L 8 on OSM14221; and FT-L 9 on two separate contigs, OSM147493 and OSM147494. OsFT was identical to FT-L 4 (Kobayashi et al. 1999). The recently registered genes Hd3a and RFT1 were identical to FT-L 2 and FT-L 3, respectively. mRNA expression of these FT homologs was detected in rice leaves, except that of FT-L 8.
RT-PCR analysis
For cDNA synthesis by reverse transcriptase (Superscript II, BioRad), 5 μg of total RNA was used. A cDNA template corresponding to ∼50–100 ng of total RNA was used for one RT-PCR reaction. The primers used for FTL, Hd3a, and RFT1 were as follows: FTL, 5′-GGAGTACCTGCACTGGCTGGTCACCG-3′, 5′-TCTGGCGCCACCCCGGTGCGTACAC-3′; Hd3a, 5′-G GAGTATCTACATTGGTTGGTCACTG-3′, 5′-TGGATGAT GATAGTGAGCATGCAGC-3′; RFT1, 5′-GGAGTATCTACA CTGGCTGGTCACCG-3′, 5′-GCCTGCATGCATATACAGC TAGGCAG-3′. After PCR amplification, samples were subjected to electrophoresis to obtain the DNA blots. The DNA blot was hybridized with specific probes, and radioactive signals were quantified using the BAS 2000 system, as described above.
Generation of UBQ::FTL plants
FTL cDNA was introduced into a Ti binary vector containing a maize Ubq1 promoter (Christensen et al. 1992). To examine flower development, >60 regenerated transgenic rice plants were obtained by Agrobacterium-mediated transformation from ∼25 hygromycin-resistant calli.
Acknowledgments
We thank M. Nobuhara for technical help in rice transformation and S. Yamamoto for providing the OsLHY cDNA sequence. The Monsanto rice database was kindly provided by Monsanto. This research was supported by a Grant-in-Aid for Scientific Research on Priority Areas (No. 10182102) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL tizawa@nias.affrc.go.jp; FAX 81-298-38-7468.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.999202.
References
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Blazquez MA, Weigel D. Independent regulation of flowering by phytochrome B and gibberellins in Arabidopsis. Plant Physiol. 1999;120:1025–1032. doi: 10.1104/pp.120.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bognar LK, Hall A, Adam E, Thain SC, Nagy F, Millar AJ. The circadian clock controls the expression pattern of the circadian input photoreceptor, phytochrome B. Proc Natl Acad Sci. 1999;96:14652–14657. doi: 10.1073/pnas.96.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997;275:80–83. doi: 10.1126/science.275.5296.80. [DOI] [PubMed] [Google Scholar]
- Bünning E. Circadian rhythms and the time measurement in photoperiodism. Cold Spring Harbor Symp Quant Biol. 1960;25:249–256. doi: 10.1101/sqb.1960.025.01.029. [DOI] [PubMed] [Google Scholar]
- Carre IA. Day-length perception and the photoperiodic regulation of flowering in Arabidopsis. J Biol Rhythms. 2001;16:415–423. doi: 10.1177/074873001129002006. [DOI] [PubMed] [Google Scholar]
- Casal JJ. Phytochromes, cryptochromes, phototropin: Photoreceptor interactions in plants. Photochem Photobiol. 2000;71:1–11. doi: 10.1562/0031-8655(2000)071<0001:pcppii>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH. Maize polyubiquitin genes: Structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell. 2000;12:2499–2510. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Circadian photoreception. Annu Rev Physiol. 2001;63:677–694. doi: 10.1146/annurev.physiol.63.1.677. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PR, Patel SR, Goosey L, Sharrock RA, Whitelam GC. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberg K, Baurle I, Paulo N, Sharrock RA, Rudiger W, Schafer E. Arabidopsis phytochromes C and E have different spectral characteristics from those of phytochromes A and B. FEBS Lett. 2000;470:107–112. doi: 10.1016/s0014-5793(00)01301-6. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- Elich TD, Chory J. Biochemical characterization of Arabidopsis wild-type and mutant phytochrome B holoproteins. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner WW, Allard HA. Effect of the relative length of day and night and other factors of the environment on growth and reproduction in plants. J Agric Res. 1920;18:553–606. [Google Scholar]
- Goto N, Kumagai T, Koornneef M. Flowering responses to night breaks in photomorphogenic mutants of Arabidopsis. Physiol Plant. 1991;83:209–215. [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Toth R, Nagy F, Millar AJ. Conditional circadian regulation of PHYTOCHROME A gene expression. Plant Physiol. 2001;127:1808–1818. [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang H-S, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K. Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant) Plant J. 2000;22:391–399. doi: 10.1046/j.1365-313x.2000.00753.x. [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. Photoresponse of light grown phyA mutants of Arabidopsis. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. Activation tagging of the floral inducer FT. Science. 1999;286:1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Krall L, Reed JW. The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc Natl Acad Sci. 2000;97:8169–8174. doi: 10.1073/pnas.140520097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy YY, Dean C. Control of flowering time. Curr Opin Plant Biol. 1998;1:49–54. doi: 10.1016/s1369-5266(98)80127-1. [DOI] [PubMed] [Google Scholar]
- Lin C. Photoreceptor and regulation of flowering time. Plant Physiol. 2000;123:39–50. doi: 10.1104/pp.123.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Bogorad L. A rice cab gene promoter contains separate cis-acting elements that regulate expression in dicot and monocot plants. Plant Cell. 1992;4:971–981. doi: 10.1105/tpc.4.8.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden PJ, Furuya M. Evidence for two actions of light in the photoperiodic induction of flowering in Pharbitis nil. Plant Cell Physiol. 1986;27:1541–1551. [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Mazzella MA, Cerdan PD, Staneloni RJ, Casal JJ. Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development. 2001;128:2291–2299. doi: 10.1242/dev.128.12.2291. [DOI] [PubMed] [Google Scholar]
- McCormac AC, Wagner D, Boylan MT, Quail PH, Smith H, Whitelam G. Photoresponses of transgenic Arabidopsis seedlings expressing introduced phytochrome B-encoding cDNAs: Evidence that phytochrome A and phytochrome B have distinct photoregulatory functions. Plant J. 1993;4:19–27. [Google Scholar]
- Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci. 1996;93:15491–15496. doi: 10.1073/pnas.93.26.15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory CA, Chua N-H, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126:2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- Monna L, Lin HX, Kojima S, Sasaki T, Yano M. Genetic dissection of a genomic region for a quantitative trait locus, Hd3, into two loci, Hd3a and Hd3b, controlling date in rice. Theor Appl Genet. 2002;104:772–778. doi: 10.1007/s00122-001-0813-0. [DOI] [PubMed] [Google Scholar]
- Neff MM, Frankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Okumoto Y, Tanisaka T, Yamagata H. Heading-time genes of rice varieties grown in South-West-Warm region in Japan. (In Japanese) Jpn J Breed. 1991;41:135–152. [Google Scholar]
- Onouchi H, Igeno MI, Perilleux C, Graves K, Coupland G. Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell. 2000;12:885–900. doi: 10.1105/tpc.12.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittendrigh CS, Minis DH. The entrainment of circadian oscillations by light and their role as photoperiodic clocks. Am Nat. 1964;108:261–295. [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Quail PH. The phytochrome family: Dissection of functional roles and signalling pathways among family members. Phil Trans R Soc Lond B. 1998;353:1399–1403. doi: 10.1098/rstb.1998.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Samach A, Coupland G. Time measurement and the control of flowering in plants. BioEssays. 2000;22:38–47. doi: 10.1002/(SICI)1521-1878(200001)22:1<38::AID-BIES8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Samach A, Gover A. Photoperiodism: The consistent use of CONSTANS. Curr Biol. 2001;11:R651–R654. doi: 10.1016/s0960-9822(01)00384-0. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Uchida K, Furuya M. Elementary processes of photoperception by phytochrome A for high-irradiance response of hypocotyl elongation in Arabidopsis. Plant Physiol. 2000;122:147–156. doi: 10.1104/pp.122.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Igeno MI, Coupland G. Activation of floral meristem identity genes in Arabidopsis. Nature. 1996;384:59–62. doi: 10.1038/384059a0. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;99:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Somers DE, Kay SA. Genetic approaches to the analysis of circadian rhythms in plants. In: Lumsden P, Millar AJ, editors. Biological rhythms and photoperiodism in plants. Oxford: BIOS Scientific; 1998. pp. 81–98. [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- Sugiyama N, Izawa T, Oikawa T, Shimamoto K. Light regulation of circadian clock-controlled gene expression in rice. Plant J. 2001;26:607–615. doi: 10.1046/j.1365-313x.2001.01063.x. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang H-S, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. Photoperiodism: An overview. In: Lumsden P, Millar AJ, editors. Biological rhythms and photoperiodism in plants. Oxford: BIOS Scientific; 1998. pp. 151–165. [Google Scholar]
- ————— Photoreceptors and photoperiodism. Flowering News Lett. 1999;27:10–15. [Google Scholar]
- Thomas B, Vince-Prue D. Photoperiodism in plants. London: Academic Press; 1997. [Google Scholar]
- Toth R, Kevei EE, Hall A, Millar AJ, Nagy F, Kozma-Bognar L. Circadian clock-regulated expression of phytochrome and cryptochrome genes in Arabidopsis. Plant Physiol. 2001;127:1607–1616. doi: 10.1104/pp.010467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Patel S, Devlin P F. Phytochromes and photomorphogenesis in Arabidopsis. Phil Trans R Soc Lond B. 1998;353:1445–1453. doi: 10.1098/rstb.1998.0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Kojima S, Takahashi Y, Lin H, Sasaki T. Genetic control of flowering time in rice, a short-day plant. Plant Physiol. 2001;127:1425–1429. [PMC free article] [PubMed] [Google Scholar]
- Yanovsky MJ, Mazzella MA, Casal JJ. A quadruple photoreceptor mutant still keeps track of time. Curr Biol. 2000;10:1013–1015. doi: 10.1016/s0960-9822(00)00651-5. [DOI] [PubMed] [Google Scholar]